Abstract
Expression of the trpEDCFBA operon of Bacillus subtilis is regulated by transcription attenuation and translation control mechanisms. We recently determined that the B. subtilis trp leader readthrough transcript can adopt a Mg2+-dependent tertiary structure that appears to interfere with TRAP-mediated translation control of trpE. In the present study, sequence comparisons to trp leaders from three other Bacillus sp. were made, suggesting that RNA secondary and tertiary structures are phylogenetically conserved. To test this hypothesis, experiments were carried out with the trp leader transcript from Bacillus stearothermophilus. Structure mapping experiments confirmed the predicted secondary structure. Native gel experiments identified a faster mobility species in the presence of Mg2+, suggesting that a Mg2+-dependent tertiary structure forms. Mg2+-dependent protection of residues within the first five triplet repeats of the TRAP binding target and a pyrimidine-rich internal loop were observed, consistent with tertiary structure formation between these regions. Structure mapping in the presence of a competitor DNA oligonucleotide allowed the interacting partners to be identified as a single-stranded portion of the purine-rich TRAP binding target and the large downstream pyrimidine-rich internal loop. Thermal denaturation experiments revealed a Mg2+- and pH-dependent unfolding transition that was absent for a transcript missing the first five triplet repeats. The stability of several mutant transcripts allowed a large portion of the base-pairing register for the tertiary interaction to be determined. These data indicate that RNA secondary and tertiary structures involved in TRAP-mediated translation control are conserved in at least four Bacillus species.
Keywords: Phylogenetic conservation, RNA secondary structure, RNA tertiary structure, RNA folding, translation control
INTRODUCTION
Expression of the Bacillus subtilis tryptophan biosynthetic trpEDCFBA operon is regulated by the trp RNA-binding attenuation protein (TRAP) by transcription attenuation (Shimotsu et al. 1986; Kuroda et al. 1988; Gollnick et al. 1990; Babitzke and Yanofsky 1993) and translation control mechanisms (Kuroda et al. 1988; Merino et al. 1995; Du and Babitzke 1998). TRAP also regulates translation of the pabA (trpG) and yhaG genes, involved in tryptophan biosynthesis and transport, respectively (Babitzke et al. 1994; Yang et al. 1995; Du et al. 1997; Sarsero et al. 2000). TRAP consists of 11 identical subunits arranged in a single ring (Antson et al. 1994, 1995). Cooperative binding of 11 tryptophan molecules activates TRAP to bind single-stranded RNAs that contain multiple (G/U/A)AG triplet repeats with the RNA wrapping around the periphery of TRAP (Otridge and Gollnick 1993; Babitzke et al. 1994; Babitzke and Yanofsky 1995; Babitzke et al. 1996; Antson et al. 1999).
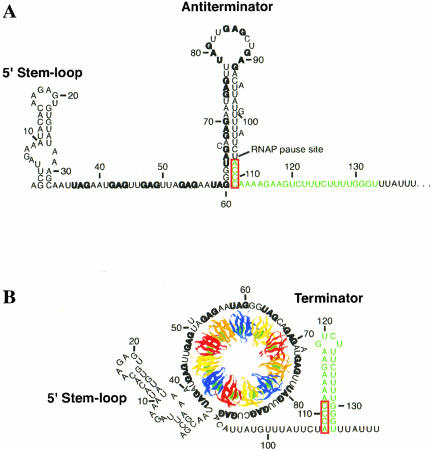
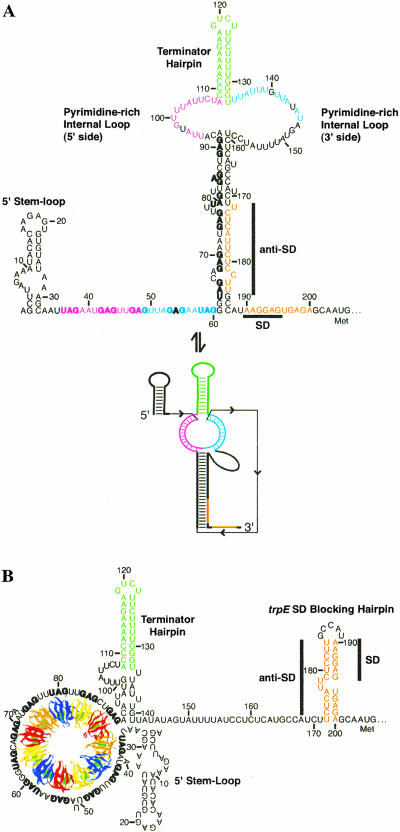
Several secondary structures and a tertiary structure have been shown to form in the trp operon leader transcript of B. subtilis. Under limiting tryptophan conditions (no TRAP binding), an antiterminator structure forms that sequesters the last six of the 11 (G/U)AG triplet repeats (Fig. 1A ▶; Babitzke and Yanofsky 1993; Babitzke et al. 1994; Babitzke et al., in press). This antiterminator structure allows transcription readthrough into the trp operon structural genes by preventing formation of an overlapping intrinsic terminator (Kuroda et al. 1988; Babitzke and Yanofsky 1993; Otridge and Gollnick 1993; Babitzke et al., in press). Tryptophan-activated TRAP can bind to the 11 (G/U)AG repeats and prevents formation of the antiterminator. Thus, the overlapping terminator hairpin forms and transcription terminates in the trp leader region (Fig. 1B ▶). In addition, NusA-stimulated RNA polymerase pausing at U107, the nucleotide that just precedes the critical overlap between the antiterminator and terminator structures, provides additional time for TRAP to bind RNA and promote termination (Yakhnin and Babitzke 2002). Once transcription has proceeded into the trp operon structural genes, the trp leader RNA can adopt a secondary structure such that the trpE Shine-Dalgarno (SD) sequence is single-stranded and available for ribosome binding (Fig. 2A ▶). If TRAP is bound to a readthrough transcript, however, a different secondary structure forms that sequesters the trpE SD sequence in the trpE SD blocking hairpin (Fig. 2B ▶; Kuroda et al. 1988; Merino et al. 1995; Du and Babitzke 1998). Furthermore, a long-distance Mg2+-dependent tertiary structure forms in this transcript that appears to interfere with TRAP-mediated trpE translation control by inhibiting TRAP binding (Fig. 2 ▶; Schaak et al. 2003).
FIGURE 1.
Model of the transcription attenuation mechanism of the B. subtilis trpEDCFBA operon. (A) Under tryptophan limiting conditions, TRAP is not activated. During transcription the 5′ stem–loop and antiterminator structures form. Antiterminator formation prevents formation of the overlapping intrinsic terminator, resulting in transcription readthrough into the trp operon structural genes. (B) Under excess tryptophan conditions, TRAP is activated. During transcription TRAP can interact with the 5′ stem–loop and bind to the (G/U)AG repeats as they are synthesized. TRAP binding prevents formation of the antiterminator, thereby allowing formation of the terminator, resulting in termination of transcription before RNA polymerase can reach the trp operon structural genes. NusA stimulates RNA polymerase, pausing at U107, which allows additional time for TRAP to bind. The (G/U)AG repeats are indicated in bold type, and the terminator is shown in green. The mutually exclusive antiterminator and terminator structures overlap by four nucleotides (outlined in red). The 11 subunits of TRAP are shown in blue, yellow, red, and orange; the tryptophan molecules are shown in green. The structure of TRAP and wrapping of the RNA around the periphery of the protein are based on crystal structures (Antson et al. 1995, 1999). Numbering is from the start of transcription.
FIGURE 2.
Model of the trpE translation control mechanism. (A) In the absence of TRAP binding, trp operon readthrough transcripts can adopt a secondary structure in which the trpE SD sequence is single-stranded and available for ribosome binding. This secondary structure was predicted from mfold v3.1 using experimental constraints determined herein. The tertiary structure model (lower structure) predicts that 24 nt in the 5′ (magenta) and the 3′ (blue) sides of the pyrimidine-rich internal loop base pair with the same colored residues between nt 33 and 60 containing the single-stranded (G/U)AG repeats (the secondary structure defects are omitted for clarity; Schaak et al. 2003). (B) When TRAP binds to trp operon readthrough transcripts, a structural rearrangement occurs in the RNA such that the trpE SD sequence becomes sequestered in a stable hairpin (trpE SD blocking hairpin) by pairing with the anti-SD sequence. Formation of this structure inhibits ribosome binding. The (G/U)AG repeats are shown in bold type; the sequences involved in the trpE SD blocking hairpin are shown in orange; the terminator hairpin that forms in trp operon readthrough transcripts is shown in green. Numbering is from the start of transcription.
Previous genetic, biochemical, and RNA structural studies indicated that the transcription attenuation mechanism is conserved in several Bacillus species (Kuroda et al. 1988; Shiratsuchi and Sato 1991; Hoffman and Gollnick 1995; Merino et al. 1995; Chen et al. 1999; Sudershana et al. 1999; Babitzke and Gollnick 2001). In the present study, a phylogenetic comparison of the trp leaders in B. subtilis, Bacillus stearothermophilus, Bacillus pumilus, and Bacillus caldotenax was carried out, suggesting that each RNA has the potential to form a tertiary interaction between the single-stranded TRAP binding target and a large, downstream pyrimidine-rich internal loop. Native gels of the B. stearothermophilus trp leader transcript revealed a Mg2+-dependent species with faster mobility, consistent with a Mg2+-dependent tertiary structure. To test whether this was the tertiary structure predicted from the phylogenetic comparison, the B. stearothermophilus trp leader RNA was probed using a variety of techniques. RNA structure mapping experiments established that the Mg2+-dependent tertiary structure could be disrupted by a competitor DNA oligonucleotide that base pairs to nucleotides 36–61, making the internal loop susceptible to nuclease cleavage. Furthermore, thermal denaturation experiments revealed a Mg2+-dependent transition that was stabilized by lowering the pH. Monitoring of this transition in wild-type and several mutant transcripts allowed the base-pairing register of the majority of this tertiary structure to be determined.
RESULTS AND DISCUSSION
The potential to form secondary and tertiary structures in trp operon readthrough transcripts is phylogenetically conserved
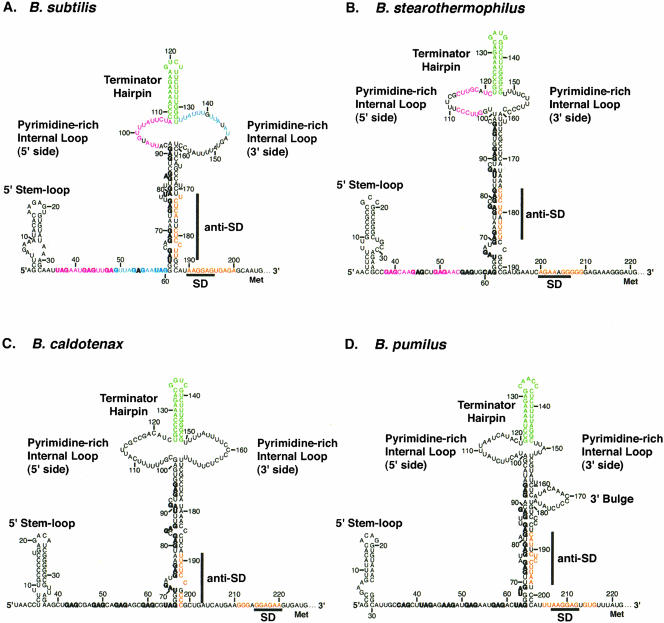
A comparative analysis of the B. subtilis, B. stearothermophilus, B. pumilus, and B. caldotenax trp leader RNA sequences and possible RNA structures is shown in Figure 3 ▶. Previous work had shown that the 5′ stem–loop is conserved in all four organisms (Sudershana et al. 1999). Each of the trp leader transcripts also contains multiple NAG triplet repeats, as well as overlapping antiterminator and terminator structures, suggesting that all four organisms regulate transcription of the trp operon by a similar attenuation mechanism (Babitzke and Gollnick 2001). We examined the trp leader sequences to see whether they share translation control features as well. Although there is very little sequence conservation, all four trp leader RNAs share common structural features known to be involved in regulating trpE translation in B. subtilis. Eleven appropriately spaced triplet repeats are present downstream from the 5′ stem–loop in the trp leaders of B. subtilis (seven GAG, four UAG) and B. caldotenax (eight GAG, three UAG), whereas 12 repeats are contained in the trp leaders of B. stearothermophilus (nine GAG, two UAG, one CAG) and B. pumilus (seven GAG, three UAG, one AAG, and one CAG). In each case, the first five to six triplet repeats are predicted to be single-stranded, and the remaining six to seven repeats are predicted to be involved in formation of the lower stem of a large secondary structure with the terminator hairpin at its apex (Fig. 3 ▶). These structures are similar to what was verified experimentally in the B. subtilis trp operon leader (Schaak et al. 2003). For simplicity, we refer to the entire secondary structure below the pyrimidine-rich internal loop as the lower stem, despite the presence of several bulged residues. Each of these large secondary structures also contains an anti-SD sequence near its 3′ end that is capable of sequestering the trpE SD sequence in a hairpin. In each case the 6–7-nucleotide SD sequence is embedded within flanking nucleotides that lengthen the predicted SD:anti-SD interaction to 9–11 base pairs. Each predicted SD:anti-SD base-pairing register also contains one or two bulges or single-base mismatches, although their sequence is not conserved. The occasional helix defects may help prevent this pairing from being too strong, thereby allowing it to switch conformations with reasonably fast kinetics.
FIGURE 3.
Comparison of trp leader RNA secondary and tertiary structures for (A) B. subtilis, (B) B. stearothermophilus, (C) B. caldotenax, and (D) B. pumilus. Conserved primary and secondary structural features include a 5′ stem–loop, 11 or 12 NAG triplet repeats (bold), a terminator hairpin (green), an anti-SD sequence that is capable of sequestering the trpE SD sequence (orange), and a pyrimidine-rich internal loop and/or bulge. In addition, each leader is predicted to form a long-range tertiary interaction between a pyrimidine-rich internal loop and/or bulge and the single-stranded triplet repeats. Identically colored interacting segments (magenta or blue) are shown for B. subtilis and B. stearothermophilus. A weak interaction of nt 55–59 and the 3′ side of the pyrimidine-rich internal loop may also occur for B. stearothermophilus (see text), although its register is uncertain and therefore not shown. The base-pairing details for the tertiary structures have been worked out for B. subtilis (Schaak et al. 2003) and B. stearothermophilus (this report); however, formation of the tertiary structure has not been examined in B. pumilus or B. caldotenax, and the register of base-pairing is uncertain.
As previously mentioned, the secondary structures of all four trp leader transcripts contain a large pyrimidine-rich internal loop at the base of the terminator hairpin (Fig. 3 ▶). Large regions of unstructured nucleotides are relatively rare in RNA, suggesting that this conserved feature may be important. In each case, the internal loop appeared capable of forming a long-range tertiary structure with the purine-rich single-stranded residues contained in the upstream repeats of the TRAP binding site. This tertiary structure was verified for B. subtilis and shown to participate in the trpE translation control mechanism (Schaak et al. 2003). Thus, it appeared that the trpE translation control mechanism might be conserved in these four Bacillus species. We studied formation of this long-range tertiary structure in the thermophile B. stearothermophilus, as the structure might be particularly stable (see below). It should be noted that the details of the pairings in B. pumilus and B. caldotenax have not been determined experimentally; therefore, no base-pairing registers are shown for these organisms, although clear possibilities exist (Fig. 3 ▶).
The trp leader of B. stearothermophilus adopts a Mg2+-dependent tertiary structure: Native gel analysis
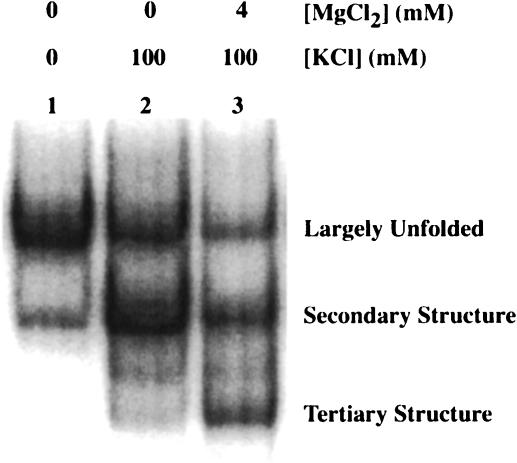
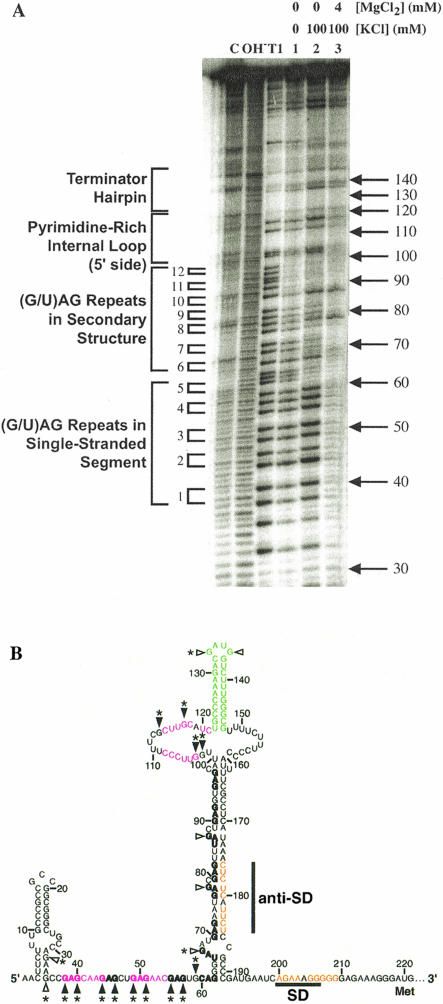
The presumed B. stearothermophilus trp leader tertiary structure contains a higher GC content than that of B. subtilis and was predicted to be more stable. To test the B. stearothermophilus trp leader for a tertiary structure, 5′ end-labeled trp leader RNA containing nt 1–232 was renatured under various salt conditions and fractionated on a native polyacrylamide gel. When the RNA was renatured in TK0M0 (40 mM Tris-HCl, pH 8.0, 0 mM KCl, 0 mM MgCl2; Fig. 4 ▶, lane 1), a major species (78%) with slower mobility and a minor species (21%) with faster mobility were observed. Because secondary structures are more compact than unfolded structures, they tend to have faster electrophoretic mobility. Thus, the minor population of RNA may have weak or partial secondary structure interactions in the absence of added K+ and Mg2+ (see below). Upon renaturation in TK100M0 (40 mM Tris-HCl, pH 8.0, 100 mM KCl, 0 mM MgCl2; Fig. 4 ▶, lane 2), the major and minor species were reversed with the percentages of faster and slower mobility species being 58% and 32%, respectively, consistent with monovalent ions promoting secondary structure formation. In addition, a third, faster mobility band was observed under these conditions, accounting for about 10% of the sample. Because tertiary structures are more compact than secondary structures, they tend to have faster electrophoretic mobility. Thus, this population of RNA may have weak or partial tertiary structure interactions in the absence of Mg2+ ions.
FIGURE 4.
Native gel mobility of the B. stearothermophilus trp leader transcript. 5′ 32P-labeled RNA (1–232) was renatured under various salt conditions and fractionated on a 6% native polyacrylamide gel. Lane 1, RNA renatured in TK0M0; lane 2, RNA renatured in TK100M0; lane 3, RNA renatured in TK100M4. Assignments for each RNA species, based on structure mapping under these conditions (see Fig. 5 ▶), are indicated.
When the RNA was renatured in TK100M4 (40 mM Tris-HCl, pH 8.0, 100 mM KCl, 4 mM MgCl2; Fig. 4 ▶, lane 3), the mobility of the fastest species increased further and was more abundant (31%), consistent with formation of Mg2+-dependent tertiary structure. Thus, by comparison to the salt dependence of structure formation observed for B. subtilis trp leader RNA (Schaak et al. 2003), we initially assigned the three species in the native gel as largely unfolded (slow mobility), secondary structure (intermediate mobility), and tertiary structure (fast mobility; Fig. 4 ▶). These assignments were confirmed by salt-dependent structure mapping (see below). Even though this long-range interaction appears to involve primarily base-pairing (see below), we refer to it as a tertiary structure, because it has an I-type pseudoknot topology and involves compaction of the structure (Pleij 1993). Both the slow and intermediate mobility species were also observed in TK100M4, possibly due to the dissociation of ions and refolding of the RNA upon gel loading or during electrophoresis. Adding 4 mM MgCl2 to the gel or increasing the MgCl2 concentration in the renaturation step to 25 mM did not significantly increase the fraction of the fastest migrating species (data not shown). It should be noted that similar native gel experiments with the B. subtilis trp leader transcript did not reveal any Mg2+-dependent bands in native gels (data not shown), even though a Mg2+-dependent structure was shown to form by structure mapping and thermal denaturation experiments (Schaak et al. 2003). The absence of a Mg2+-dependent band in B. subtilis was likely due to the extensive A-U and G-U content of the base-pairing, compared to the higher G-C base-pairing content in B. stearothermophilus.
Structure mapping experiments
RNase T1 was used to probe the accessibility of G residues in the B. stearothermophilus trp leader RNA as a function of salt concentration. 5′ end-labeled trp leader RNA containing nt 1–232 in TK0M0 was found to be relatively unstructured (Fig. 5A ▶, lane 1). With the exception of the G residues in the last two triplet repeats and those predicted to be base-paired in the terminator hairpin, the G residues were cleaved with nearly equal efficiency. Upon renaturation in TK100M0 (favoring secondary structure; Fig. 5A ▶, lane 2), the majority of the G residues between 62 and 97 were protected from RNase T1 cleavage, consistent with formation of the lower stem between nt 62 and 191 (Fig. 5B ▶). Within this stretch, G67, G78, and G87 remained accessible to RNase T1 cleavage, consistent with their location in bulges (Fig. 5B ▶). Furthermore, G113, G117, G133, and G136 remained accessible to RNase T1 cleavage, consistent with their locations in the 5′ side of the pyrimidine-rich internal loop or the hairpin loop at the apex of the structure. The G residues between 36 and 61 also remained accessible to RNase T1 cleavage in TK100M0 (Fig. 5A ▶, lane 2), consistent with the first five triplet repeats remaining single-stranded. Although predicted to be base-paired, G102 and G103 were also cleaved by RNase T1. These data suggest that either these bases do not pair with C158 and C159 under these conditions, creating a slightly larger internal loop, or that being adjacent to the internal loop causes them to be dynamic.
FIGURE 5.
Ion dependence of B. stearothermophilus trp leader RNA structure formation. (A) 5′ 32P-labeled RNA (1–232) was subjected to partial RNase T1 digestion to probe RNA structure under various salt conditions. C is the untreated RNA, OH− is a limited alkaline hydrolysis ladder, and T1 is a limited RNase T1 digest under RNA denaturing conditions. Lane 1, RNA renatured in TK0M0; lane 2, RNA renatured in TK100M0; lane 3, RNA renatured in TK100M4. Important secondary structural features are given on the left side of the gel, including triplet repeat number. (B) Sites of RNase T1 cleavage superimposed on the secondary structure. Filled and open triangles indicate sites of strong and weak cleavage, respectively, in TK100M0. Asterisks (*) indicate sites of reduced cleavage upon the addition of 4 mM Mg2+. Numbering is from the start of transcription.
Renaturation in TK100M4 (favoring tertiary interactions) altered the structure further, with G residues between 36 and 61, as well as G102, G103, G113, and G117 in the 5′ side of the pyrimidine-rich internal loop, being protected from RNase T1 cleavage (Fig. 5A ▶, lane 3; Fig. 5B ▶, asterisks). Protection of these residues is consistent with their involvement in a Mg2+-dependent tertiary structure. Also, the 5′ stem–loop residues G32 and G35 are more protected from RNase T1 cleavage in TK100M4, consistent with the known ability of Mg2+ to strengthen RNA secondary structure as well as promote tertiary structure (Laing et al. 1994). The cleavage patterns of the bulged nucleotides G78 and G87 were relatively unchanged upon the addition of Mg2+, consistent with the absence of substantial secondary structure rearrangement upon formation of tertiary structure. Importantly, the G78 and G87 data also provide an internal control establishing that reduction in RNase T1 cleavage is not simply due to inhibition of enzyme activity by Mg2+ (Schaak et al. 2003). Although not predicted to pair in the tertiary interaction, G67 became protected upon the addition of Mg2+, perhaps because formation of the tertiary structure sterically hinders the access of RNase T1 to the base of the lower stem or because Mg2+ promotes formation of a non-Watson–Crick secondary structure. Overall, these results support formation of a tertiary structure in the B. stearothermophilus trp leader RNA which is similar to that predicted from the phylogenetic alignment.
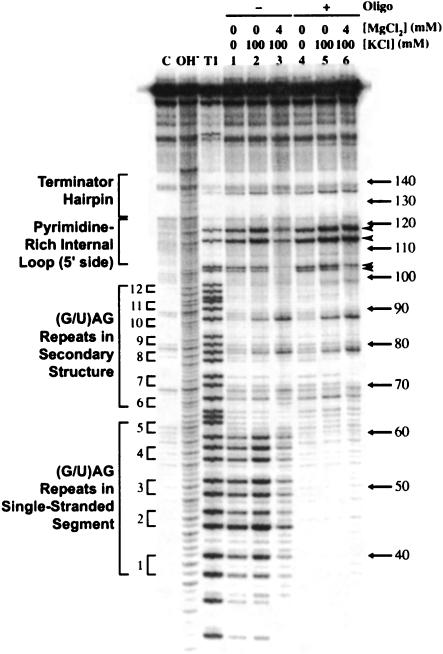
To determine the interacting partners of the Mg2+-dependent tertiary structure, RNA structure mapping experiments were repeated in the presence of a competitor DNA oligonucleotide complementary to nt 36–61 (Fig. 6 ▶). The competition experiments were performed with a transcript containing nt 1–191 to remove the possibility of the competing SD:anti-SD pairing. Quantitation of lanes 3 (no competitor) and 6 (with competitor; Fig. 6 ▶) in TK100M4 revealed a fourfold increase in cleavage of G113 and G117 and a twofold increase in cleavage of G102 and G103 in the presence of the oligonucleotide competitor. As all four of these residues are located in the pyrimidine-rich internal loop, these data show that hybridization of the competitor oligonucleotide to nt 36–61 affects residues in the distant internal loop, consistent with a long-distance tertiary interaction between the single-stranded segment containing the first five triplet repeats and the pyrimidine-rich internal loop. Note that loss of cleavage of all G residues between nt 36 and 61 in lanes 4, 5, and 6 of Figure 6 ▶ confirmed that the competitor DNA hybridized to the expected RNA segment, and gain of cleavage of G residues in the internal loop (Fig. 6 ▶, lane 6) confirmed that RNase T1 activity was not inhibited by TK100M4. It should also be noted that the 1–191 transcript appeared to have secondary structure in the absence of added monovalent ions (TK0M0; Fig. 6 ▶, lane 1). This contrasts with observations for the 1–232 transcript (Fig. 5A ▶, lane 1) for which the competing SD:anti-SD pairing was possible. Thus, the absence of competing structures might allow this transcript to be structured even in the absence of added monovalent ions.
FIGURE 6.
B. stearothermophilus trp leader RNA structure formation in the absence and presence of a competing oligonucleotide. 5′ 32P-labeled trp leader RNA (1–191) was subjected to partial RNase T1 digestion without or with a DNA oligonucleotide complementary to nt 36–61. Samples in lanes 1–3 were renatured in the absence of the competing oligonucleotide; samples in lanes 4–6 were renatured in the presence of 10 μM oligonucleotide. C, OH−, and T1 are as in Figure 5 ▶. Important secondary structural features are given on the left side of the gel, including the triplet repeat number. Arrowheads indicate sites of increased cleavage in the presence of competing oligonucleotide and TK100M4. Numbering at the right is from the start of transcription.
Thermal denaturation studies
Thermal denaturation experiments were carried out with the B. stearothermophilus trp leader RNA renatured under various conditions to test whether a unique transition was observable in the presence of Mg2+. Transcripts that contained nt 1–191 of the B. stearothermophilus trp leader were renatured in either TK100M0 (pH 8.0) or TK100M4 (pH 8.0) and melted from 5°C–95°C with a step of 0.1°C. For RNA renatured in TK100M0, transitions were observed with Tm values of 61°C and 85°C (Fig. 7A ▶, black). On the basis of the large change in hypochromicity and the lower melting temperature, we tentatively assigned the transition at 61°C to the lower stem (Fig. 3 ▶). Similarly, the transition at 85°C was assigned to the terminator hairpin on the basis of the Tm calculated from nearest-neighbor thermodynamic parameters using mfold (Mathews et al. 1999; Zuker et al. 1999), and an empirical correction for ionic strength (Tinoco and Schmitz 2000), which predicted a Tm of 80°C, in good agreement with the experimental value.
FIGURE 7.
Thermal denaturation curves for B. stearothermophilus trp leader RNA. RNAs were melted from 5°C–95°C at pH 7.0 and 8.0, and from 20°C–60°C at pH 6.0 at a step of 0.1°C, and the data were smoothed over 3°C before taking the first derivative. No transitions were found from 5°C–20°C, so these data are not shown. (A) 1–191 renatured in either TK100M0, pH 8.0 (black) or TK100M4, pH 8.0 (red). (B) 1–191 (black), 36–191 (red), or 62–191 (green) renatured in TK100M4, pH 8.0. (C) 1–191 renatured in TK100M4, pH 8.0 (black), MoK100M4, pH 7.0 (red), or MeK100M4, pH 6.0 (green).
In the presence of 4 mM Mg2+ the two transitions at 61°C and 85°C were further stabilized to 74°C and 93°C, respectively, and a new transition appeared at 42°C (Fig. 7A ▶, red). The unfolding transition of the lower stem in TK100M4 moved to a Tm of ~74°C and also contained a low-temperature (65°C) shoulder, suggesting that the lower stem unfolds in at least two steps. That the 65°C and 74°C transitions in TK100M4 have a combined area similar to the 61°C transition in TK100M0 further supports this notion. The details of this portion of the folding were not analyzed further. The new Mg2+-dependent transition at 42°C was therefore likely due to the long-range tertiary interaction between the pyrimidine-rich internal loop and the single-stranded segment containing the first five triplet repeats. The Tm of this transition was higher than that observed for the AU/GU-rich base-pairing in B. subtilis (39°C) under similar conditions (Schaak et al. 2003), as expected, and the difference was even greater at pH 7.0 and lower (see below).
To confirm that the Mg2+-dependent transition was due to the melting of tertiary interactions, 5′-deletion transcripts containing only nt 36–191 or 62–191 were renatured in TK100M4 and melted (Fig. 7B ▶). According to the model depicted in Figure 3B ▶, the 36–191 transcript would still be capable of forming the tertiary structure, whereas the 62–191 transcript would not. The melting profiles of all three transcripts were nearly identical above 60°C (Fig. 7B ▶), consistent with formation of similar secondary structural elements. Importantly, between 20°C and 60°C the melting profiles of the 1–191 (black) and 36–191 (red) transcripts were virtually identical with a transition near 42°C, whereas the 62–191 transcript (green) lacked this Mg2+-dependent transition. These data demonstrate that the 42°C transition is not due to melting of the 5′ stem–loop. Instead, the data strongly support assignment of this transition to melting of a Mg2+-dependent tertiary structure similar to that shown in Figure 3B ▶. As a further control, a synthetic RNA oligonucleotide containing nt 36–61 was melted in TK100M4 to test whether the 42°C transition was possibly due to self-structure of 36–61; no discernible transition was found for this transcript (data not shown). Melting of the long-range tertiary structure before secondary structure experimentally supports a hierarchical folding pathway for this RNA (Brion and Westhof 1997; Tinoco and Bustamante 1999; Schaak et al. 2003).
RNA tertiary structures are often stabilized at lower pH because proton binding can facilitate non-Watson–Crick base pair formation, such as A+•C wobble pairs, as well as the close approach of negative charge (Wang et al. 1991; Connell and Yarus 1994; Legault and Pardi 1997; Ravindranathan et al. 2000). Thus, transcripts containing nt 1–191 were melted at pH 6.0, 7.0, and 8.0 in the presence of 100 mM KCl and 4 mM MgCl2 (Fig. 7C ▶). The melting profiles for all samples were nearly identical above 60°C (data not shown), consistent with the absence of protonated base pairs in the secondary structure (Fig. 3B ▶). However, the Mg2+-dependent tertiary transition was found to be further stabilized with decreasing pH. The Tms were approximately 42, 44, and 47°C at pH 8.0, 7.0, and 6.0, respectively, consistent with a strong pH dependence. Melts at pH 6.5 and 7.5 also fit the observed trend (data not shown). These data show that the tertiary structure contains at least one nucleotide that has a significantly upward-shifted pKa, suggesting that there may be a base pair within the tertiary interaction that requires protonation, such as an A+•C base pair (Wang et al. 1991; Connell and Yarus 1994; Legault and Pardi 1997; Ravindranathan et al. 2000).
Models for the tertiary interaction
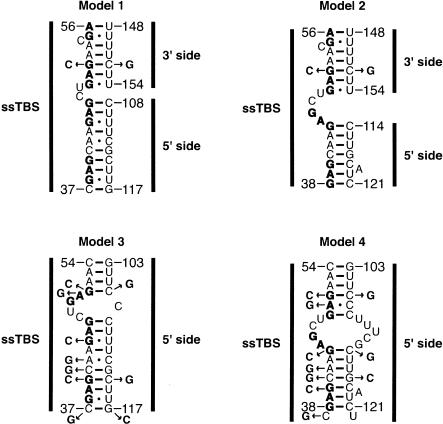
Four base-pairing models were considered for the long-range tertiary interaction (Fig. 8 ▶). These models were developed by applying a combination of phylogenetic comparison and structure prediction (Mathews et al. 1999; Zuker et al. 1999). Because mfold does not predict pseudoknot interactions, this modeling was accomplished by connecting nt 36–61 with 100–121 via a stable UUCG tetraloop to create a hairpin, with the UUCG residues prohibited from base-pairing in the calculation. Potential interactions to the 3′ side of the internal loop were modeled by inspection. Models 1 and 2 involve interactions between the first four single-stranded triplet repeats and both sides of the internal loop, whereas models 3 and 4 involve interactions between the same four triplet repeats and the 5′ side of the internal loop only. Moreover, models 1 and 2 have similar pairings at the top of the structure, as do models 3 and 4, whereas models 1 and 3 have similar pairings at the base of the structure, as do models 2 and 4. A series of single mutations as well as compensatory changes were introduced to test these models, and their effects on the stability of the tertiary interaction were examined by thermal denaturation.
FIGURE 8.
Base-pairing models for tertiary interactions in B. stearothermophilus trp leader RNA. Models 1 and 2 contain interactions between the single-stranded TRAP binding site (ssTBS) and both the 5′ and 3′ sides of the pyrimidine-rich internal loop (See Fig. 3 ▶). In contrast, models 3 and 4 contain only interactions between the ssTBS and the 5′ side of the pyrimidine-rich internal loop. Models 3 and 4 may also have interactions to the 3′ side of the internal loop, but are not shown here. Models 1 and 2 have similar pairings at the top of the structure, as do models 3 and 4, whereas models 1 and 3 have similar pairings at the base of the structure, as do models 2 and 4. Note that models 1, 3, and 4 have the potential for an A+•C base-pairing interaction, which is stabilized by lower pH. Mutants used to test the models are shown, and numbering is from the start of transcription.
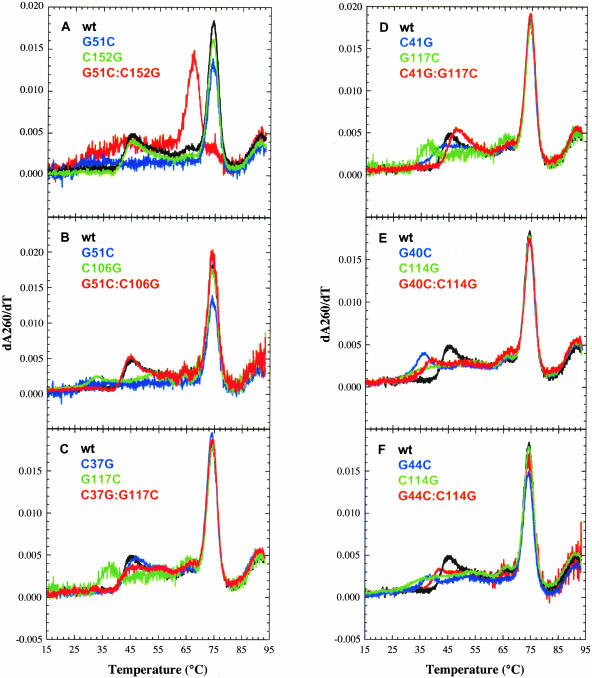
To distinguish between models 1 and 2 versus models 3 and 4, the top of the structure was varied. G51 as well as its possible pairing partners, C152 (models 1 and 2) or C106 (models 3 and 4) were mutated and melts were conducted. It should be noted that all melts were at pH 6.5 to favor tertiary structure formation (see above). Changing G51 to a C would destabilize the interaction in all four models. However, mutating C152 to a G would only have an effect if model 1 or 2 was correct, whereas mutating C106 to a G would only have an effect if model 3 or 4 was correct. Finally, the compensatory change G51C:C152G would only restore the interaction if model 1 or 2 was correct, whereas the compensatory G51C:C106G would only restore the interaction if model 3 or 4 was correct. Changing G51 to a C resulted in loss of a distinct tertiary structure melting transition (Fig. 9A,B ▶, blue), consistent with all four models. Importantly, the C152G mutation had no effect on the tertiary transition (Tm = 45°C; Fig. 9A ▶, green), whereas C106G significantly destabilized the tertiary structure (Tm = 33°C; Fig. 9B ▶, green). The compensatory G51C:C152G double mutation did not restore the tertiary transition; instead, a very broad transition between ~ 30°C and 40°C was observed (Fig. 9A ▶, red). These results are inconsistent with models 1 and 2. In contrast, the G51C:C106G compensatory mutation (Fig. 9B ▶, red) fully restored the tertiary transition, with a Tm of 45°C equivalent to that of wild-type (Fig. 9B ▶, black). These data support both models 3 and 4, in which the majority of the interactions are to the 5′ side of the internal loop. It should be noted that the fourth triplet repeat involving G55 and G57, as well as G59, might interact weakly with the U-rich 3′ side of the internal loop, because these G residues showed Mg2+-dependent protection from RNase T1 cleavage (Fig. 5 ▶). An interaction with both sides of the pyrimidine-rich internal loop would be consistent with what was found for B. subtilis (Schaak et al. 2003) and could coexist with either model 3 or 4. However, any such interactions do not appear to involve C152, and because there are a number of redundant base-pairing possibilities to the 3′ side of the internal loop, the base-pairing register of this portion of the interaction was not further pursued.
FIGURE 9.
Thermal denaturation curves for wild-type and mutant B. stearothermophilus trp leader RNAs. All RNAs (36–191) were melted from 5°C–95°C at pH 6.5 with a step of 0.1°C, and the data were smoothed over 3°C before taking the first derivative. Data from 15°C–95°C are provided. (A) Plots for wild-type (black), G51C (blue), C152G (green), and G51C:C152G (red). (B) Wild-type (black), G51C (blue), C106G (green), and G51C:C106G (red). (C) Wild-type (black), C37G (blue), G117C (green), and C37G:G117C (red). (D) Wild-type (black), C41G (blue), G117C (green), and C41G:G117C (red). (E) Wild-type (black), G40C (blue), C114G (green), and G40C:C114G (red). (F) Wild-type (black), G44C (blue), C114G (green), and G44C:C114G (red).
To differentiate between models 3 and 4, the base of the structure was altered. The potential pairing partners of G117 (C37 in model 3 and C41 in model 4) and C114 (G40 in model 3 and G44 in model 4) were tested. Consistent with both models, changing G117 to a C destabilized the tertiary interaction, lowering the Tm to 37°C (Fig. 9C,D ▶, green). Mutating C37 to a G actually led to a slight stabilization of the tertiary transition, resulting in a Tm of 47°C with a sharp transition (Fig. 9C ▶, blue). In contrast, mutating C41 to a G produced a broad transition for the tertiary interaction with a slightly lower Tm of 44°C (Fig. 9D ▶, blue). Inspection of model 4 revealed the possibility of the C37G mutation creating a wobble pair between G37 and U122, which may be available to the 5′ side of the internal loop instead of the terminator hairpin, on the basis of mfold predictions (data not shown). The C37G:G117C compensatory mutation exhibited a Tm of 45°C (Fig. 9C ▶, red) that was similar to wild-type (Fig. 9C ▶, black), but with a somewhat broader transition. In contrast, the C41G:G117C compensatory mutation resulted in a slightly higher Tm of 47°C with a sharp transition (Fig. 9D ▶, red), comparable to wild-type (Fig. 9D ▶, black). These data largely support model 4 as being most stable under these conditions (pH 6.5 and 4 mM MgCl2), although they do not entirely exclude model 3.
Next, the base-pairing partners of C114 were tested. The C114G single mutation resulted in a lowered Tm of 39°C and a very broad melting transition (Fig. 9E,F ▶, green). The G40C mutation had a significantly lower Tm of 36°C, although it retained a rather sharp transition (Fig. 9E ▶, blue). The G44C mutation also resulted in a lower Tm of 41°C and a very broad melting transition (Fig. 9F ▶, blue). Both the G40C:C114G (Fig. 9E ▶, red) and the G44C:C114G (Fig. 9F ▶, red) compensatory mutations partially restored the tertiary interaction, with observed Tm values of ~ 39°C and 42°C, respectively. The Tms of both compensatory mutants were slightly lower than the wild-type Tm of 45°C (Fig. 9E ▶, black, Fig. 9F ▶, black), suggesting that although the structure depicted in model 4 may dominate over that shown in model 3, an equilibrium may exist between the two structures.
The potential contribution of the A+•C base pair in models 3 and 4 to the stability of the tertiary structure was tested with A42G and A50G mutations, respectively. If model 3 was correct, the A42G mutation was expected to stabilize the tertiary interaction and eliminate the pH dependence of the transition, while having little effect on the tertiary interaction and not eliminating the pH dependence if model 4 was correct (A-U base pair converted to a G•U wobble). The A50G mutation, on the other hand, was expected to stabilize the tertiary structure in both models 3 and 4, but only if model 4 was correct would the pH dependence be eliminated. Both single mutations led to significant stabilization of the tertiary interaction (Tm = 53°C) and eliminated the pH dependence for this unfolding transition (Fig. 10 ▶), with all other transitions remaining identical above 60°C (data not shown). These data strongly support the pH dependence of the melts coming from an A+•C base pair. Moreover, stabilization of the tertiary interaction and elimination of pH dependence for both mutant transcripts further support an equilibrium between the pairings in models 3 and 4, which can be shifted by the introduction of particular mutations. As both pairings contain an A+•C wobble pair, it is possible that the population of these states in the wild-type sequence is a function of pH.
FIGURE 10.
Thermal denaturation curves for A+•C to GC mutant B. stearothermophilus trp leader RNAs. All RNAs (36–191) were melted from 5°C–95°C with a step of 0.1°C, and the data were smoothed over 3°C before taking the first derivative. Plots for (A) A42G and (B) A50G melted in TK100M4, pH 8.0 (black), MoK100M4, pH 7.0 (red), or CK100M4, pH 6.5 (green).
Conclusions
Although the trp operon transcription attenuation and translation control mechanisms have been most extensively studied in B. subtilis (Babitzke and Gollnick 2001; Gollnick et al. 2002), it is now apparent that these mechanisms are conserved in other Bacillus species. Our phylogenetic comparisons suggest that the secondary structural features critical to regulation are conserved, as is the tertiary interaction. The results herein indicate that a Mg2+-dependent tertiary structure forms in the trp operon leader transcript of B. stearothermophilus. This tertiary interaction is more stable than in B. subtilis, as evidenced by a compact Mg2+-dependent species on a native gel, as well as a more stable unfolding transition, especially at lower pH. Because the tertiary structure appears to interfere with the ability of TRAP to regulate translation of trpE in B. subtilis, the tertiary structure may regulate translation of trpE in other Bacillus species as well.
MATERIALS AND METHODS
Plasmids
Plasmid pJS9 was constructed by PCR using Bsttrpl (Chen et al. 1999) as the template. The PCR product was digested with EcoRI and BamHI and ligated into the same sites of the pTZ18U polylinker (U.S. Biochemical), resulting in a plasmid containing nt 1–232 of the B. stearothermophilus trp leader region. This plasmid was used to generate transcripts for the initial Mg2+-dependent RNase T1 structure mapping experiments. Plasmids pJS15 and pJS17 were constructed by PCR using pJS9 as the template. The PCR products were digested with EcoRI and BamHI and ligated into pTZ18U, resulting in plasmids containing nt 1–191 and 36–191 of the B. stearothermophilus trp leader region, respectively. Plasmid pJS16 was constructed similarly except that the digested PCR fragment was ligated into the EcoRI-BamHI sites of the pUC19 polylinker (New England Biolabs), resulting in a plasmid containing nt 62–191 of the B. stearothermophilus trp leader region. Plasmids pJS15, pJS16, and pJS17 were designed to eliminate the potential for formation of various trp leader RNA structures and were used to generate transcripts for thermal denaturation experiments. Plasmid pJS15 was also used to generate transcripts for the oligonucleotide competition studies. Plasmids pJS27 (G51C), pJS28 (G44C), pJS29 (C41G), pJS30 (G40C), pJS31 (C37G), pJS32 (C114G), pJS33 (G117C), pJS34 (C152G), pJS54 (A42G), pJS55 (C106G), and pJS57 (A51G) were generated using pJS17 as template, mutagenic primers, and the QuikChange kit (Stratagene). All compensatory mutations were generated using the same method. Plasmid pJS45 (G51C:C152G) used pJS34 (C152G) as template; plasmids pJS46 (G44C:C114G) and pJS50 (G40C:C114G) used pJS32 (C114G) as template; plasmids pJS51 (C37G:G117C) and pJS52 (C41G:G117C) used pJS33 (G117C) as template; and pJS56 (G51C:C106G) used pJS27 (G51C) as template. These plasmids were also used to generate transcripts for thermal denaturation experiments. All mutations were confirmed by automated DNA sequencing.
RNA structure prediction and phylogenetic comparison
Prediction of trp leader RNA structures for B. pumilus, B. caldotenax, and B. stearothermophilus used a combination of the structures predicted by mfold (Mathews et al. 1999; Zuker et al. 1999) and visual comparison with the structural features determined for B. subtilis (Figs. 1 ▶, 2 ▶). RNA structure predictions were carried out using mfold v3.1, unless a Tm prediction was required, in which case v2.3 was used.
Preparation, purification, and radiolabeling of RNA
For in vitro structure mapping (pJS9 or pJS15) and UV melting (pJS28–34, pJS45–46, pJS50–52, and pJS54–57), the respective purified plasmid DNA (QIAGEN) was digested with BamHI and subjected to run-off transcription using the MegaScript in vitro transcription kit (Ambion). The RNA was gel-purified, concentrated by ethanol precipitation, dissolved in TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA), and stored at −20°C. The RNA concentration was determined spectrophotometrically. The 5′ end-labeled RNA used for structure mapping was prepared by dephosphorylation of the gel-purified transcript with calf intestinal alkaline phosphatase and subsequent labeling with polynucleotide kinase and [γ-32P]ATP. Radiolabeled transcripts were gel-purified as described for unlabeled RNA. The RNA concentration was determined by scintillation counting.
Native gel mobility assay
5′ end-labeled trp leader RNA (4 nM) containing nt 1–232 was denatured in TE buffer at 90°C for 1 min to eliminate potential dimerization products, and allowed to cool to room temperature for 10 min. The RNA was then renatured in TK0M0 (40 mM Tris-HCl, pH 8.0), TK100M0 (40 mM Tris-HCl, pH 8.0, 100 mM KCl), or TK100M4 (40 mM Tris-HCl, pH 8.0, 100 mM KCl, 4 mM MgCl2) at 50°C for 10 min, and allowed to cool to room temperature for 10 min. Glycerol was added to 10% final concentration, and the samples were fractionated on a 6% (29:1 acrylamide/bis)/0.5X TBE native gel at 330 V for 4–5 h. The temperature of the apparatus was controlled by an external bath that flowed into a heat exchanger located behind the gel plates. The external bath temperature was 12°C, and the actual gel temperature was measured at 16°C by insertion of a thermocouple probe into the gel (Bevilacqua and Bevilacqua 1998). Gels were dried, visualized using a PhosphorImager (Molecular Dynamics), and quantified using ImageQuant software (Molecular Dynamics).
Structure mapping
Structure mapping experiments were carried out using 5′ end-labeled RNA (4 nM final concentration) and G-specific RNase T1 (Sigma). Titrations were carried out to establish conditions under which ~90% of the RNA remained uncleaved after treatment (0.017 U/μL of RNase T1). The RNA was denatured in TE buffer at 90°C for 1 min and allowed to cool for 10 min at room temperature. The RNA was then renatured in TK0M0, TK100M0, or TK100M4 at 50°C for 10 min, and allowed to cool to room temperature for 10 min. Samples were heated to the reaction temperature for 10 min prior to adding RNase T1. RNase T1 reactions were carried out at 37°C for 10 min, and subsequently stopped by adding an equal volume of 95% formamide loading buffer containing 20 mM EDTA and immediate placement on dry ice. Samples were quantified by scintillation counting so that an equal number of counts were loaded per lane. Samples were fractionated by 8.3 M urea/6% polyacrylamide gel electrophoresis. Sequencing lanes for G residues were prepared by limited digestion with RNase T1 (0.2 U/μL) under RNA denaturing conditions of 50% urea at 50°C, and limited hydrolysis ladders were prepared by treating RNA with alkali. Control samples were prepared in the same way except that RNase T1 was omitted. Reactions that were carried out in the presence of the DNA oligonucleotide complementary to positions 36–61 of the trp leader transcript were renatured in 1X TE at 90°C for 1 min in the presence of 10 μM oligonucleotide. All other steps were carried out as described above. Gels were dried and visualized using a PhosphorImager.
Thermal denaturation
Transcripts used in thermal denaturation experiments were prepared by in vitro transcription as described above. RNAs were renatured at 90°C for 1 min in TE, followed by slow cooling at room temperature for 10 min. RNAs were further renatured in TK100M0 or TK100M4 at 50°C for 10 min, followed by slow cooling at room temperature for 10 min. To test the pH dependence of tertiary structure formation, RNAs were renatured in TK100M4 (Tris-HCl buffer, pH 8.0), HK100M4 (HEPES buffer, pH 7.5), MoK100M4 (MOPS buffer, pH 7.0), CK100M4 (cacodylate buffer, pH 6.5), or MeK100M4 (MES buffer, pH 6.0) at 50°C for 10 min, followed by slow cooling at room temperature for 10 min. Mutant trp leader RNAs were renatured in CK100M4 (pH 6.5) at 50°C for 10 min, followed by slow cooling at room temperature for 10 min. Thermal denaturation profiles were obtained at 260 nm in 10-mm pathlength cuvettes using a Gilford Response II spectrophotometer equipped with a temperature controller. Samples were heated at a rate of ~0.1°C/min over a range of 5°C–95°C (pH 8.0, 7.0, and 6.5) or 20°C–60°C (pH 7.0 and 6.0), and readings were taken every 0.1°C. Absorbance data was smoothed over a 3° range prior to taking the first derivative to determine the melting temperatures; larger temperature windows distorted the Tm of closely spaced transitions, and so were not used.
Acknowledgments
We thank members of the Babitzke and Bevilacqua labs for helpful comments. This work was supported by N.I.H. grant GM52840 to P.B. and National Sciences Foundation CAREER Grant MCB-9984129 to P.C.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ``advertisement'' in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5149603.
REFERENCES
- Antson, A.A., Brzozowski, A.M., Dodson, E.J., Dauter, Z., Wilson, K.S., Kurecki, T., Otridge, J., and Gollnick, P. 1994. 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J. Mol. Biol. 244: 1–5. [DOI] [PubMed] [Google Scholar]
- Antson, A.A., Otridge, J., Brzozowski, A.M., Dodson, E.J., Dodson, G.G., Wilson, K.S., Smith, T.M., Yang, M., Kurecki, T., and Gollnick, P. 1995. The structure of trp RNA-binding attenuation protein. Nature 374: 693–700. [DOI] [PubMed] [Google Scholar]
- Antson, A.A., Dodson, E.J., Dodson, G., Greaves, R.B., Chen, X., and Gollnick, P. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401: 235–242. [DOI] [PubMed] [Google Scholar]
- Babitzke, P. and Gollnick, P. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J. Bacteriol. 183: 5795–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke, P. and Yanofsky, C. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc. Natl. Acad. Sci. 90: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1995. Structural features of L-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis. J. Biol. Chem. 270: 12452–12456. [DOI] [PubMed] [Google Scholar]
- Babitzke, P., Stults, J.T., Shire, S.J., and Yanofsky, C. 1994. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J. Biol. Chem. 269: 16597–16604. [PubMed] [Google Scholar]
- Babitzke, P., Yealy, J., and Campanelli, D. 1996. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: Effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J. Bacteriol. 178: 5159–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke, P., Schaak, J.E., Yakhnin, A.V., and Bevilacqua, P.C. 2003. Role of RNA structure in transcription attenuation in Bacillus subtilis: The trpEDCFBA operon as a model system. Methods Enzymol. (in press). [DOI] [PubMed]
- Bevilacqua, J.M. and Bevilacqua, P.C. 1998. Thermodynamic analysis of an RNA combinatorial library contained in a short hairpin. Biochemistry 37: 15877–15884. [DOI] [PubMed] [Google Scholar]
- Brion, P. and Westhof, E. 1997. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26: 113–137. [DOI] [PubMed] [Google Scholar]
- Chen, X., Antson, A.A., Yang, M., Li, P., Baumann, C., Dodson, E.J., Dodson, G.G., and Gollnick, P. 1999. Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus. J. Mol. Biol. 289: 1003–1016. [DOI] [PubMed] [Google Scholar]
- Connell, G.J. and Yarus, M. 1994. RNAs with dual specificity and dual RNAs with similar specificity. Science 264: 1137–1141. [DOI] [PubMed] [Google Scholar]
- Du, H. and Babitzke, P. 1998. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 273: 20494–20503. [DOI] [PubMed] [Google Scholar]
- Du, H., Tarpey, R., and Babitzke, P. 1997. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 179: 2582–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick, P., Ishino, S., Kuroda, M.I., Henner, D.J., and Yanofsky, C. 1990. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc. Natl. Acad. Sci. 87: 8726–8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick, P., Babitzke, P., Merino, E., and Yanofsky, C. 2002. Aromatic amino acid metabolism in Bacillus subtilis. In Bacillus subtilis and its closest relatives: From genes to cells, pp. 233–244. ASM Press, Washington, DC.
- Hoffman, R.J. and Gollnick, P. 1995. The mtrB gene of Bacillus pumilus encodes a protein with sequence and functional homology to the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis. J. Bacteriol. 177: 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, M.I., Henner, D., and Yanofsky, C. 1988. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J. Bacteriol. 170: 3080–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing, L.G., Gluick, T.C., and Draper, D.E. 1994. Stabilization of RNA structure by Mg ions. Specific and nonspecific effects. J. Mol. Biol. 237: 577–587. [DOI] [PubMed] [Google Scholar]
- Legault, P. and Pardi, A. 1997. Unusual dynamics and pK(a) shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 119: 6621–6628. [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Merino, E., Babitzke, P., and Yanofsky, C. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 177: 6362–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otridge, J. and Gollnick, P. 1993. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc. Natl. Acad. Sci. 90: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij, C.W. 1993. Appendix 2: RNA pseudoknot. In The RNA world: The nature of modern RNA suggests a prebiotic RNA world, pp. 609–613. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ravindranathan, S., Butcher, S.E., and Feigon, J. 2000. Adenine protonation in domain B of the hairpin ribozyme. Biochemistry 39: 16026–16032. [DOI] [PubMed] [Google Scholar]
- Sarsero, J.P., Merino, E., and Yanofsky, C. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 182: 2329–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaak, J.E., Yakhnin, H., Bevilacqua, P.C., and Babitzke, P. 2003. A Mg2+-dependent RNA tertiary structure forms in the Bacillus subtilis trp operon leader transcript and appears to interfere with trpE translation control by inhibiting TRAP binding. J. Mol. Biol. 32: 555–574. [DOI] [PubMed] [Google Scholar]
- Shimotsu, H., Kuroda, M.I., Yanofsky, C., and Henner, D.J. 1986. Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon. J. Bacteriol. 166: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratsuchi, A. and Sato, S. 1991. Nucleotide sequence of trpE, anthranilate synthase I gene, of Bacillus caldotenax. Biochim. Biophys. Acta 1090: 348–350. [DOI] [PubMed] [Google Scholar]
- Sudershana, S., Du, H., Mahalanabis, M., and Babitzke, P. 1999. A 5′ RNA stem-loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol. 181: 5742–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco Jr., I. and Bustamante, C. 1999. How RNA folds. J. Mol. Biol. 293: 271–281. [DOI] [PubMed] [Google Scholar]
- Tinoco Jr., I. and Schmitz, M. 2000. Thermodynamics of formation of secondary structure in nucleic acids. In Thermodynamics in biology, pp. 131–176. Oxford University Press, New York.
- Wang, C., Gao, H., Gaffney, B.L., and Jones, R.A. 1991. Nitrogen-15-labeled oligodeoxynucleotides. 3. Protonation of the adenine N1 in the A.C and A.G mispairs of the duplexes (D[Cg(N-15(1))Agaattcccg])2 and (D[Cgggaattc(N-15(1)Acg])2. J. Am. Chem. Soc. 113: 5486–5488. [Google Scholar]
- Yakhnin, A.V. and Babitzke, P. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc. Natl. Acad. Sci. 99: 11067–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M., de Saizieu, A., van Loon, A.P., and Gollnick, P. 1995. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP). J. Bacteriol. 177: 4272–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M., Mathews, D.H., and Turner, D.H. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA biochemistry and biotechnology, NATO ASI Series, Kluwer Academic Publishers, Boston.