Abstract
Eukaryotic poly(A) binding protein (PABP) is a ubiquitous, essential cellular factor with well-characterized roles in translational initiation and mRNA turnover. In addition, there exists genetic and biochemical evidence that PABP has an important nuclear function. Expression of PABP from Arabidopsis thaliana, PAB3, rescues an otherwise lethal phenotype of the yeast pab1Δ mutant, but it neither restores the poly(A) dependent stimulation of translation, nor protects the mRNA 5′ cap from premature removal. In contrast, the plant PABP partially corrects the temporal lag that occurs prior to the entry of mRNA into the decay pathway in the yeast strains lacking Pab1p. Here, we examine the nature of this lag-correction function. We show that PABP (both PAB3 and the endogenous yeast Pab1p) act on the target mRNA via physically binding to it, to effect the lag correction. Furthermore, substituting PAB3 for the yeast Pab1p caused synthetic lethality with rna15-2 and gle2-1, alleles of the genes that encode a component of the nuclear pre-mRNA cleavage factor I, and a factor associated with the nuclear pore complex, respectively. PAB3 was present physically in the nucleus in the complemented yeast strain and was able to partially restore the poly(A) tail length control during polyadenylation in vitro, in a poly(A) nuclease (PAN)-dependent manner. Importantly, PAB3 in yeast also promoted the rate of entry of mRNA into the translated pool, rescued the conditional lethality, and alleviated the mRNA export defect of the nab2-1 mutant when overexpressed. We propose that eukaryotic PABPs have an evolutionarily conserved function in facilitating mRNA biogenesis and export.
Keywords: PABP, mRNA biogenesis, mRNA export
INTRODUCTION
Poly(A) binding protein (PABP) is highly conserved in eukaryotes, and its function is essential for viability in Saccharomyces cerevisiae (Sachs et al. 1987), Aspergillis nidulans (Marhoul and Adams 1996), Caenorhabditis elegans (A. Petcherski and J. Kimble, pers. comm.) and Drosophila melanogaster (Sigrist et al. 2000). PABP participates in at least three major post-transcriptional events, for example, mRNA biogenesis, regulation of mRNA turnover, and initiation of protein synthesis. However, the exact reasons why the PABP function is essential remain incompletely understood.
PABP is an abundant protein [4 μM in Hela cells (Gorlach et al. 1994)], and the majority of PABP in the cell is cytoplasmic at steady state. PABP is the key factor responsible for the poly(A) tail stimulated pathway of translational initiation (for reviews, see Sachs 2000; Schwartz and Parker 2000). Both yeast (Tarun and Sachs 1996) and human (Imataka et al. 1998) PABPs interact with the translation initiation factor eIF4G, thereby causing circularization of the mRNA via bridging its 5′ and 3′ termini (closed loop model [Jacobson 1996]). Such an interaction could facilitate formation of the 48S translation initiation complex de novo, the 60S ribosomal subunit joining, and/or translational reinitiation (Munroe and Jacobson 1990; Tarun and Sachs 1995; Kahvejian et al. 2001; Searfoss et al. 2001). However, the mechanism of translational stimulation may be more complex than just an interaction between PABP and eIF4G, as these two phenomena could be genetically uncoupled (Kessler and Sachs 1998). Moreover, observations made in the yeast strains conditionally defective in poly(A) tail synthesis suggest the possibility that yeast PABP (Pab1p) also interacts directly with ribosomes (Proweller and Butler 1996). In addition, recent evidence in yeast and mammalian cells links PABP to translational termination and eukaryotic release factor 3 (eRF3; Cosson et al. 2001; Uchida et al. 2002). Thus, PABP may participate in translation in multiple ways.
In addition, PABP participates in the control of the mRNA degradation in the cytoplasm, although the exact way in which PABP regulates mRNA decay may vary between species and between different transcripts. In yeast, the major mRNA degradation route occurs via deadenylation-dependent decapping (for reviews, see Schwartz and Parker 2000; Wilusz et al. 2001b). The yeast PABP, Pab1p, impedes mRNA decapping until the process of deadenylation progresses to the point when only 12–15 Å residues are left, and the last Pab1p binding site on the mRNA poly(A) tail has been eliminated. It can be envisioned that dissociation of the last molecule of Pab1p causes a significant rearrangement in the structure of the mRNP, and leads to a loss of the 3′ end–5′ end association, enabling the decapping enzyme to attack the 5′ cap. A competition between the translation initiation and mRNA decay factors plays an important role in the control of deadenylation and decapping, for example, mutations in yeast translation initiation factors promote both decapping and deadenylation rates (Schwartz and Parker 1999). Inhibition of decapping by PABP was also observed in mammalian cell extracts, although in that case, it seems to be independent of the eIF4E/eIF4G interaction with PABP (Gao et al. 2001). Moreover, the circularization of the mRNA via the eIF4E/eIF4G/Pab1p interaction in yeast extracts also accounts for only a part of the inhibitory effect of Pab1p on decapping, as a partial inhibition of decapping by Pab1p could still be observed when eIF4E was prevented from interacting with the 5′ cap (Wilusz et al. 2001a). Furthermore, a deletion of the eIF4G-interacting domain from the yeast PABP that was tethered to the mRNA in a poly(A)-independent manner did not affect its ability to inhibit mRNA decay (Coller et al. 1998).
The role of PABP in deadenylation is complex. PABP acts as a deadenylation inhibitor in mammalian cell extracts (Bernstein et al. 1989; Ford et al. 1997) and when overexpressed in Xenopus oocytes (Wormington et al. 1996). In the case of the α-globin mRNA, PABP interacts with the αCP complex that binds to the 3′-UTR of this transcript, and slows the rate of its deadenylation (Wang et al. 1999). A more complex picture arises in the case of the c-fos mCRD (major coding region determinant), a sequence element that specifies rapid deadenylation and decay of the c-fos transcript in a manner that is dependent on translation (Grosset et al. 2000). The mCRD binding complex interacts with PABP, thus bridging the mCRD and the poly(A) tail. This interaction prevents deadenylation prior to the initial round(s) of translation, but promotes it after the complex has been displaced by the ribosomes traversing the mCRD, possibly because the removal of mCRD binding complex also promotes dissociation of PABP from the poly(A) tail, making it vulnerable to exonucleolytic attack (Grosset et al. 2000). Yeast Pab1p also inhibits deadenylation in vitro (Wilusz et al. 2001a). On the other hand, Pab1p is also required for the proper rate of deadenylation in yeast in vivo (Caponigro and Parker 1995). A possible resolution of this paradox can be envisioned if Pab1p actually promotes the entry of the mRNA into the decay pathway, rather than accelerates deadenylation per se. Yeast strains lacking Pab1p (but viable due to bypass suppressor mutations) exhibit a temporal lag before mRNA decay commences, which likely reflects a role of Pab1p in efficient mRNA biogenesis (Caponigro and Parker 1995).
In this study, we provide evidence supporting and extending the view that PABP is important for the mRNA biogenesis. Using cross-species complementation of the yeast pab1 null mutant by the Arabidopsis PAB3 cDNA, we have shown previously that rescue of viability of the yeast pab1Δ mutant required neither the restoration of poly(A)-dependent translation nor the protection of the 5′ cap from premature removal (Chekanova et al. 2001). However, plant PABP significantly reduced or eliminated the lag prior to mRNA decay in yeast (Chekanova et al. 2001). In this work, this system is further exploited to examine the nature of the lag prior to mRNA decay in pab1Δ cells and its correction by PABP. We have found that the Arabidopsis PAB3 accelerated the entry of the mRNA into the degradation pathway, as well as its entry into the translated pool when expressed in yeast. PAB3 was also able to partially restore the poly(A) tail length control during polyadenylation reaction, in the PAN-dependent manner. Furthermore, a fraction of PAB3 was physically present in the nucleus in the complemented yeast strain. The substitution of the plant PAB3 for the endogenous yeast Pab1p caused synthetic lethality with rna15-2, an allele of the gene encoding the Rna15p subunit of the pre-mRNA cleavage factor CFIA, and with gle2-1, a mutant allele of the gene encoding the NPC-associated protein. Finally, overexpression of PAB3 in yeast rescued the cold sensitivity and alleviated the mRNA export block of the nab2-1 mutant strain, which is defective in one of the shuttling hnRNP proteins required for mRNA export. We propose that eukaryotic PABPs have an evolutionarily conserved function in facilitating mRNA biogenesis and export.
RESULTS
PAB3 and Pab1p act directly on the target mRNA to correct the lag prior to the entry of mRNA into the decay pathway in yeast
To ascertain that PABPs (the heterologous Arabidopsis PAB3, as well as the endogenous yeast Pab1p) reduce or eliminate the lag prior to the entry of the mRNA into the degradation pathway directly (that is, by physically binding to the target mRNAs) a yeast strain was constructed that simultaneously expressed two nearly identical MFA2-based reporter mRNAs that differed mainly in the presence or absence of the poly(A) tail—the binding site for the PABP. The first reporter was the same GAL1 promoter-driven MFA2pG mRNA (Caponigro and Parker 1995) that we used previously in the transcriptional pulse-chase analysis in the presence or absence of PAB3 in yeast (Chekanova et al. 2001). The second MFA2-based reporter (MFA2-MS2-RZ) contained the ribozyme derived from the hepatitis δ virus in place of the normal polyadenylation signal. This latter construct gives rise to a transcript that undergoes autocatalytic processing in vivo in yeast cells, resulting in nonadenylated mRNA (Quadt et al. 1995; Coller et al. 1998). This construct was also expressed from the GAL1 promoter. Other differences between these two reporter transcripts, namely the presence of the poly(G) tract in the 3′-UTR of the first reporter, and the MS2 protein binding sites in the second reporter, have been demonstrated previously to have no effect on the mRNA turnover properties of the respective transcripts (Decker and Parker 1993; Coller et al. 1998).
Because of the lethality associated with the pab1Δ mutation, the rate of decay of these two reporters was analyzed in the spb2Δ suppressor background, expressing either yeast Pab1p, Arabidopsis PAB3, or no PABP (strains YDB246, YDB256, and YRP881, respectively; genotypes of the yeast strains used in this work are given in Table 1 ▶), by use of a transcriptional shutoff of the GAL1 promoter. The spb2Δ mutation is a bypass suppressor of pab1Δ, which causes a loss of the 60S ribosomal subunit protein RPL39 (Sachs and Davis 1989), but does not have any direct effect on mRNA turnover (Caponigro and Parker 1995). The suppressor phenotype is probably due to the resulting underaccumulation of the ribosomal 60S subunits, which are thus less able to sequester the 40S subunits into empty 80S couples (Sachs and Davis 1989). The resulting excess of the free 40S subunits (which are normally limiting for initiation) may allow the cell to translate mRNA more efficiently, thereby indirectly compensating for the loss of Pab1p. In support of this view, certain mutations that exacerbate the 40S subunit deficit are partially suppressed by decreasing the 60S subunit levels (Finley et al. 1989). In addition, the binding of the charged tRNA to the ribosome A site, as well as the kcat of the peptidyltransferase in spb2Δ mutant are increased by 40%–70% relative to the wild-type ribosomes (Dresios et al. 2000, 2001) at the expense of translational fidelity.
TABLE 1.
Genotypes of the yeast strains used in this work
| Strain | Relevant genotype | Reference |
| YRP881 | trp1 ura3 leu2 his4 cup1:: LEU2pm spb2:: URA3 pab1:: URA3 | Caponigro and Parker (1995) |
| YDB203 | ade2 his3 leu2 trp1 ura3 pab1:: HIS3, pDB419 (TRP1/CEN/pGAL1-PAB3) | Chekanova et al. (2001) |
| YDB220 | PDB464 (TRP1/CEN/PAB1) in YRP881 | This work |
| YDB221 | trp1 ura3 leu2 his4 cup1:: LEU2pm spb2:: URA3, pab1:: URA3, pDB463 (TRP1/2μ/pADHI-PAB3) | Chekanova et al. (2001) |
| YDB227 | pMM2-6 in YRP881 | This work |
| YDB236-8 | trp1 ura3 leu2 his4 cup1:: LEU2pm spb2:: URA3 pab1:: URA3 pan3:: TRP, pDB488 (G418r/2μ/pADHI-PAB3) | Chekanova et al. (2001) |
| YDB246 | pMM2-6 and pDB490 in YRP881 | This work |
| YDB256 | pMM2-6 and pDB493 in YRP881 | This work |
| YJA223 | his3 leu2 trp1 ura3 nab2:: LEU2 TRP1:: nab2-21 | Hector et al. (2002) |
| YRH201C | ade2 his3 leu2 trp1 ura3 can1 nab2:: HIS3 pGAL:: NAB2 | Hector et al. (2002) |
| ACY429-1038 | trp1 ura3 leu2 his3 NAB2:: HIS3, pAC1038 (LEU2/CEN/nab2-1) | Green et al. (2002) |
| BJ5464 | ura3 trp1 leu2 his3 pep4:: HIS3 prbD1.6R can1 | Jones (1991) |
| rna14-1W | ura3 trp1 ade2 leu2 his3 rna14-1 | Amrani et al. (1997) |
| rna15-2W | ura3 trp1 ade2 leu2 his3 rna15-2 | Amrani et al. (1997) |
| SWY1022 | ade2 ade3 ura3 his3 leu2 trp1 can1-100 nup100:: HIS3 | Murphy et al. (1996) |
| SWY1024 | ade2 ade3 ura3 his3 leu2 lys2 can1-100 nup100:: HIS3 | Murphy et al. (1996) |
| YCH130 | ade2 ade3 ura3 his3 leu2 lys2 can1-100 | Murphy et al. (1996) |
| W303α | ade2 ade3 ura3 his3 leu2 trp1 can1-100 | Murphy et al. (1996) |
| SWY1136 | ade2 ura3 his3 leu2 trp1 can1-100 gle2-1 | Murphy et al. (1996) |
| mtr2-9 | ura3 ade2 leu2 his3 trp1 mtr2:: HIS3, pRS315/mtr2-9 | Santos-Roza et al. (1998) |
| MTR2 | ura3 ade2 leu2 his3 trp1 mtr2:: HIS3, pRS316/MTR2 | Santos-Roza et al. (1998) |
| mex67-5 | ade2 his3 leu2 trp1 ura3 mex67:: HIS, pUN100/mex67-5 | Segref et al. (1997) |
| MEX67 | ade2 his3 leu2 trp1 ura3 mex67:: HIS3, pUN100/MEX67 | Segref et al. (1997) |
| FY23 | ura3 leu2 trp1 | Winston et al. (1995) |
| FY86 | his3 leu2 ura3 | Winston et al. (1995) |
| CCY282 | ura3 leu2 trp1 rat2-1 | Heath et al. (1995) |
| OLY101 | his3 leu2 ura3 rat3-1 | Li et al. (1995) |
| LGY101 | his3 leu2 ura3 rat7-1 | Gorsch et al. (1995) |
| TDY105 | trp1 leu2 ura3 nupI45-10 | Dockendorff et al. (1997); Snay-Hodge et al. (1998) |
| CSY550 | trp1 leu2 ura3 rat8-2 | Snay-Hodge et al. (1998) |
Mutant and corresponding isogenic wild-type strains used in synthetic lethal tests are separated into the bottom portion of the table and are shaded in gray.
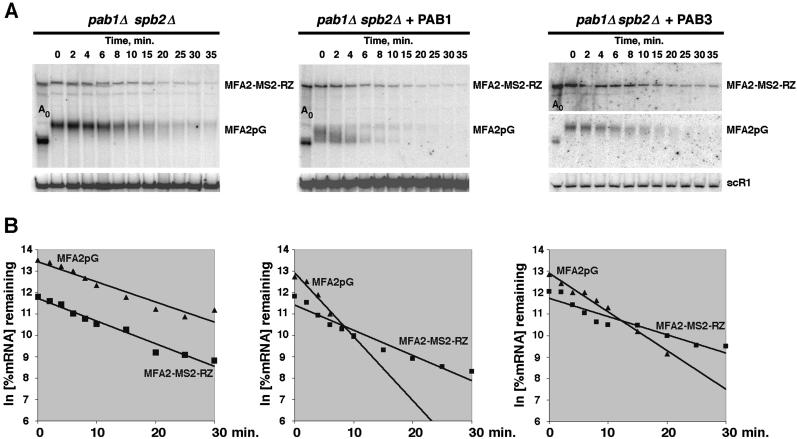
The decay rates of the MFA2pG and MFA2-MS2-RZ mRNAs in the absence of PABP in the cell were virtually indistinguishable (Fig. 1 ▶, T½ = 7 min in both cases). Introduction of either the Pab1p-expressing construct or PAB3-expressing construct resulted in selective acceleration of the decay of the MFA2pG mRNA, but not of the MFA2-MS2-RZ mRNA. Because we have shown previously that PAB3 accelerates mRNA decay in yeast chiefly via acceleration of the rate of entry of the mRNA into the decay pathway, rather than via activating the mRNA decay per se, these data indicate that the endogenous yeast Pab1p, as well as the plant PAB3 expressed in yeast (Chekanova et al. 2001), must act on the target mRNAs directly to be able to correct the lag prior to the onset of the mRNA decay that is observed in the PABP-deficient strains. In other words, these PABP proteins promoted the rate of entry into the decay pathway of only those mRNA to which they were able to bind physically. Two possible explanations for this result are: (1) that the MFA2-MS2-RZ mRNA that could not bind PABP was normally exported from the nucleus, but was somehow shielded from the cytoplasmic decay; or (2) that its dwelling time in the nucleus was longer than in the case of the MFA2pG mRNA. We favor the latter explanation, because the mRNAs that cannot bind PABP are usually less stable, rather than more stable (Bernstein et al. 1989; Wormington et al. 1996; Ford et al. 1997; Wang et al. 1999; Grosset et al. 2000), and because the results of a recent FISH study using an analogous ribozyme-containing construct in yeast, strongly suggest that its export from the nucleus is compromised (Libri et al. 2002).
FIGURE 1.
PAB3 and Pab1p act to correct the lag by binding to the target mRNAs, rather than by acting indirectly. (A) Decay of the MFA2pG and MFA2-MS2-RZ mRNAs analyzed by Northern blotting in the pab1Δ spb2Δ yeast cells expressing Pab1p (YDB246), PAB3 (YDB256), or no PABP (YRP881), as indicated above the respective panels, after the transcriptional shutoff by glucose of the GAL1 promoter that drives the expression of the MFA2pG and MFA2-MS2-RZ transcripts. Time points after the glucose addition are shown above the panels. RNA in the lane marked Ao was a 0-min time point sample that was deadenylated by the oligo(dT)/RNaseH treatment. Blots were probed sequentially with the oligonucleotide probes specific for MFA2-MS2-RZ and MFA2pG. Signals were normalized relative to the scR1 RNA (RNA pol III transcript) signals (bottom). (B) Quantitation of the decay profiles shown in A.
Lagging mRNA population retains an intact 5′ cap
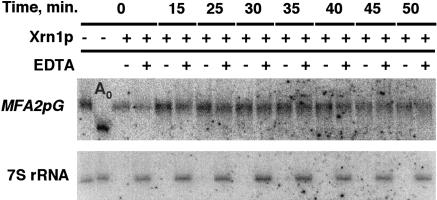
In an effort to shed additional light on the mechanistic nature of the temporal lag prior to the entry of mRNA into the decay pathway in the pab1Δ strains, we examined the 5′ cap status of transcripts experiencing the lag. This was prompted by a recent discovery that some of the decapping and the 5′–3′ degradation steps of the major mRNA decay pathway in yeast occur in the discrete cytoplasmic processing bodies (P bodies; Sheth and Parker 2003). Importantly, mRNA decay intermediates that lack the 5′ cap appear to be concentrated in P bodies, whereas the intact full-length mRNAs are distributed throughout the cytoplasm (Sheth and Parker 2003). This raises the possibility that transcripts experiencing the lag were retained in the P bodies, but were somehow resistant to further decay. To assess the cap status of the mRNAs experiencing the lag, the RNA samples from the transcriptional pulse-chase experiment conducted in the YRP881 (pab1Δ spb2Δ) strain were subjected to the exhaustive treatment by recombinant Xrn1p in vitro. Xrn1p is a Mg++-requiring enzyme that readily degrades uncapped RNA, but is inactive toward RNA containing the 5′ cap structure. Degradation of the endogenous 7S rRNA precursor, which is naturally uncapped, served as an internal control for the completeness of the Xrn1p digestion. As shown in Figure 2 ▶, MFA2pG mRNA experiencing the lag prior to its decay was completely resistant to the Xrn1p activity. These data demonstrate that the lagging population of mRNA retains its 5′ cap, and thus rule out the possibility that the observed lag is due to a sequestration of the messenger RNA and shielding it from the 5′–3′ exonucleolytic degradation after it has undergone decapping.
FIGURE 2.
Assay of the 5′ cap status of the MFA2pG mRNA during the lag in the YRP881 (pab1Δ spb2Δ) strain. Total RNA samples corresponding to the time points of the transcriptional pulse-chase experiment indicated on the top of the figure were treated with the recombinant 5′–3′ exonuclease Xrn1p in the presence or absence of the EDTA, as indicated above the lanes. RNA in the lane marked Ao was deadenylated by the oligo(dT)/RNaseH treatment. Sequential Northern blot hybridizations with the MFA2pG (top) and 7S rRNA precursor (bottom) specific probes are shown.
Substitution of the PAB3 for the yeast Pab1p causes synthetic lethality in combination with the rna15-2 and gle2-1
A genetic approach was also used to try to gain further insight into the nature of the lag. We have demonstrated previously that PAB3 is no longer capable of supporting cell viability in yeast in the absence of the PAN3 gene, which encodes a subunit of the poly(A) nuclease (PAN) that is indispensable for the catalytic function of this enzyme. This situation is essentially equivalent to a synthetic lethal interaction, if one assumes that the substitution of the plant PAB3 for the endogenous yeast Pab1p is equivalent to a partial loss of function of Pab1p. This assumption is justified, because the plant PAB3 cannot perform two of the known functions of the yeast Pab1p, that is, it neither protects the mRNA 5′ cap from the premature removal in yeast, nor interacts with the yeast eIF4G. As a consequence, it is incapable of supporting the poly(A)-dependent stimulation of translation, as well as the synergy in translational stimulation that is normally observed between the effects of the 5′ cap and the poly(A) tail in yeast cells. In addition, PAB3 corrects the temporal lag prior to the entry of mRNA into the decay pathway in the pab1Δ strains partially, rather than perfectly (Chekanova et al. 2001).
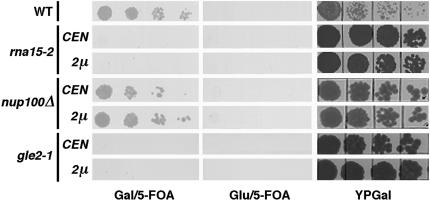
The finding that pan3Δ mutation is synthetically lethal with the substitution of PAB3 for Pab1p prompted us to conduct a series of binary synthetic lethal tests between the substitution of PAB3 for Pab1p and mutations in various factors involved in mRNP biogenesis, processing, and export, including rna14-1, rna15-2, hrp1-3, nup100Δ, nup145-10, rat2-1, rat3-1, rat7-1, rat8-2, mex67-5, mtr2-9, and gle2-1 (see Table 1 ▶ and Materials and Methods). Two additional synthetic lethal interactions were found among the 12 mutants tested. Importantly, we have observed no correlation between the degree of the growth defect in the various mutants tested and their tendency to exhibit synthetic lethality in combination with the substitution of PAB3 for Pab1p. One of the synthetic lethal interactions was with the rna15-2 allele, and the other with gle2-1. Rna15p is a component of CFIA, a complex of four polypeptides that is required for the pre-mRNA cleavage and polyadenylation (Gross and Moore 2001). The notion of the evolutionarily conserved functional link between the PABP and Rna15p is also corroborated by the earlier finding that the yeast Pab1p suppresses the rna15-2 allele when overexpressed, and that it interacts physically with Rna15p in the pulldown experiments in vitro (Amrani et al. 1997). Gle2p is an NPC-associated protein. The gle2-1 mutation leads to an arrest of mRNA export at nonpermissive temperature, as well as to herniation and clustering of NPCs (Murphy et al. 1996). At the permissive temperature, ~25% of gle2-1 cells exhibit accumulation of the poly(A)+ RNA in the nucleus, but no ultrastructural aberrations are evident (Murphy et al. 1996). Furthermore, it was proposed recently that Gle2p may act to help deliver an export complex to NPC (Blevins et al. 2003). Taken together with the biochemical data presented below, these findings lead to a view that PAB3 participates in the nuclear functions of Pab1p in yeast. Specifically, the genetic interactions with rna15-2 and gle2-1 may suggest the links between the evolutionarily conserved nuclear function of PABP and early and/or late nuclear steps of mRNA biogenesis.
PAB3 enters the yeast cell nucleus and partially rescues the poly(A) tail length control during polyadenylation in vitro
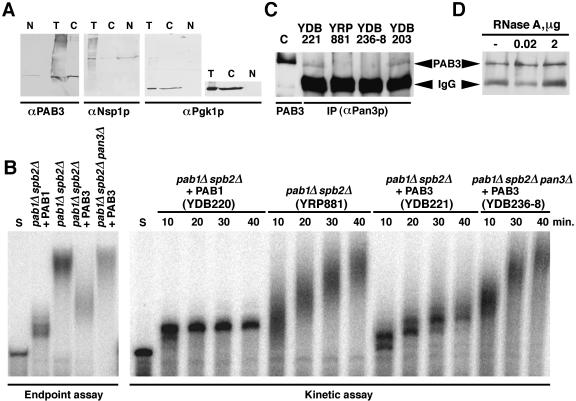
A fraction of the endogenous yeast Pab1p has been shown previously to localize in the nucleus and associate with CFIA (Minvielle-Sebastia et al. 1997). Genetic links between the PAB3 function in yeast and nuclear proteins (above) imply that PAB3 may also be present physically in the nucleus of the yeast cell. To address this possibility, subcellular fractionation of the PAB3-complemented YDB203 cells (pab1Δ spb2Δ + PAB3) was undertaken. Cells from the YDB203 strain were converted to spheroplasts, lysed, fractionated by Percoll gradient centrifugation, and the nuclear and the cytoplasm-enriched fractions probed with the PAB3-specific antibodies. The data (Fig. 3 ▶) demonstrate that a fraction of PAB3 is present in the nucleus in the PAB3-complemented yeast strain. Densitometric scanning and quantitation of the immunoblots suggest that at least 7% of the total cellular PAB3 in yeast is nuclear at steady state, although this figure is likely to be an underestimate, because the subcellular fractionation protocol causes partial loss of the cytoplasmic material, as well as contamination of the cytoplasmic fraction with the content of the lysed nuclei (as can be seen from the distribution of the Nsp1p signal, Fig. 3 ▶). On the other hand, the extent of contamination of the nuclear fraction with cytoplasmic material was estimated by probing for an abundant cytoplasmic protein Pgk1p (Fig. 3 ▶) and for eIF4G (data not shown). No detectable cytoplasmic contamination was observed.
FIGURE 3.
Example of the plasmid shuffle test for the synthetic lethality between the substitution of PAB3 for Pab1p and mutations in various factors involved in mRNP biogenesis, processing, and export (see Table 1 ▶ for the full listing). The respective mutant strains (rna15-2, nup100Δ, gle2-1, and the wild-type control are shown) with their chromosomal PAB1 gene disrupted and the functional copy of PAB1 provided on CEN/URA3 plasmid, were transformed with either centromeric or high-copy (2 μ) plasmid-bearing pGAL-PAB3 cassette, as indicated at left. Transformants were grown in synthetic complete medium with uracil and dilution series plated onto galactose or glucose-based synthetic medium with 5-FOA to select for the cells that have lost the CEN/URA3/PAB1 plasmid. Only cells that grew on Gal/5-FOA, but not on Glu/5-FOA, resulted from complementation of the pab1 deletion by the pGAL-PAB3 expression cassette. Plating on the rich YPGal medium (to provide the total viable cell number) is also shown. All plates were grown for 6 d.
The genetic link between PAB3 function in yeast and the endogenous Rna15p, a subunit of the cleavage factor I, suggests that PAB3 may be able to function in cleavage and/or polyadenylation of pre-mRNA in yeast. To test this possibility, we assayed cleavage, polyadenylation, as well as a coupled cleavage/polyadenylation of the GAL7 pre-mRNA-derived artificial substrate in extracts prepared from the YDB220 (pab1Δ spb2Δ + Pab1p), YRP881 (pab1Δ spb2Δ) and YDB221 (pab1Δ spb2Δ + PAB3) cells. We saw no evidence of any influence of PAB3 on pre-mRNA cleavage (data not shown). This was expected, because the yeast Pab1p also has no effect on the pre-mRNA cleavage step of the 3′ end processing (Amrani et al. 1997; Kessler et al. 1997; Minvielle-Sebastia et al. 1997; Brown and Sachs 1998). Therefore, all subsequent assays were performed using the precleaved substrate. In the extracts YDB220 (pab1Δ spb2Δ + Pab1p; Fig. 1B ▶, left), as well as in the wild-type extracts (data not shown), the precleaved substrate was extended by ~90 Å residues, which closely parallels the extent of polyadenylation observed in vivo. In contrast, in the pab1Δ spb2Δ cells (YRP881), control of the extent of polyadenylation was lost, resulting in the synthesis of the abnormally long poly(A) tails (~150 Å residues). This phenomenon has been observed previously in extracts prepared from the Pab1p-deficient, as well as PAN-deficient cells (Fig. 1B ▶; Amrani et al. 1997; Kessler et al. 1997; Minvielle-Sebastia et al. 1997; Brown and Sachs 1998). Importantly, in extracts prepared from the YDB221 (pab1Δ spb2Δ + PAB3) cells, control of the extent of polyadenylation was partially restored, resulting in the maximum of the distribution of the poly(A) synthesized at the 30-min time point of ~120 As.
Because in the previous studies PAN function has been shown to be important for proper control of the polyadenylation in vitro (Brown and Sachs 1998), its possible requirement for the PAB3-mediated effect was also examined. Extracts from the YDB236-8 (pab1Δ spb2Δ pan3Δ + PAB3) were also prepared and assayed side-by-side with the extracts from the YDB220 (pab1Δ spb2Δ + Pab1p), YRP881 (pab1Δ spb2Δ), and YDB221 (pab1Δ spb2Δ + PAB3) cells in the 30-min endpoint assay, as well as in the kinetic assay, to follow the dynamic pattern of changes in the poly(A) tail length. Two main observations could be made. First, partial restoration of the poly(A) tail length control by PAB3 was completely dependent on the PAN function, as it was abolished in the pan3Δ genetic background. Thus, the PABP-stimulated, PAN-dependent control of the extent of the poly(A) tail addition may be an evolutionarily conserved phenomenon. Second, the kinetic behavior of the polyadenylation reaction was different in the Pab1p containing (strain YDB220, pab1Δ spb2Δ + Pab1p), PABP-deficient (strain YRP881, pab1Δ spb2Δ), and PAB3 containing (strain YDB221, pab1Δ spb2Δ + PAB3) extracts. In the YDB220 extracts, the maximal extent of polyadenylation was reached in 10 min, and no further changes in the poly(A) tail length occurred thereafter. In contrast, although the initial rate of polyadenylation in the pab1Δ spb2Δ (strain YRP881) extracts was similar, the poly(A) addition continued throughout the time course of the experiment. In the extracts from the PAB3-expressing cells (YDB221), although polyadenylation also continued past the 10-min time point, it proceeded with the much slower apparent rate. Importantly, in the extracts of YDB236-8 (pab1Δ spb2Δ pan3Δ + PAB3), the dynamic pattern of changes of the poly(A) tail length distribution was similar to those observed in the pab1Δ spb2Δ (strain YRP881) extracts.
Yeast Pab1p interacts directly with Pan3p (D. Mangus and A. Jacobson, pers. comm.). The PAN requirement for the partial restoration of the control the poly(A) tail length by PAB3 in vitro, together with the synthetic lethality between the PAB3 substitution for the yeast Pab1p and the loss of Pan3p (Chekanova et al. 2001), prompted us to test whether PAB3 and Pan3p might also interact physically. To this end, immunoprecipitation with anti Pan3p antibody was conducted from the total extract of the strain expressing PAB3 in the pab1Δ spb2Δ background (strain yDB221), as well as from an isogenic control strain YDB236-8 that lacked the PAN3 gene (Fig. 4C ▶). In addition, a YDB203 strain, which was complemented by PAB3, and therefore, required its expression for growth, and pab1Δ spb2Δ PAN3 strain YRP881 were also tested. PAB3 was detected in the immunoprecipitates from the yDB221 and YDB203 cells, but not from the YDB236-8 (pab1Δ spb2Δ pan3Δ + PAB3) cells (Fig. 4C ▶). Moreover, this interaction was not bridged by RNA, because it was insensitive to RNase A treatment (Fig. 4D ▶). Thus, we conclude that PAB3 is able to partially control the extent of the poly(A) tail addition in yeast extracts in the PAN-dependent manner, and is able to interact with the PAN subunit in yeast.
FIGURE 4.
Evidence that PAB3 functions in the nucleus in yeast. (A) Results of the subcellular fractionation of the extract of the PAB3-complemented yeast strain YDB203. Equivalent amounts of the total unfractionated extract (T), as well as the nuclear (N) and cytoplasmic (C) fractions were immunoblotted and probed for PAB3, Nsp1p (nuclear marker) and Pgk1p (cytoplasmic marker). Overexposure of the immunoblotting for Pgk1p is shown at right to demonstrate the absence of the cytoplasmic contamination in the nuclear fraction. (B) Results of the endpoint (left) and kinetic (right) polyadenylation assays in vitro using the precleaved substrate. Relevant genotypes of the strains from which the extracts were prepared are shown at top. Lanes marked S contain the precleaved substrate RNA only. (C) PAB3 copurifies with the yeast Pan3p in a coimmunoprecipitation assay. Extracts from the yDB221 (pab1Δ spb2Δ + PAB3), yDB236-8 (pab1Δ spb2Δ pan3Δ + PAB3), and YDB203 (PAN3 strain complemented by PAB3) cells were incubated with the antiPan3p antiserum immunoprecipitates captured on protein A agarose, extensively washed, and the bound material immunoblotted for PAB3. Recombinant PAB3 was loaded in the leftmost lane (marked C) as a positive control. Positions of the signals corresponding to PAB3 and IgG heavy chain are indicated by arrowheads. (D) Communoprecipitation of PAB3 and Pan3p from YDB203 extracts is resistant to treatment with 0.02–2 μg of RNase A for 40 min at room temperature.
PAB3 accelerates the entry of the newly synthesized mRNA into the translated pool
In considering possible mechanisms of complementation of the pab1Δ phenotype by PAB3, it seemed counterintuitive that just the acceleration of the mRNA decay, which results from the lag correction by PAB3, would be sufficient for the cell viability in the absence of the endogenous Pab1p. Rather, we reasoned that the expression of the plant PAB3 protein in the PABP-deficient yeast cells might have additional consequences on gene expression, other than just the correction of the lag prior to the entry of the mRNA into the decay pathway. We hypothesized that an acceleration of the rate of mRNA biogenesis and/or export might be a part of such a mechanism, because PAB3 functions in the 3′ end processing in yeast (above), and because the endogenous yeast Pab1p also has been implicated in mRNA biogenesis, and particularly in the 3′ end processing (Caponigro and Parker 1995; Amrani et al. 1997; Kessler et al. 1997; Minvielle-Sebastia et al. 1997; Brown and Sachs 1998; Mangus et al. 1998; Morrissey et al. 1999), which in turn is important for mRNA export in yeast (Brodsky and Silver 2000; Hilleren et al. 2001; Dower and Rosbash 2002; Hammell et al. 2002). One expected consequence of such an activity of PABP in facilitating mRNP biogenesis would be an acceleration of the rate of entry of the mRNA into the translated pool.
To test this possibility, we have compared the kinetics of accumulation of the protein encoded by the reporter mRNA after its transcriptional induction in the PABP-deficient and PAB3-expressing cells, as an indirect measure of the kinetics of the entry of the reporter mRNA into the translated pool. An advantage of using PAB3-expressing strain in this analysis is that the Arabidopsis PAB3 supports neither the poly(A)-dependent enhancement of translation nor the poly(A)/cap synergy in yeast (Chekanova et al. 2001). Thus, any effect of PAB3 on the kinetics of the accumulation of the protein that is encoded by the reporter mRNA toward its steady state would be largely due to an effect of PAB3 on the kinetics of the entry of the mRNA into the translation cycle, rather than due to effects on the mRNA translation efficiency.
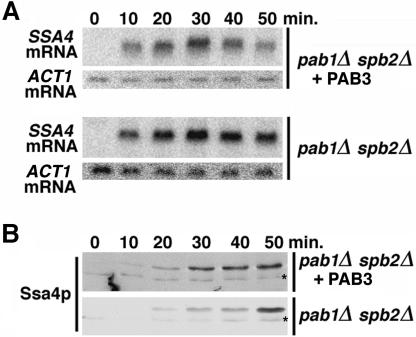
Because the very small size and an extreme hydrophobicity of the MFA2-encoded peptide made quantitative Western analyses difficult, a different reporter was used in this assay. Yeast SSA4 mRNA, which encodes a Hsp70-type heat-shock protein Ssa4p, is virtually undetectable in cells grown at 28°C, but is rapidly induced after the shift to 42°C. In the YDB221 (pab1Δ spb2Δ + PAB3) cells, SSA4 mRNA level peaked at a ~30-min time point after its induction by heat shock and then declined, which reflected its decay (Fig. 5A ▶). In contrast, in the YRP881 (pab1Δ spb2Δ) cells, SSA4 message was stable throughout the time course of the experiment. Thus, SSA4 mRNA is also subject to the lag prior to the onset of mRNA decay, and this lag was corrected by the Arabidopsis PAB3 in yeast at 42°C. The critical observation was that PAB3 also reproducibly accelerated the rate of accumulation of the Ssa4p protein toward its steady-state level (Fig. 5B ▶), so that the Ssa4p steady state was reached faster in the YDB221 (pab1Δ spb2Δ + PAB3) cells than in the YRP881 cells lacking PABP (15 min versus 30 min). Because PAB3 supports neither the poly(A)-dependent stimulation of the initiation of the protein synthesis in yeast, nor the 5′ cap/poly(A) translational synergy (Chekanova et al. 2001), we conclude that the PAB3-dependent acceleration of the rate of approach of the Ssa4p protein level toward its steady state reflects the acceleration of the entry of SSA4 mRNA into the translated mRNA pool by PAB3.
FIGURE 5.
Kinetics of the accumulation of the SSA4 mRNA (A, Northern blot) and Ssa4p heat-shock protein encoded by it (B, Western blot) in the YRP881 (pab1Δ spb2Δ) and YDB221 (pab1Δ spb2Δ + PAB3) strains after the shift to 37°C. ACT1 mRNA (A) and nonspecific crossreacting asterisk (B) were used as loading controls.
PAB3 overexpression rescues the cold-sensitive phenotype and mRNA export defect of the yeast nab2-1 mutant
The above observation that PAB3 was able to accelerate the rate of entry of the SSA4 mRNA into the translated pool in yeast, as well as the known role of the yeast Pab1p in mRNA biogenesis, led us to test the possibility that the Arabidopsis PAB3 could affect the mRNA export in yeast. It has been demonstrated recently that overexpression of the yeast Pab1p, or targeting of the Pab1p into the nucleus, lead to the suppression of the cold-sensitive lethal phenotype and the mRNA export block of the nab2-21 mutant (Hector et al. 2002). The Nab2p protein is one of the shuttling hnRNP proteins in yeast that associates with the poly(A)+ mRNA in the nucleus, and plays an important role in facilitating the assembly of the export-competent mRNP (Green et al. 2002; Hector et al. 2002). In addition, Nab2p has a second function in the poly(A) tail length control during nuclear polyadenylation, and the nab2 mutant cells accumulate hyperadenylated mRNA (Hector et al. 2002). Overexpression or nuclear targeting of Pab1p suppressed the cold-sensitive lethality and mRNA-export defect, although it had no effect on the poly(A) tail length control defect in the nab2-21 cells (Hector et al. 2002). This suggests that Pab1p may have a role in the biogenesis of the export-competent mRNP that is distinct from the poly(A) tail length control per se.
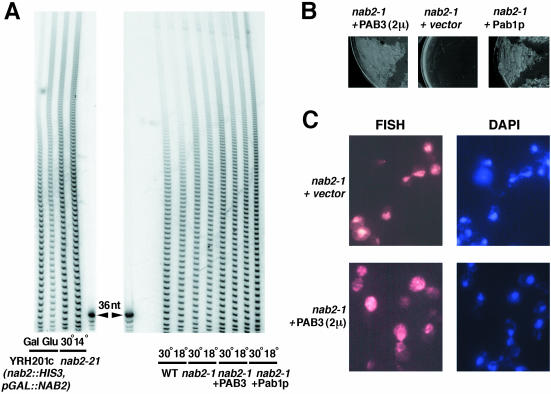
We have asked whether the heterologous PAB3 would be able to restore the mRNA export in the nab2 mutant cells. We chose a different nab2 allele, nab2-1, as it showed much lower frequency of spontaneous suppressors and/or revertants than nab2-21. Also, nab2-1 had a considerably less-pronounced poly(A) tail length-control defect, compared with nab2-21 (Fig. 6A ▶). However, much like with nab2-21 allele, this poly(A) tail length defect was equally evident at both permissive and nonpermissive temperatures (Fig. 6A ▶). The nab2-1 mutant is also cold sensitive, and shows an accumulation of the poly(A)+ mRNA in the nucleus at the nonpermissive temperature (Fig. 6C ▶; Green et al. 2002).
FIGURE 6.
Consequences of the overexpression of PAB3 in the nab2-1 mutant background. (A) A comparison of the poly(A) tail phenotypes of the nab2-21 (left) and nab2-1, as well as nab2-1 strain overexpressing PAB3 or Pab1p (right) at normal (30°C) and low (14 or 18°C, as indicated) temperatures. Total poly(A) tail length distributions in the YRH201c cells, in which Nab2p is expressed from GAL promoter, on galactose, and after the shift into glucose medium are also shown (left). A 36-nucleotide long DNA oligonucleotide was used as a size marker. (B) Phenotypic rescue of the cold sensitivity of the nab2-1 mutant by overexpression of Pab1p or PAB3. The strains shown above the respective panels were streaked onto the selective synthetic medium and incubated at 18°C for 12 d. (C) FISH with the (dT)70 probe of the nab2-1 mutant cells transformed with the 2μ/PAB3 construct or an empty 2μ vector, as indicated, after the shift to the nonpermissive temperature (18°C) for 16 h. Staining for nuclei with DAPI is shown in the two right panels.
We overexpressed the plant PAB3 from the ADH1 promoter construct borne on a high-copy plasmid (this resulted in ~2.5–3-fold higher levels of PAB3 compared with the amount of Pab1p in the wild-type strain; data not shown), and also added one extra copy of the PAB1 gene on a single-copy plasmid [approximately twofold overexpression of PAB1 was sufficient to rescue the mRNA export defect and cold sensitivity of nab2-21 mutant (Hector et al. 2002)], in the nab2-1 strain. In either case, the poly(A) tail length distribution was not changed noticeably (Fig. 6A ▶). However, PAB3 reproducibly rescued the cold sensitivity of the nab2-1 cells at 14°C, even though the rate of growth was still slower than in the cells containing one extra copy of PAB1 (Fig. 6B ▶). The effect of PAB3 on the mRNA export defect at 18°C was then tested by FISH with the oligo(dT) probe. Importantly, PAB3 partially reversed the nuclear accumulation of the poly(A)+ mRNA observed in the nab2-1 cells at 18°C (Fig. 6C ▶), suggesting that PAB3 can stimulate mRNA biogenesis and/or export in yeast.
DISCUSSION
Evidence presented in this work strongly suggests that eukaryotic PABP has an evolutionarily conserved function in facilitating mRNA biogenesis and export. First, the Arabidopsis PABP, PAB3, accelerated the entry of the mRNA into the degradation pathway, as well as its entry into the translated pool when expressed in yeast. Second, plant PABP was present physically in the yeast cell nucleus in the complemented yeast strain, and it partially restored the poly(A) tail length control during polyadenylation reaction, in the PAN-dependent manner. Third, the substitution of the plant PAB3 for the endogenous yeast Pab1p resulted in synthetic lethality with rna15-2, an allele of the gene encoding the Rna15p subunit of the nuclear factor CFI, which is required for the pre-mRNA cleavage, as well as polyadenylation, and with gle2-1, a mutant allele of the gene encoding the NPC-associated protein. Fourth, overexpression of the plant PABP in yeast rescued the cold sensitivity and alleviated the mRNA export block of the nab2-1 mutant strain, which is defective in one of the shuttling hnRNP proteins required for mRNA export.
The possible role of the mRNA poly(A) tails, and by implication, PABP, in the nucleocytoplasmic mRNA transport has been a subject of a number of studies in a variety of eukaryotic systems. Early experiments by Krowczynska et al. (1985) showed that all mRNA that is exported to the cytoplasm is polyadenylated, whereas the cytoplasmic poly(A)− species are derived from those that are poly(A)+ during transport. On the other hand, a study that used the polyadenylation inhibitor cordycepine, suggested that poly(A) is not strictly required for transport, although it might have a stimulatory role (Zeevi et al. 1982). The polyadenylated SV40 transcripts synthesized in vivo from the transfected templates were found to preferentially partition into the cytoplasm in Xenopus oocytes (Wickens and Gurdon 1983) and human cells (Connelly and Manley 1988), relative to the poly(A)− ones. Similarly, appending a RNA pol II 3′ end processing signal onto a reporter gene of bacterial origin resulted in an increase in its cytoplasmic-to-nuclear steady-state ratio in COS cells (Eckner et al. 1991). RNA microinjection studies in Xenopus oocytes also suggested that poly(A) is stimulatory, although it may not be absolutely required for export (Jarmolowski et al. 1994). The NS1 protein of influenza virus inhibits polyadenylation of cellular transcripts, and thereby prevents their export (Nemeroff et al. 1998). Evidence obtained in the COS cells argues that poly(A) tails promote export, but it appears that it is the dynamic process of the 3′ end formation, rather than just the mere presence of a monotonous run of Å residues near the 3′ end, is what is required for the efficient mRNA export (Huang and Carnichael 1996), because a poly(A) tract encoded in the body of the transcript was not sufficient.
In yeast, the RNA pol II transcripts that lack a polyadenylation signal were shown to be retained in the nucleus, as visualized by in situ hybridization (Long et al. 1995). Using another technology, on the basis of the tethering of GFP to the reporter transcripts, Brodsky and Silver (2000) have observed mRNA export defects in the rna14-1, rna15-2, hrp1-3, and pap1-1 strains, which are conditionally defective in the components of CFI and PAP, respectively. Furthermore, in a screen for a failure to export the SSA4 mRNA at an elevated temperature, Hammell et al. (2002) have found several alleles of RNA15, RNA14, FIP1, and PAP1. Dower and Rosbash (2002) have found that the T7 RNA polymerase transcripts accumulate in the nucleus, unless they are cleaved and polyadenylated by the machinery that normally processes the RNA pol II transcripts. All of these findings indicate the link between the mRNA 3′ end processing, polyadenylation, and export. Interestingly, the rna15-1 mutant was found to have a very slow mRNA decay rate (Gonzalez et al. 2000). This could be due to a lag prior to the mRNA export, similar to the one that is observed in the pab1 mutant strains, occurring because of improper maturation of the 3′ terminal domain of the mRNP.
Yeast Pab1p has been functionally linked previously to Pbp1p, a predominantly nuclear, Pab1p-interacting protein that facilitates proper polyadenylation (Mangus et al. 1998). However, the role of Pab1p in the nuclear steps of mRNA biogenesis may be broader than just the control of the length of the newly made poly(A) tails. This view is consistent with the results of Hector et al. (2002) that nuclear Pab1p can suppress the mRNA export defect of the nab2-21 cells in a way that can be genetically uncoupled from the control of the poly(A) tail length by Pab1p. Rather, these findings, together with the ones presented here, suggest that the dynamic process of formation of the proper architecture of the 3′ domain of mRNP may be essential.
Our results may seem at odds with the results of Kadowaki et al. (1992), who found that inactivation of the temperature-sensitive pab1-F364L allele did not visibly change the nucleocytoplasmic distribution of the poly(A)+ RNA, as visualized by in situ hybridization. However, the pab1-F364L allele that was used in that study has an extremely slow turnover rate (Sachs and Davis 1989). Thus, in the absence of definitive information as to whether it is temperature sensitive for function of the existing protein, or for the de novo synthesis of the functional polypeptide, these findings should be interpreted with caution. Second, this experiment, as well as many other studies examining the possible role of poly(A) and PABP in mRNA export, relied exclusively on the assays that are static in nature (such as FISH), and as such, did not directly address the possibility of more subtle changes in the rate of mRNA export. On the other hand, our data presented in this work suggest that PABP facilitates mRNA export, rather than being strictly required for it.
Interestingly, in Schizosaccharomyces pombe, PABP shuttles between the nucleus and the cytoplasm, and importantly, its overexpression could suppress conditional lethality, as well as an associated mRNA export defect of a rae1-167 nup184-1 synthetic lethal strain (Thakurta et al. 2002). It should be pointed out that RAE1 of S. pombe is a homolog of the Gle2p of S. cerevisiae, which is functionally implicated in the evolutionarily conserved PABP function by our finding of synthetic lethality between the gle2-1 allele and a substitution of the plant PAB3 for the yeast Pab1p. The ability of the S. pombe PABP to shuttle is directly related to its ability to rescue this mRNA export defect (Thakurta et al. 2002). However, this function of PABP in mRNA export is apparently redundant with other factors in S. pombe, as the loss of the S. pombe PABP does not lead to mRNA export defects, and, in fact, has no discernible phenotype at all. It is also significant in this context that the S. cerevisiae Pab1p interacts with Xpo1p, a nucleocytoplasmic transport receptor, and shuttles between the nucleus and the cytoplasm in a Xpo1p-dependent manner (Hammell et al. 2002). The nucleocytoplasmic shuttling ability seems to be conserved in PABP from other eukaryotic species, including human (Afonina et al. 1998) and Leishmania (Bates et al. 2000). Moreover, human PABP is also subject to arginine methylation (Lee and Bedford 2002), which is a modification that is characteristic of many shuttling proteins that are involved in nucleocytoplasmic transport (McBride and Silver 2001).
The findings presented here and elsewhere (Thakurta et al. 2002) of the genetic links between the nuclear function of PABP and Gle2p, a NPC associated factor, suggest the inner face of the NPC as another possible site of PABP action in the nucleus. On the other hand, a series of recent studies (Burkard and Butler 2000; Hilleren et al. 2001; Jensen et al. 2001a,b; Andrulis et al. 2002; Libri et al. 2002; Zenklusen et al. 2002) have led to a view of a release of the pre-mRNA from the transcription site as a distinct, regulated step in the biogenesis of functional mRNP that involves the nuclear exosome (for reviews, see Neugebauer 2002; Jensen et al. 2003). These results suggest an additional possibility (which is not mutually exclusive with the possible role in the NPC-associated step), that the role of PABP as an evolutionarily conserved facilitator of mRNA biogenesis may be linked to a release of the mRNP from the site of transcription. Future experiments will attempt to resolve, or reconcile these possibilities.
MATERIALS AND METHODS
Plasmid constructs
The MFA2-MS2-RZ construct used for the experiments shown in Figure 1 ▶ was pMM2-6 (Coller et al. 1998), kindly provided by Jeff Coller (University of Arizona). The constructs used for expressing Pab1p and PAB3, pDB464 (TRP1/CEN/PAB1), pDB419 (TRP1/CEN/pGAL1-PAB3), pDB463 (TRP1/2μ/pADHI-PAB3), and pDB488 (G418r/TRP1/2μ/pADHI-PAB3) were described previously (Chekanova et al. 2001). The construct pDB493 is equivalent to pDB488, except that its TRP1 marker was disrupted via linearization at the EcoRV site and limited digestion with ExoIII and mung bean nucleases, followed by religation. The construct pDB490 (G418r/CEN/PAB1) was similarly derived from the pDB489 (G418r/TRP1/CEN/PAB1; Chekanova et al. 2001) via disabling the TRP1 marker.
Yeast strains and techniques
To construct the yeast strains for the synthetic lethal tests, strains bearing mutations in the various factors involved in mRNA processing or export (shaded gray in the Table 1 ▶) were first transformed with the pAS77, a CEN/URA3/PAB1 plasmid (Sachs and Davis 1989). The chromosomal copy of the PAB1 gene was then disrupted by transformation with linear DNA fragments containing HIS3, LEU2, or TRP1 marker (depending on the strain used), flanked by the 300 bp from the native 5′ and 3′ flanking regions of PAB1. Chromosomal integrants were selected on the basis of the PCR assay using the pair of primers corresponding to the PAB1 sequence, one within and one outside of the sequence present in the integrated fragment, and subsequently verified by Southern analysis. Then, the PAB3-expressing construct was introduced, which consisted of a pGAL1-PAB3 cassette identical to that of pDB419, on both CEN and 2μ-based plasmids with either LEU2, HIS3, or Geneticin resistance marker, as dictated by the auxotrophies of the respective strains. Transformants were grown in synthetic medium containing uracil, and dilution series spotted onto YPD, SC galactose + 5 FOA, and SC glucose + 5-FOA plates. The carbon-source dependence of growth was used as a criterion to exclude the spontaneously arising extragenic suppressors of pab1Δ mutation. Other yeast strains used in this work are listed in Table 1 ▶. General yeast genetic methods were according to Guthrie and Fink (1991).
Purification of Xrn1p and cap status assays
The His-tagged Xrn1p was expressed in the yeast strain BJ5464 from the construct pAJ95 (2μ/LEU2/pGAL10-XRN1-HA-His6, a kind gift from Arlen Johnson, University of Texas, Austin) as described previously (Johnson and Kolodner 1991), and protein purified on a Ni++ column as suggested by the manufacturer (QIAGEN). Treatment of total RNA with the purified Xrn1p was as per Boeck et al. (1998).
Subcellular fractionation
The YDB203 cells grown in YPGal to OD600 = 0.5 were converted to spheroplasts, lysed, and washed as described by Azad et al. (2001) and fractionated on Ficoll gradient according to Aris chapter (Dove et al. 1998). Equivalent proportions of total, nuclear, and cytoplasmic material were analyzed by immunoblotting as shown in Figure 4 ▶.
RNA analyses
RNA decay analyses, transcriptional pulse-chase experiments, and total poly(A) tail length analysis were done as described previously (Chekanova et al. 2001). The 7S rRNA precursor probe (oligo 020) and hybridization conditions were according to Mitchell et al. (1997).
Immuprecipitation and immunonoblotting
Antibodies were used in immunoblotting experiments in the following dilutions: against PAB3 (rabbit polyclonal), at 1:1000; against Pgk1p (mouse monoclonal, Molecular Probes), at 0.5 μg/mL; against Nsp1p (mouse monoclonal [Tolerico et al. 1999], a kind gift from John Aris, University of Florida, Gainesville), at 1:10000; and against Ssa4p (rabbit polyclonal, a kind gift from Elizabeth Craig, University of Wisconsin), at 1:3000. Immunoblots were quantitated using GEL Quant v. 1.0 (Multiplexed Biotechnologies, Inc.). Immunoprecipitation was conducted in 100 μL of buffer using a 1:1000 dilution of the rabbit antiserum raised against the Pan3p (Brown and Sachs 1998). The immunoprecipitates were captured on 20 μL of the protein A agarose beads (Santa Cruz), washed seven times with 500 μL of PBS, and bound material eluted into SDS-PAGE loading buffer, and probed with PAB3 antiserum. Immunoprecipitates shown in Figure 4D ▶ were treated with indicated amounts of RNase A for 40 min at room temperature prior to washes.
In vitro polyadenylation assays
Extracts were prepared by the liquid nitrogen homogenization method, and assays performed as described previously by Brown and Sachs (1998).
Fluorescent in situ hybridization
FISH with a Cy3-labeled oligo(dT)70 probe was carried out as described in Vainberg et al. (2001).
Acknowledgments
We thank Ken Dower and Michael Rosbash for help with FISH. We also thank John Aris, Scott Butler, Charles Cole, Jeff Coller, Anita Corbett, Elizabeth Craig, Ken Dower, Michael Henry, Anita Hopper, Ed Hurt, Arlen Johnson, Claire Moore, Roy Parker, Michael Rosbash, Alan Sachs, Pam Silver, Maurice Swanson, Karsten Weis, Susan Wente, and Marvin Wickens for constructs, strains, antibodies, and advice; Andrei Petcherski, Judith Kimble, David Mangus, and Allan Jacobson for communicating unpublished data; James Dutko and Sergei Reverdatto for their help with densitometry; and Henry Tedeschi for his support. This work was funded by the Basic Biosciences Minigrant Program and USDA. We thank Anita Corbett and Michael Rosbash for critically reading the manuscript. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
CFI, cleavage factor I
FISH, fluorescent in situ hybridization
PABP, poly(A) binding protein
PAN, poly(A) nuclease
PAP, poly(A) polymerase
RRM, RNA recognition motif
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5128903.
REFERENCES
- Afonina, E., Stauber, R., and Pavlakis, G.N. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273: 13015–13021. [DOI] [PubMed] [Google Scholar]
- Amrani, N., Minet, M., Le Gouar, M., Lacroute, F., and Wyers, F. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol.. 17: 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis, E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841. [DOI] [PubMed] [Google Scholar]
- Azad, A.K., Stanford, D.R., Sarkar, S., and Hopper, A.K. 2001. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell 12: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, E.J., Knuepfer, E., and Smith, D.F. 2000. Poly(A)-binding protein I of Leishmania: Functional analysis and localization in trypanosomatid parasites. Nucleic Acids Res. 28: 1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, P., Peltz, S.W., and Ross, J. 1989. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 9: 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins, M.B., Smith, A.M., Phillips, E.M., and Powers, M.A. 2003. Complex formation among the RNA export proteins Nup98, Rae1/Gle2 and TAP. J. Biol. Chem. 278: 20979–20988. [DOI] [PubMed] [Google Scholar]
- Boeck, R., Lapeyre, B., Brown, C.E., and Sachs, A.B. 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18: 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, A.S. and Silver, P.A. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.E. and Sachs, A.B. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18: 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard, K.T. and Butler, J.S. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., and Parker, R. 1995. Multiple functions for the poly(A) binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 9: 2421–2432. [DOI] [PubMed] [Google Scholar]
- Chekanova, J.A., Shaw, R.J., and Belostotsky, D.A. 2001. Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr. Biol. 11: 1207–1214. [DOI] [PubMed] [Google Scholar]
- Coller, J.M., Gray, N.K., and Wickens, M.P. 1998. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes & Dev. 12: 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, S. and Manley, J.L. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 2: 440–452. [DOI] [PubMed] [Google Scholar]
- Cosson, B., Couturier, A., Chabelskaya, S., Kiktev, D., Inge-Vechtomov, S., Philippe, M., and Zhouravleva, G. 2001. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 22: 3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- Dockendorff, T.C., Heath, C.V., Goldstein, A.L., Snay, C.A., and Cole, C.N. 1997. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol. Cell. Biol. 17: 906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove, J.E., Brockenbrough, J.S., and Aris, J.P. 1998. Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. Methods Cell. Biol. 53: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower, K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresios, J., Derkatch, I.L., Liebman, S.W., and Synetos, D. 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39: 7236–7244. [DOI] [PubMed] [Google Scholar]
- Dresios, J., Panopoulos, P., Frantziou, C.P., and Synetos, D. 2001. Yeast ribosomal protein deletion mutants possess altered peptidyltransferase activity and different sensitivity to cycloheximide. Biochemistry 40: 8101–8108. [DOI] [PubMed] [Google Scholar]
- Eckner, R., Ellmeier, W., and Birnstiel, M.L. 1991. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10: 3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, D., Bartel, B., and Varshavsky, A. 1989. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401. [DOI] [PubMed] [Google Scholar]
- Ford, L., Bagga, P.S., and Wilusz, J. 1997. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 17: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Wilusz, C.J., Peltz, S.W., and Wilusz, J. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20: 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C.I., Ruiz-Echevarria, M.J., Vasudevan, S., Henry, M.F., and Peltz, S.W. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5: 489–499. [DOI] [PubMed] [Google Scholar]
- Gorlach, M., Burd, C.G., and Dreyfuss, G. 1994. The mRNA poly(A)-binding protein: Localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211: 400–407. [DOI] [PubMed] [Google Scholar]
- Gorsch, L.C., Dockendorff, T.C., and Cole, C.N. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell. Biol. 129: 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.M., Marfatia, K.A., Crafton, E.B., Zhang, X., Cheng, X., and Corbett, A.H. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277: 7752–7760. [DOI] [PubMed] [Google Scholar]
- Gross, S. and Moore, C. 2001. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. 98: 6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosset, C., Chen, C.Y., Xu, N., Sonenberg, N., Jacquemin-Sablon, H., and Shyu, A.B. 2000. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103: 29–40. [DOI] [PubMed] [Google Scholar]
- Guthrie, C. and Fink, G.R. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- Hammell, C.M., Gross, S., Zenklusen, D., Heath, C.V., Stutz, F., Moore, C., and Cole, C.N. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22: 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, C.V., Copeland, C.S., Amberg, D.C., Del Priore, V., Snyder, M., and Cole, C.N. 1995. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell. Biol. 131: 1677–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector, R.E., Nykamp, K.R., Dheur, S., Anderson, J.T., Non, P.J., Urbinati, C.R., Wilson, S.M., Minvielle-Sebastia, L., and Swanson, M.S. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Carnichael, G.G. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka, H., Gradi, A., and Sonenberg, N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17: 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, A. 1996. Poly(A) metabolism and translation: The closed-loop model. In Translational control (eds. J.W.B Hersheyet al.), pp. 451–480. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jarmolowski, A., Boelens, W.C, Izaurralde, E, amd Mattaj, I.W. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell. Biol. 124: 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.H., Boulay, J., Rosbash, M., and Libri, D. 2001a. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11: 1711–1715. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001b. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Dower, K., Libri, D., and Rosbash, M. 2003. Early formation of mRNP. License for export or quality control? Mol. Cell 11: 1129–1138. [DOI] [PubMed] [Google Scholar]
- Johnson, A.W. and Kolodner, R.D. 1991. Strand exchange protein 1 from Saccharomyces cerevisiae. A novel multifunctional protein that contains DNA strand exchange and exonuclease activities. J. Biol. Chem. 266: 14046–14054. [PubMed] [Google Scholar]
- Jones, E.W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Meth. Enzymol. 194: 428–453. [DOI] [PubMed] [Google Scholar]
- Kadowaki, T., Zhao, Y., and Tartakoff, A.M. 1992. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc. Natl. Acad. Sci. 89: 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Roy, G., and Sonenberg, N. 2001. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66: 293–300. [DOI] [PubMed] [Google Scholar]
- Kessler, M., Henry, M.F., Shen, E., Zhao, J., Gross, S., Silver, P.A., and Moore, C.L. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 11: 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, S.H. and Sachs, A.B. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krowczynska, A., Yenofsky, R., and Brawerman, G. 1985. Regulation of messenger RNA stability in mouse erythroleukemia cells. J. Mol. Biol. 181: 231–239. [DOI] [PubMed] [Google Scholar]
- Lee, J. and Bedford, M.T. 2002. Related articles, links abstract PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 3: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, O., Heath, C.V., Amberg, D.C., Dockendorff, T.C., Copeland, C.S., Snyder, M., and Cole, C.N. 1995. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell 6: 401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Elliott, D.J., Stutz, F., Rosbash, M., and Singer, R.H. 1995. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA 1: 1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Mangus, D.A., Amrani, N., and Jacobson, A. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18: 7383–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhoul, J.F. and Adams, T.H. 1996. Aspergillus fabM encodes an essential product that is related to poly(A)-binding proteins and activates development when overexpressed. Genetics 144: 1463–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, A.E. and Silver, P.A. 2001. State of the arg: Protein methylation at arginine comes of age. Cell 106: 5–8. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Preker, P.J., Wiederkehr, T., Strahm, Y., and Keller, W. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 94: 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466. [DOI] [PubMed] [Google Scholar]
- Morrissey, J.P., Deardorff, J.A., Hebron, C., and Sachs, A.B. 1999. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast 15: 687–702. [DOI] [PubMed] [Google Scholar]
- Munroe, D. and Jacobson, A. 1990. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10: 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, R., Watkins, J.L., and Wente, S.R. 1996. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell 7: 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff, M.E., Barabino, S.M., Li, Y., Keller, W., and Krug, R.M. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1: 991–1000. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being co-transcriptional. J. Cell. Sci. 115: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Proweller, A. and Butler, J.S. 1996. Ribosomal association of poly(A)-binding protein in poly(A)-deficient Saccharomyces cerevisiae. J. Biol. Chem. 271: 10859–10865. [DOI] [PubMed] [Google Scholar]
- Quadt, R., Ishikawa, M., Janda, M., and Ahlquist, P. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. 92: 4892–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 447–465. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sachs, A.B. and Davis. R.W. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translational initiation. Cell 58: 857–867. [DOI] [PubMed] [Google Scholar]
- Sachs, A.B., Davis, R.W., and Kornberg, R.D. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7: 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Pante, N., and Hurt, E. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18: 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C. and Parker, R. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae. In Translational control of gene expression (eds, N. Sonenberg et al.), pp. 807–825. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Searfoss, A., Dever, T.E., and Wickner, R. 2001. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: Relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21: 4900–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Luhrmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, S.J., Thiel, P.R., Reiff, D.F., Lachance, P.E., Lasko, P., and Schuster, C.M. 2000. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405: 1062–1065. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge, C.A., Colot, H.V., Goldstein, A.L., and Cole, C.N. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, S. and Sachs, A.B. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes & Dev. 9: 2997–3007. [DOI] [PubMed] [Google Scholar]
- ———. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thakurta, A.G., Yoon, J., and Dhar, R. 2002. Schizosaccharomyces pombe spPABP, a homologue of Saccharomyces cerevisiae Pab1p, is a non-essential, shuttling protein that facilitates mRNA export. Yeast 19: 803–810. [DOI] [PubMed] [Google Scholar]
- Tolerico, L.H., Benko, A.L., Aris, J.P., Stanford, D.R., Martin, N.C., and Hopper, A.K. 1999. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 151: 57–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, N., Hoshino, S., Imataka, H., Sonenberg, N., and Katada, T. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 277: 5982–5987. [DOI] [PubMed] [Google Scholar]
- Vainberg, I.E., Dower, K., and Rosbash, M. 2000. Nuclear export of heat shock and non-heat-shock mRNA occurs via similar pathways. Mol. Cell. Biol. 20: 3996–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Day, N., Trifillis, P., and Kiledjian, M. 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 19: 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens, M.P. and Gurdon, J.B. 1983. Posttranscriptional processing of simian virus 40 late transcripts in injected frog oocytes. J. Mol. Biol. 163: 1–26. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Gao, M., Jones, C.L., Wilusz, J., and Peltz, S.W. 2001a. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 7: 1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wormington, M., and Peltz, S.W. 2001b. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2: 237–246. [DOI] [PubMed] [Google Scholar]
- Winston, F., Dollard, C., and Ricupero-Hovasse, S.L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Wormington, M., Searfoss, A.M., and Hurney, C.A. 1996. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 15: 900–909. [PMC free article] [PubMed] [Google Scholar]
- Zeevi, M., Nevins, J.R., and Darnell Jr., J.E. 1982. Newly formed mRNA lacking polyadenylic acid enters the cytoplasm and the polyribosomes but has a shorter half-life in the absence of polyadenylic acid. Mol. Cell. Biol. 2: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen, D., Vinciguerra, P., Wyss, J.C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22: 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]