Abstract
An important aspect of the assembly of RNPs, and in particular of spliceosomes, is the succession of proteins bound to any given site on the RNA. Protein–RNA cross-linking is a well-established technique for investigating this, but the identification of a cross-linked protein has so far relied upon the availability of antibodies for immunoprecipitation or Western blot studies. To facilitate identification of proteins independent of these techniques, site-specific protein–RNA cross-links were purified in a large scale, which were then used for mass spectrometry (MS). This approach was carried out by the use of a minimal pre-mRNA construct containing a single photoactivatable azidophenacyl group and an adjacent biotin-dT tag for affinity purification of the cross-linked product. To test the feasibility of the method, we purified cross-links to nucleotide 9 downstream of the 5′ splice site of pre-mRNA in the spliceosomal complexes A (“pre-spliceosome”) and H. By this method, we were able to identify several proteins by MS; the hnRNP proteins A2/B1 were cross-linked to the pre-mRNA in complex A, and FUSE 2/FBP (a homolog of the intronic splicing enhancer KSRP) was cross-linked in complex H.
Keywords: Site-specific cross-linking, RNP, mass spectrometry, MALDI-MS, RNA–protein interactions, biotin
INTRODUCTION
Assembly of ribonucleoprotein particles (RNPs) involves the binding of proteins or protein complexes to the RNA molecule. As such, protein–RNA, protein–protein, and RNA–RNA interactions are of fundamental importance for structural and functional integrity of an RNP. Many RNPs, such as ribosomes, snoRNPs, the SRP, and the telomerase RNP, perform central cellular functions such as the translation or modification of RNA, the translocation of proteins, or the synthesis of telomeric DNA. The highly complex spliceosome, involved in the processing of pre-mRNA, consists of several separate small nuclear (sn)RNPs and up to 300 proteins (for review, see Burge et al. 1999; Jurica and Moore 2003). For this reason, the spliceosome is an interesting system to study complex protein–RNA interactions.
To find out which nucleotides of the RNA molecule a particular protein is bound to, the standard method is that of cross-linking, in which a position-specific cross-linker fixes a point on the RNA to the protein(s) in the immediate vicinity. After treatment of the RNA with nucleases, a radioactive cross-linked protein can be characterized by electrophoretic molecular-weight determination. However, its identification in a multicomponent complex requires the availability of specific antibodies for immunoprecipitation or Western blotting. In this study, we show that this requirement can be circumvented by the use of a suitable isolation method for the cross-linked protein(s), which enriches and purifies the cross-linked proteins from the vast majority of non-cross-linked proteins, and their subsequent identification by mass spectrometry. We illustrate the practicability of this technique by describing its application in defining the proteins surrounding a specific site of the intron in two protein–RNA complexes, namely spliceosomal complex A and the unspecific complex H, complexes which can be rapidly and efficiently assembled in vitro (see below).
Protein–RNA cross-linking is frequently used to study protein–RNA interactions. For example, direct (zero-length) protein–RNA contacts can be analyzed by UV cross-linking using site-specifically incorporated 32P or 4-thiouridine (for review, see Reed and Chiara 1999). A broader view of proteins lying in the direct vicinity of a particular position can be obtained using the cross-linking reagents p-azidophenacyl bromide (APB, ~9 Å) or benzophenone-iodacetamide, (~15 Å). Both are introduced at single positions in the RNA through site-specifically incorporated phosphorothioates (for review, see Konarska 1999). We chose APB to obtain knowledge about proteins surrounding a distinct site of the RNA, because of its high-specific cross-linking and modification efficiency (Konarska 1999), properties essential for our studies.
The isolation and purification of the cross-linked protein–RNA species was performed by using a biotin–streptavidin selection. This has the advantage of being very selective and efficient (KD = 10−15), whereas biotin, being a very small molecule, does not introduce major structural perturbations. Biotin-containing cross-linkers such as sulpho-SBED could, in principle, also be used (Geselowitz and Neumann 1995), but because of its length (~22 Å), they render it difficult to make meaningful statements about the protein composition around a given point on the RNA. Our approach was therefore to introduce the modified biotin-containing nucleotide biotin-dT at a defined site into a synthetic oligoribonucleotide, so that protein–RNA cross-links could be isolated via the RNA-bound biotin and purified in high yield for analysis by mass spectrometry.
The spliceosome is formed de novo upon each pre-mRNA molecule that is to be spliced. If pre-mRNA is incubated with nuclear extract at 0°C in the absence of ATP, then the splicing-unspecific complex H is formed, in which the pre-mRNA is largely covered by proteins; these particles are termed heterogeneous nuclear (hn)RNPs. Some of the hnRNP proteins are also known to act as alternative splicing factors (for review, see Dreyfuss et al. 1993; Krecic and Swanson 1999). If ATP is added and the temperature is raised to 30°C, then the consensus sequences of the pre-mRNA are recognized by snRNPs and by certain proteins (so-called splicing factors). This initiates the splicing process, in which the spliceosome is assembled in discrete steps, passing successively through gel-detectable complexes termed A, B, C, mRNP, and the post-spliceosomal complex (for review, see Burge et al. 1999). During this process, numerous proteins are exchanged at the consensus sequences of the pre-mRNA. This protein exchange has been confirmed by cross-linking studies, for example at the branch point (MacMillan et al. 1994), or in an artificial trans-splicing system, at the 5′ splice site (Sha et al. 1998).
Position C9 of the intron, chosen for our first experiments with pre-mRNA, lies several nucleotides downstream of the positions at which base-pairing occurs with the U1 snRNA, which is replaced by U6 snRNA after entry of the U4/U6.U5 tri-snRNP (Mount et al. 1983; Wassarman and Steitz 1992). Our choice of this site was guided by its proximity to the base-pairing, and also by the fact that the modification gave a workable yield in cross-linking, while appearing not to interfere substantially with the splicing reaction. We were able to demonstrate cross-links between this position and proteins in the pre-splicing “complex H” and in the early spliceosomal “complex A”, also termed the pre-spliceosome. Mass-spectrometric analysis of the cross-linked proteins in complex H revealed the presence of the FUSE 2/FBP protein (a homolog of the intronic splicing enhancer KSRP; Min et al. 1997). In complex A, we demonstrate the involvement of the hnRNP proteins A2 and B1, proteins shown to play a role in alternative splicing (Mayeda et al. 1994).
RESULTS AND DISCUSSION
Our method for the identification of purified protein–RNA cross-links by mass spectrometry combines (1) preparation of site-specifically modified pre-mRNA containing cross-linker and biotin; (2) photo-affinity cross-linking; (3) purification of protein–RNA cross-links by biotin–streptavidin affinity selection; and (4) their analysis by mass spectrometry. The strategy, as applied here to the spliceosomal complexes H and A, is outlined in Figures 1 ▶ and 2 ▶.
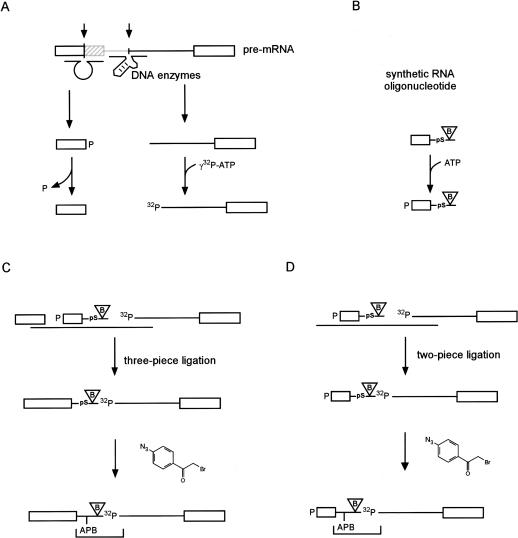
FIGURE 1.
Preparation of site-specifically modified pre-mRNA. (A) Generation of RNA fragments. Full-length Minx pre-mRNA was prepared by transcription in vitro and 5′ and 3′ fragments were generated from it by site-directed cleavage with RNA-cleaving DNA enzymes, as shown. The 5′ fragment was dephosphorylated and the 3′ fragment was 5′ labeled with 32P. (B) 5′ phosphorylation (nonradioactive) of [pS+9B]-nt21. The 21-nt oligonucleotide contains a phosphorothioate at the intron position C9 (pS+9) and an adjacent biotin (B). (C) Three pieces of RNA, as shown, were hybridized to a splint oligonucleotide and ligated with T4 DNA Ligase, yielding [pS+9B]-pre-mRNA. The product was modified with APB at the phosphorothioate (pS), leading to [APB+9B]-pre-mRNA. After RNase T1 digest, a radioactive RNA fragment remains containing APB and biotin as indicated by the bracket. (D) As in C, but two pieces of RNA were used, resulting in the 5′ truncated product [pS+9B]-pre-mRNA or [APB+9B]-pre-mRNA-S after modification with APB.
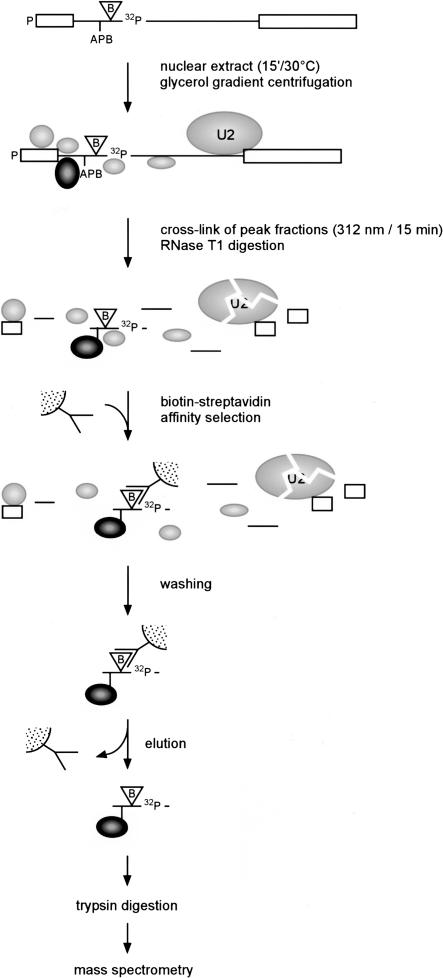
FIGURE 2.
Purification of protein–RNA cross-links by biotin–streptavidin selection. Pre-mRNA was prepared as shown in Figure 1 ▶ and incubated with nuclear extract for 15 min at 30°C. Subsequently, complex A was purified by glycerol gradient centrifugation. U1 snRNP was not included in the figure, as it is not investigated whether U1 was base-paired with the 5′ splice site in this complex (Das et al. 2000). The fractions containing complex A were pooled, irradiated, and digested with RNase T1. Cross-linked proteins that were covalently bound to the nucleotide next to the biotin-containing residue were affinity purified with streptavidin beads. After stringent washing, the proteins were eluted from the beads for analysis by mass spectrometry.
Preparation of site-specifically modified pre-mRNA
Site-specifically modified pre-mRNA was constructed by two- or three-piece ligation (Moore and Sharp 1992), creating either full-length pre-mRNA ([pS+9B]-pre-mRNA), or a shortened form truncated by 48 nt at its 5′ end and bearing a 5′-phosphate ([pS+9B]-pre-mRNA-S; see Fig. 1 ▶). The 5′ and 3′ fragments were prepared by cleavage of in vitro transcribed pre-mRNA with RNA-cleaving DNA enzymes (Santoro and Joyce 1997,Santoro and Joyce 1998). The enzymes were designed to cleave after nts 48 and 69 of the pre-mRNA.
The synthetic 21-nt oligonucleotide [pS+9B]-nt21 contains (1) a single phosphorothioate at the intron position C9 for modification with the photoaffinity cross-linker; (2) a stabilizing 2`-deoxyribose (Gish and Eckstein 1988); and (3) the nucleotide biotin-dT for subsequent selection with streptavidin (Fig. 1B ▶). The design was such that after RNase T1 cleavage, the resulting 5-nt fragment contained the cross-linker, the biotin, and the radioactive phosphate. The 5′ fragment, [pS+9B]-nt21 and the 3′ fragment were then assembled into the pre-mRNA-derived construct to be used for cross-linking, either by three-piece ligation (Fig. 1C ▶), or with omission of the 5′ fragment, by two-piece ligation (Fig. 1D ▶).
These constructs were then modified with APB as described in Materials and Methods.
Photo-affinity cross-linking in complexes H and A
Splicing of [APB+9B]-pre-mRNA was analyzed by following the time-course of splicing in comparison with unmodified pre-mRNA. The splicing efficiency (in terms of both yield and rate) was only slightly reduced by the modification (data not shown). This pre-mRNA was then used to analyze protein–RNA cross-links in the distinct spliceosomal complexes A and H. In parallel reactions, complex H was assembled on ice, and complex A at 30°C (see Fig. 3A ▶ for gradient profiles), both for 10 min, and separated by glycerol gradient centrifugation. The content of each reaction mixture was determined by native gel electrophoresis. Figure 3B ▶ shows the fractions from the 30°C incubation; the products seen are complex A, unreacted complex H, and a trace of complex B (the next splicing intermediate after complex A). Complexes H and A could not be separated cleanly, so the complex A fractions contain a minor amount of contaminating complex H. The latter is found predominantly in gradient fractions 3 and 4, whereas complex A predominates in fractions 5 and 6 (Fig. 3B ▶). Fractions 7 and higher contain a mixture of complexes A and B. If the incubation with ATP at 30°C was prolonged to 30 min, fully assembled spliceosomes appeared (data not shown).
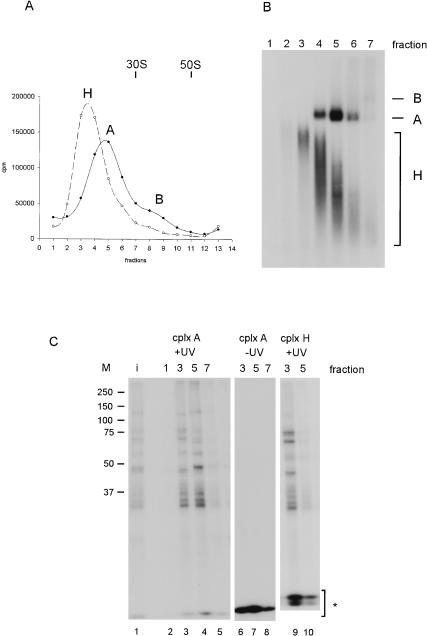
FIGURE 3.
Analysis of cross-links in complexes H and A with [APB+9B]-pre-mRNA. (A) Gradient profiles of complex H (incubation with [APB+9B]-pre-mRNA for 10 min on ice) and complex A (10 min at 30°C) after centrifugation in a 1.5 mL S55-S rotor. Complex H sediments at around 12 S, complex A at around 20 S, and complex B, which appears as a minor product, at around 40 S. (B) Nondenaturing gel analysis of gradient fractions 1–7 from the 30°C incubation. A total of 10 μL from each fraction was applied to a 2% nondenaturing agarose gel. The complexes are identified at right. (C) Photoaffinity cross-linking at 312 nm after gradient centrifugation. Aliquots from fractions 1, 3, 5, and 7 of B were cross-linked, and parallel aliquots from fractions 3, 5, and 7 were subjected to identical procedures, but without UV irradiation. Radioactive proteins were then separated on a 10% SDS–polyacrylamide gel. (Lane 1) Products of the 30°C incubation cross-linked without prior centrifugation (input, “i”); (lanes 2–5) fractions 1, 3, 5, and 7 from B after cross-linking; (lanes 6–8) as in lanes 3–5, but in a mock cross-linking reaction without UV light applied. (M) Prestained molecular-weight markers (Bio-Rad) are indicated at left. The asterisk indicates the RNase T1-digested non-cross-linked RNA.
Aliquots of each gradient fraction from the 30°C incubation were taken for cross-linking by UV-irradiation (see Materials and Methods). The RNA was then digested with RNase T1 under mildly denaturing conditions at 50°C, and the radioactive cross-linked species were analyzed on an SDS–polyacrylamide gel (Fig. 3C ▶).
Lanes 2–5 of Figure 3C ▶ show the cross-links obtained from fractions 1, 3, 5, and 7 in Figure 3A ▶. In lane 3, cross-links from complex H only are seen (Fig. 3B ▶, cf. lane 3). In lane 4, (fraction 5 of the gradient) cross-links from complex A are present together with those from complex H (Fig. 3B ▶, cf. lane 5); thus, only cross-links that appear in lane 4 and are absent in lane 3 can be assigned to complex A. In all, we observed several strong cross-links in the A complex fraction 5 (corresponding to proteins of molecular weight ca. 30 and 50 kD) and some weaker ones (ca. 60 and 120 kD). Only the 50-kD cross-link seems to be specific for complex A; all of the others were found in complex H as well. Some more cross-linked proteins were found in complex H (three of ~75 kD and one of ~45 kD; lane 3) and to a lesser extent in complex A.
The controls (-UV-irradiation, lanes 6–8) confirmed that the radioactive bands were genuine products of irradiation. In further comparisons, the incubation at 0°C gave a cross-linking pattern identical to that seen in Figure 3C ▶, lane 3, confirming the assignment to complex H (lanes 9 and 10). The patterns of cross-links obtained with pre-mRNA, that did not contain biotin, were also identical to those shown here; also, no cross-links were observed when the digestion was performed with both RNase T1 and proteinase K, therefore, the radioactive bands were not due to undigested RNA (data not shown).
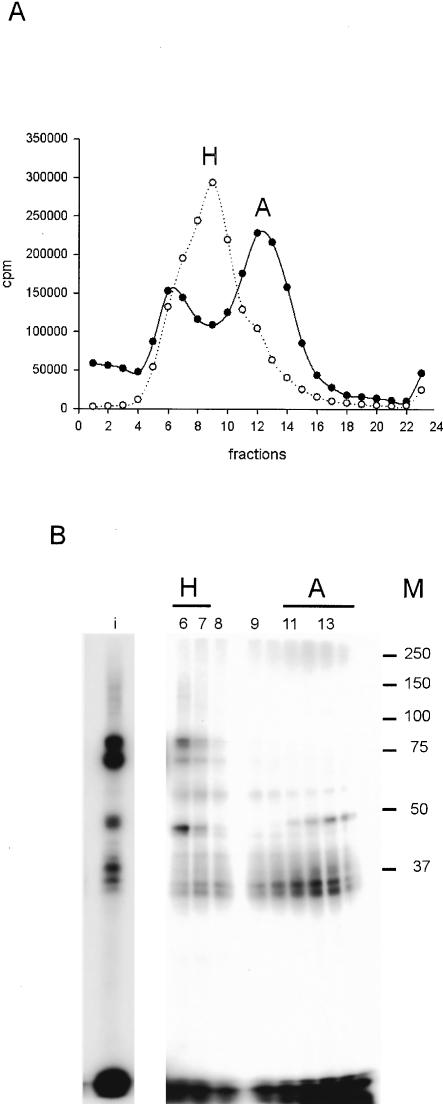
For purification of cross-links for mass spectrometry, the reaction has to be scaled up. For this reason, we first investigated whether the two-piece ligated [APB+9B]-pre-mRNA-S (Fig. 1D ▶) could replace the full-length pre-mRNA. The single, rather than double ligation was expected to result in a higher ligation yield and also to reduce cost and time, because the 5′ fragment was not required. Although the splicing efficiency was slightly reduced compared with [APB+9B]-pre-mRNA, splicing complex formation was not affected (data not shown; see also Krainer et al. 1984). We therefore used [APB+9B]-pre-mRNA-S to check whether we could observe the same cross-links using this construct. In preliminary experiments, a 15-min incubation time was found to lead to the greatest yield of complex A. Under the different gradient conditions, we were now able to separate complexes H and A as distinct peaks (Fig. 4A ▶). However, the fractionated complex A seemed to dissociate during native gel electrophoresis (data not shown). The protein cross-linking pattern of the large-scale cross-linking reaction was similar to that with the full-length pre-mRNA. As complexes H and A are better separated, no coinciding cross-links of the H complex are found in the A complex regions from the preparative gradient (cf. Fig. 4B ▶ and Fig. 3C ▶). In complex H, all cross-links found in the small-scale analysis are also seen with the shorter pre-mRNA in the large-scale preparation, namely, the proteins around 75, 60, 45, and 30 kD (Fig. 4B ▶, cf. fractions 6 and 7 with Fig. 3C ▶, fraction 3). Proteins assigned to cross-link in complex A (fraction 5, Fig. 3C ▶) were also found in fractions 11–14, the peak corresponding to complex A. These were the 60-, 50-, and 30-kD proteins. The proteins around 75 and 120 kD were absent, indicating that they belong only to complex H. However, the intensities of the observed cross-links vary slightly. For example, the 120-kD cross-link is found as a weak shadow only (see input and fractions 6 and 7). The lower amount of this cross-link is probably due to experimental variations and not to the use of the shorter pre-mRNA, as it was also found in other experiments when [APB+9B]-pre-mRNA-S was used. The amount of the 30-kD protein appears to be lower in complex H than in complex A; this may be because of higher efficiency of cross-linking to this protein in complex A, or to its displacement in complex H by other proteins.
FIGURE 4.
Analysis of cross-links in complexes A and H using [APB+9B]-pre-mRNA-S. (A) Gradient profile of complexes H and A. Incubation at 0°C or 30°C was stopped after 15 min, and the reaction mixture was centrifuged in a TST41.14 rotor. Complex H was found in fractions 7–10 from the 0°C incubation, and narrowed to fractions 6–7 when incubation conditions (30°C) produced complex A as well; the latter was found in fractions 11–14. (B) Cross-linking profile of the gradient fractions. Twenty microliters of a cross-linked and T1-digested 500 μL gradient fraction or 10 μL of the starting material of one preparative reaction, cross-linked without centrifugation (input, “i”), were separated on 10% SDS–polyacrylamide gel. (M) The molecular weights of the prestained marker are indicated at right. For the input, a shorter exposure is shown.
Biotin–streptavidin selection and mass spectrometric analysis
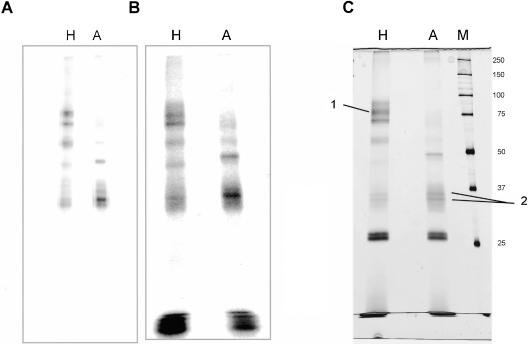
The conditions and controls as described above for the [APB+9B]-pre-mRNA-S were now applied for the biotin–streptavidin selection. A total of five gradients were used for purification of cross-links in complex H, and because of the lower yield of assembled complex A, eight gradients for cross-links in complex A (see Table 1 ▶ for further data of the purification process and Fig. 4A ▶ for the gradient profile). Four fractions from each gradient were pooled, fractions 11–14 (complex A), and fraction 7–10 (complex H); these were cross-linked at 312 nm, digested with RNase T1, and used for the biotin–streptavidin selection. An aliquot of the cross-linked proteins was taken before selection for comparison of the selection specificity (Fig. 5A ▶). The affinity purification was performed under semidenaturing conditions with NP-40S and SDS. Low concentrations of SDS led to an increased binding efficiency of the biotin to streptavidin. The selected cross-links were washed under stringent conditions to remove unbound proteins; milder conditions led to high background of unspecific purified proteins (data not shown). Although a minor amount of biotin was found in the supernatant of the binding reaction, the washes were found to be essentially free of radioactivity. After washing, biotin-containing proteins were eluted from the beads, separated on a SDS–polyacrylamide gel, and stained with silver nitrate. The silver-stained bands (Fig. 5C ▶) match very well with the radioactive cross-linking pattern (Fig. 5B ▶), showing that the purification was specific. Two spurious bands at around 25 kD appeared in the silver stain after purification of H and A complex cross-links; these bands were also observed in a mock purification with nuclear extract alone. In addition, to show that all proteins are selected with the same efficiency, the input cross-links for the affinity selection were compared with the purified cross-links and no differences were seen (Fig. 5 ▶, cf. A and B).
TABLE 1.
Summary of affinity selection yields
| H complex | A complex | |
| Total input | 125 pmole | 185 pmole |
| No. of gradients/pooled fractions | 5/20 | 8/32 |
| Pooled gradient peaks | 73 pmole | 73 pmole |
| Percent gradient peak/total input | 60% | 40% |
| Input for affinity selection1 | 60 pmole | 57 pmole |
| Beads after washing | 34 pmole | 25 pmole |
| Eluted proteins2 | 27 pmole | 22 pmole |
| % eluted/gradient peak | 37% | 30% |
| Left on beads | 7 pmole | 3 pmole |
| Elution efficiency | 80% | 88% |
Quantities were determined by scintillation counting and are correspondingly approximate. 1Losses are due to adhesion of the sample to the walls of the 24-well plate and tubes. 2This includes a correction for quenching by the elution buffer.
FIGURE 5.
Large-scale preparation of protein–RNA cross-links. A total of 600 μL of complex H or complex A reaction mixtures containing 37 nM [APB+9B]-pre-mRNA-S were loaded on a 14-mL TST41.14 rotor. A total of 5 (H) or 8 (A) such gradients were taken, and the peaks were pooled for cross-linking and isolation of the product. (A,B) Comparison of input cross-links with eluate. For cross-linking, gradient fractions 7–10 (H) and 11–14 (A) were pooled, cross-linked, and digested with RNase T1. A 10-μL aliquot was withdrawn before the biotin–streptavidin selection, and to compare it with the eluted protein–RNA cross-links from the beads, a PhosphorImager picture was taken from the silver-stained gel in C. (Note: The gel in A ran for a longer time, so no degraded RNA is visible at the bottom.) (C) Silver-stained gel of material eluted from the beads. The 150-μL protein–RNA cross-links were fractionated on a 10% SDS–polyacrylamide gel. (M) A total of 2 μL (0.36 μg) of unstained molecular weight marker (Bio-Rad); the molecular weight is indicated at right. (1) FUSE 2; (2) hnRNP A2/B1.
Proteins identified by mass spectrometry
By using mass spectrometry, it was now possible to determine the identity of the three strongest bands. The ~70-kD cross-link in complex H was identified as the 68-kD far upstream element (FUSE)-binding protein 2/KH-type splicing regulatory protein/FBP. The protein consists of four so-called KH domains homologous with hnRNP protein K (Siomi et al. 1993). A homolog of FUSE, KSRP, has been described as an intronic enhancer (Min et al. 1997). The proteins have 70% amino acid similarity overall and 74% amino acid identity in the KH domains. Another homolog of KSRP is PSI, a splicing repressor of the Drosophila P-element IVS3 (Siebel et al. 1994, 1995). The two proteins cross-linked around 30 kD in complex A were identified as hnRNP A2 and B1 (Burd et al. 1989). They are transcribed from the same gene, but differ by a 12 amino acid insertion in hnRNP B1, due to an alternative splicing event. Apart from being hnRNP proteins, these proteins also have a function in alternative splicing (Mayeda et al. 1994).
Conclusions
In this work, we have shown that proteins bound at a specific position to RNA can be isolated and purified on a small scale (1–10 pmole for the individual protein) by using biotin–streptavidin selection, and that the cross-linked proteins can be identified by mass spectroscopy. Success with this approach has the following prerequisites:
An RNA must be produced with three modifications, that is, a position-specific cross-linker, a neighboring biotin, and a radioactive label. These three modifications must remain together during treatment with nucleolytic enzymes, and they must not change the composition or the function of the particle. With the help of DNA enzymes, RNA fragments can be generated for a large number of positions needed for the incorporation of a synthetic oligonucleotide containing all necessary modifications and a 32P-label for a specific position. In combination with ribonucleases, a minimal RNA fragment containing cross-linker, biotin, and radioactive label can be obtained and purified by biotin–streptavidin selection.
The cross-linked product must be obtainable in a quantity sufficiently large to be analyzed by MS. In our experiments, such high yields could be obtained by using a 5′-shortened pre-mRNA molecule, the two-piece ligated product.
In a first attempt, it proved possible to identify several cross-linked proteins in this way. Among the cross-linked proteins, we found hnRNP A2/B1, a component of complex H, which also has a function in alternative splicing. Furthermore, it has been found in tobramycin-purified complex A (Hartmuth et al. 2002). We found the FUSE 2 protein in complex H; this protein has not been identified previously as a constituent of this or a spliceosomal complex. FUSE 2 has four KH domains, and is thus probably an RNA-binding protein. Two proteins that show homology with FUSE 2, KSRP (Min et al. 1997) and PSI (Siebel et al. 1994, 1995), are believed to take part in alternative splicing.
Irrespective of the actual proteins found and their possible function, the described approach works in two very complex multicomponent RNPs, namely the spliceosomal complexes H and A. The approach thus has general application in the purification and identification of protein–RNA cross-links in other RNPs. In further studies, we will apply this technique to the investigation of protein–RNA interactions in higher assembly stages of the spliceosome. In combination with the recently developed methods for the purification of spliceosomal complexes (Hartmuth et al. 2002; for review, see Jurica and Moore 2002; Makarov et al. 2002) we would be able to first characterize a protein–RNA cross-link, and then assign it to a distinct spliceosomal complex. This will help to gain further insight into the assembly and function of the spliceosome.
MATERIALS AND METHODS
Preparation of site-specifically modified pre-mRNA
Pre-mRNA was prepared by PCR amplification of pMinx (Zillmann et al. 1988). The sense primer comprised a T7 promoter (GGGGTACCTAATACGACTCACTATAGGGAGACGGAATTCG AGCTCGCCC; all DNA oligonucleotides were supplied by MWG Biotech) and the antisense primer contained a BamHI cleavage site (CGCGGATCCCCACTGGAAAGACC). BamHI-restricted PCR products were transcribed in vitro by using recombinant T7 polymerase. Transcription reactions (500 μL) contained 100 nM Minx DNA, 40 mM Tris-HCl (pH 8.1), 34.5 mM MgCl2, 5 mM DTT, 0.01% Triton X-100, 0.1 mg/mL BSA, 1 mM GTP, 5 mM m7GpppG (Kedar s.c.), 7.5 mM each of CTP, UTP, and ATP, and 6-μL recombinant His-tagged T7 polymerase (4.4 mg/mL). The reaction mixture was incubated for 16 h at 37°C. The DNA template was then digested with 20 U RQ1 DNase (1 U/μL; Promega) for 1 h at 37°C.
Gel-purified pre-mRNA transcripts were cleaved with RNA-cleaving DNA enzymes (Santoro and Joyce 1997). For generation of the 5′ fragment (nt 1–48), enzyme type 10–23 was used to cleave between nt 48 and 49 (CGTTCGGAGGGGCTAGCTACA ACGACGACGGGTTTC), and enzyme type 8–17 for generation of the 3′ fragment (nt 70–223), which cleaves between nt 69 and 70 (AGTTCTACATGTCCGAGCCGGACGAAGGCTCTTACC). The hybridization temperature with the RNA was ~30–34°C for the base-paired regions on both sides of the loop of the DNA enzyme (cf. Fig. 1A, ▶top). Reactions were performed essentially as described (Santoro and Joyce 1998). Briefly, up to 2 nmole of pre-mRNA was mixed with twice the amount of both DNA enzymes, denatured for 1 min at 96°C, and cooled to room temperature. Then, 2 mM MnCl2, 25 mM N-(2-hydroxyethyl)piperazine-N`-3-propanesulfonic acid (EPPS; pH 7.5), 75 mM NaCl, and 0.05% SDS were added to give a final volume of 500 μL, and the mixture was incubated for 12–24 h at 37°C, allowing 90%–100% cleavage efficiency. The 5′ and 3′ fragments were purified by gel electrophoresis. For preparation of site-specifically modified pre-mRNAs, a synthetic oligonucleotide ([pS+9B]-nt21, derived from the Minx sequence [(nt 49–69: CCUCCGAACGGUAAGAG(dC)-pS-C(B-dT)A, the exon sequence is shown in bold and (d) deoxy, (pS) phosphorothioate, (B-dT) biotin; RNA Tec] was used.
Preparation of [pS+9B]-pre-mRNA
The analytical-scale three-piece ligation requires dephosphorylation of the 5′ fragment, labeling of the 3′ fragment, and phosphorylation of the oligonucleotide (see Results and Fig. 1 ▶). (1) A total of 180 pmole of the 5′ fragment was dephosphorylated by the method of Cameron and Uhlenbeck (1977) in a 20 μL reaction with 0.4 mM ATP, 40 U RNasin (Promega), and 40 U T4-polynucleotide kinase (PNK; Promega) in the buffer provided by the manufacturer for 75 min at 37°C. (2) A total of 60 pmole of the 3′ fragment was 5′ phosphorylated with 15 pmole [γ-32P]ATP (5000 Ci/mmole; Amersham) using 40 U of PNK in a total volume of 20 μL. (3) Phosphorylation of the 180 pmole synthetic oligomer [pS+9B]-nt21 was performed as in 2, above, except that 2 mM ATP was used. Reaction mixtures in 2 and 3 were incubated for 45 min at 37°C, 2 μL of 20 mM-unlabeled ATP was then added, and incubation was continued for another 30 min. In all preparations, the final step was inactivation of PNK by incubation for 15 min at 65°C, followed by precipitation with 2.7 M NH4OAc and 3 vol of ethanol, pooling, and repeated precipitation. A total of 120 pmole of DNA splint oligomer (GCTTGGGCTCGAGGTAACCAGTTC TACATGCTAGGCTCTTACCGTTCGGAGGCCGACGGGTTTC CGATCCAAG) was added before the second precipitation. The pellet containing all fragments was dissolved in 8.4 μL H2O, and 2 μL 10× T4 DNA ligase buffer [500 mM Tris-HCl (pH 7.5), 100 mM MgCl2, 100 mM DTT, 10 mM ATP, 250 μg/mL BSA], heated to 96°C, and then cooled to 16°C in a heating block, over 20 min. After hybridization, 0.7 μL of RNasin (40 U/μL), 0.7 μL BSA (5 mg/mL) 0.5 μL ATP (20 mM), and 6 μL T4 DNA Ligase (2 × 106 U/mL, NEB) was added and incubated for 16 h at 16°C. After gel purification, the amount of ligated product was quantified by measuring the absorption at 260 nm (18 pmole yield).
Preparation of [pS+9B]-pre-mRNA-S
Large-scale two-piece ligation (see Fig. 1 ▶) of [pS+9B]-nt21 and 3′ fragment was carried out as described above, except that a threefold volume of all reagents was used. (1) A total of 540 pmole of [pS+9B]-nt21 was phosphorylated with 120 U PNK. (2) A total of 540 pmole of 3′ fragment was labeled with 5 pmole [γ-32P]ATP and 12 U PNK. For the hybridization/ligation-reaction, 510 pmole DNA splint oligonucleotide was used. After gel purification, the amount of ligated product was quantified by measuring the absorption at 260 nm (380 pmole yield).
For modification with the photoaffinity cross-linker 6 mM p-azidophenacyl bromide (APB; Fluka), 80% DMSO in 14 mM HEPES-NH4OAc (pH 8.5) was used to modify 10–200 pmole RNA in a reaction volume of 50–750 μL. The reaction mixture was incubated for 45 min at 42°C. Excess APB was removed by two phenol/chloroform extractions, followed by chloroform extraction and precipitation of the RNA (Konarska 1999).
Assembly of spliceosomal complexes and glycerol gradient centrifugation
For analytical purposes, spliceosomal complexes were assembled on [APB+9B]-pre-mRNA in a 100 μL reaction containing 35% nuclear extract (prepared according to Dignam et al. 1983), 1.3 mM ATP, 27 mM creatine phosphate, and 2.4 mM MgCl2. Incubation for assembly of complexes H and A was performed for 10 min on ice (H) or at 30°C (A). Reactions were then supplemented with 0.125 mg/mL heparin sulphate and applied to a 1.5 mL linear 10%–30% glycerol gradient containing 100 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA, and 20 mM HEPES (pH 7.9). Centrifugation was performed in a Sorvall S55-S rotor at 55,000 rpm for 1:45 h at 4°C. Gradients were fractionated manually into 13 fractions of 110 μL each.
For preparative purposes, complexes H and A were assembled as above, except that 37 nM [APB+9B]-pre-mRNA was incubated for 15 min in a total reaction volume of 660 μL containing 30% nuclear extract on ice (complex H) or 30°C (complex A). For preparative cenrifugation, 600 μL of the reaction product was applied to a linear 10%–30% glycerol gradient (14 mL, buffer as above), and centrifugation was performed in a Centricon TST41.14 rotor at 30,000 rpm for 15.5 h at 4°C. Gradients were fractionated manually into 23 fractions of 500 μL each, and pmoles were quantified by the specific activity of the RNA.
To identify the complexes in the gradient fractions, 10 μL of one fraction was mixed with 2.5 μL 5× loading buffer (2× TBE, 30% glycerol, 0.25% xylene cyanol) and electrophoresed on a 2% native agarose gel (low-melting-point agarose; Invitrogen) in 0.5× TBE with 8 V/cm for 3.5–4 h at room temperature (Das and Reed 1999).
Cross-linking
For analytical cross-linking, 45 μL of one fraction was cross-linked in a single well of a 96-well polystyrene plate, covered with the polystyrene lid provided, for 5 min with a 312 nm germicidal lamp (Herolab) placed about 3 cm from the sample. Preparative cross-linking was performed at 4°C by irradiating 0.25–2 mL in a covered 24-well polystyrene plate for 15 min (other conditions as above). The maximum depth of the sample was 1 cm. Volumes greater than 1 mL in one well were gently mixed every 5 min to improve cross-linking efficiency. Reaction mixtures were then digested by addition of 8.25 U RNase T1 (1000 U/μL; Ambion) in a total volume of 1 mL, and incubated for 2 h at 50°C.
To analyze protein–RNA cross-links, 10 μL (preparative) or 30 μL (analytical) aliquots were electrophoresed on 10% SDS–polyacrylamide gels, and the radioactive bands were detected by autoradiography or with a PhosphorImager (Amersham). Prestained markers (BioRad) were run as molecular-weight standards.
Biotin–streptavidin affinity separation
A total of 30-μL streptavidin-agarose beads (Sigma) were preblocked essentially as described (Lamond and Sproat 1994), except that 0.1% Nonidet P-40 substitute (NP-40S; Sigma), and 100 mM PBS (pH 7.5) was used. A total of 1.5 mL cross-linked and T1-digested samples, 0.1% NP-40S, 0.07% SDS, and 133 μL IPP500 [10 mM Tris-HCl (pH 8); 500 mM NaCl, 0.1% NP-40S] were loaded successively onto the beads, with a final volume of 2 mL, and allowed to bind by end-over-end rotation for 1.5 h at 4°C. For sample volumes above 1.5 mL, the beads were exposed to successive aliquots of the sample with a single wash with IPP500 between consecutive aliquots. After the biotinylated cross-linked proteins had been coupled to the beads, these were washed four times with a 1:1 mixture of 1% SDS and IPP750 (see above, except that 750 mM NaCl was used), and then four times with IPP750, each for 1 min. Elution from the beads was performed with urea and SDS (Swack et al. 1978) with 3× 50 μL aliquots of loading-elution buffer [400 mM urea, 2% SDS, and 30 mM Tris-HCl (pH 6.8), 0.1 mM EDTA (pH 8), and 0.25% bromophenol blue] in three steps as follows: 5 min at 96°C, followed by 30 min at 50°C, and an additional denaturing step at 96°C for 5 min. The eluate (150 μL) was then loaded onto a 10% SDS–polyacrylamide gel, and stained with silver nitrate (Blum et al. 1987).
Mass spectrometry
Proteins separated by SDS–polyacrylamide gel were analyzed by matrix-assisted laser desorption/ionization MS (MALDI-MS) and identified as described in Hartmuth et al. (2002).
Acknowledgments
We thank Monika Raabe for excellent technical assistance, Heiko Manninga for preparation of T7 RNA-Polymerase, and Paul Woolley for critically reading the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Grant Lu 294/12-1), the Bundesministerium für Bildung und Forschung Grant 031U215B (to R.L.) and the Fonds der Chemischen Industrie.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5175703.
REFERENCES
- Blum, H., Beier, H., and Gross, H.J. 1987. Improved silver staining of plant proteins, RNA and DNA polyacrylamide gels. Electrophoresis 8: 93–99. [Google Scholar]
- Burd, C.G., Swanson, M.S., Görlach, M., and Dreyfuss, G. 1989. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: A diversity of RNA binding proteins is generated by small peptide inserts. Proc. Natl. Acad. Sci. 86: 9788–9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In: The RNA world (eds. R.F. Gesteland et al.), pp. 303–357. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Cameron, V. and Uhlenbeck, O.C. 1977. 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry 16: 5120–5126. [DOI] [PubMed] [Google Scholar]
- Das, R. and Reed, R. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5: 1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, R., Zhou, Z., and Reed, R. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5: 779–787. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Matunis, M.J., Piñol-Roma, S., and Burd, C.G. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62: 289–321. [DOI] [PubMed] [Google Scholar]
- Geselowitz, D.A. and Neumann, R.D. 1995. Quantitation of triple-helix formation using a photo-cross-linkable aryl azide/biotin/oligonucleotide conjugate. Bioconjugate Chem. 6: 502–506. [DOI] [PubMed] [Google Scholar]
- Gish, G. and Eckstein, F. 1988. DNA and RNA sequence determination based on phosphorothioate chemistry. Science 240: 1520–1522. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., Urlaub, H., Vornlocher, H.P., Will, C.L., Gentzel, M., Wilm, M., and Lührmann, R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 99: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S. and Moore, M.J. 2002. Capturing splicing complexes to study structure and mechanism. Methods 28: 336–345. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12: 5–14. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M. 1999. Site-specific derivatization of RNA with photocrosslinkable groups. Methods 18: 22–28. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Maniatis, T., Ruskin, B., and Green, M.R. 1984. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36: 993–1005. [DOI] [PubMed] [Google Scholar]
- Krecic, A.M. and Swanson, M.S. 1999. hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell Biol. 11: 363–371. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I. and Sproat, B.S. 1994. Isolation and characterization of ribonucleoprotein complexes. In: RNA processing: A practical approach. (eds. D. Rickwood and B.D. Hames), pp. 103–140. Oxford University Press, Oxford, UK.
- MacMillan, A.M., Query, C.C., Allerson, C.R., Chen, S., Verdine, G.L., and Sharp, P.A. 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 8: 3008–3020. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Lührmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., Munroe, S.H., Cáceres, J.F., and Krainer, A.R. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13: 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, H., Turck, C.W., Nikolic, J.M., and Black, D.L. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes & Dev. 11: 1023–1036. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. and Sharp, P.A. 1992. Site-specific modification of pre-mRNA: the 2`-hydroxyl groups at the splice sites. Science 256: 992–997. [DOI] [PubMed] [Google Scholar]
- Mount, S.M., Pettersson, I., Hinterberger, M., Karmas, A., and Steitz, J.A. 1983. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33: 509–518. [DOI] [PubMed] [Google Scholar]
- Reed, R. and Chiara, M.D. 1999. Identification of RNA-protein contacts within functional ribonucleoprotein complexes by RNA site-specific labeling and UV crosslinking. Methods 18: 3–12. [DOI] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G.F. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37: 13330–13342. [DOI] [PubMed] [Google Scholar]
- Sha, M., Levy, T., Kois, P., and Konarska, M.M. 1998. Probing of the spliceosome with site-specifically derivatized 5′ splice site RNA oligonucleotides. RNA 4: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel, C.W., Admon, A., and Rio, D.C. 1995. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes & Dev. 9: 269–283. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Kanaar, R., and Rio, D.C. 1994. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes & Dev. 8: 1713–1725. [DOI] [PubMed] [Google Scholar]
- Siomi, H., Matunis, M.J., Michael, W.M., and Dreyfuss, G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swack, J.A., Zander, G.L., and Utter, M.F. 1978. Use of avidin-sepharose to isolate and idenify biotin polypeptides from crude extracts. Anal. Biochem. 87: 114–126. [DOI] [PubMed] [Google Scholar]
- Wassarman, D.A. and Steitz, J.A. 1992. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 257: 1918–1925. [DOI] [PubMed] [Google Scholar]
- Zillmann, M., Zapp, M.L., and Berget, S.M. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 8: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]