Abstract
Expanding our previous finding of an adenosine-initiated transcription system, we now demonstrate that either the 5′ site or the N6 site of adenosine nucleotides can be modified extensively without abolishing their ability to initiate transcription under the T7 φ2.5 promoter. Two series of amino derivatives of adenosine nucleotides were synthesized. Fluorescein and biotin groups were coupled to AMP derivatives through linkers of different sizes and hydrophobicities. Both fluorescein- and biotin-conjugated (at either the 5′ or N6 site) adenosine nucleotides can act as efficient transcription initiators, producing fluorescein- and biotin-labeled RNA at the specific 5′ end by a one-step transcription procedure, eliminating posttranscriptional modification. Furthermore, N6-modified adenosine derivative-initiated transcription synthesizes 5′ end modified RNA with a free phosphate group, providing the possibility for further derivatization. The current finding makes easily available a variety of site-specifically functionalized RNA, which may be used in nucleic acid detection, RNA structural and functional investigation, and generation and isolation of novel functional RNA.
Keywords: RNA synthesis, fluorescent labeling, biotin labeling, site-specific modification, in vitro transcription
INTRODUCTION
Efficient in vitro transcription methods using T3, T7, and SP6 RNA polymerases (Krieg and Melton 1987; Milligan et al. 1987; Pokrovskaya and Gurevich 1994) have been widely used to produce RNA for a variety of applications. Commonly used in vitro transcription systems all use guanosine and its nucleotides for transcription initiation (Pokrovskaya and Gurevich 1994). We previously developed an adenosine derivative-initiated transcription system using the T7 φ2.5 promoter (Huang et al. 2000; Huang 2003). Three common coenzymes (CoA, NAD, and FAD) can serve as efficient transcription initiators to produce 5′ coenzyme-linked RNA (Huang 2003). To further explore the utility of this adenosine derivative-initiated transcription system, we wished to extend the investigation to include adenine-base modification tolerance on transcription initiation. It is shown here that either 5′- or N6-modification of adenosine may be accepted by T7 RNA polymerase, whereas 8-modification of adenosine abolishes its initiation activity. Therefore, a variety of 5′- and N6-modified derivatives of adenosine nucleotides were chemically synthesized and investigated for their ability to initiate RNA transcription under the T7 φ2.5 promoter. Efficient RNA labeling at the specific 5′ end can be achieved by either one-step transcription initiation or a two-step procedure of transcription and posttranscriptional modification.
RESULTS
The T7 class II promoter initiates transcription with ATP (Huang et al. 2000). A few 5′-modified AMP derivatives (adenosine-containing coenzymes) have been shown to act as transcription initiators for efficient incorporation into the 5′ end of RNA (Huang 2003). In the present investigation, we wished to explore (1) which site(s) of the adenine base can be modified without abolishing its ability to initiate transcription, and (2) what kinds of modifications (modifying group size, linker length, hydrophobicity, etc.) are acceptable at the permissible site(s) on the adenine base and at the 5′ site of AMP. Two series of 5′- and N6-amino derivatives of AMP as well as their fluorescein and biotin conjugates were synthesized. These compounds were then tested for their ability to label the 5′ end of RNA through transcription initiation.
Permissible modification at the N6 site of adenine for transcription initiation
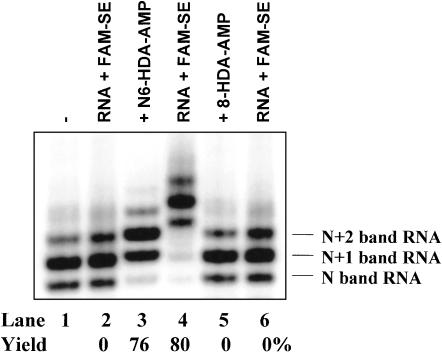
Permissible modification sites on the adenine base for transcription initiation were first investigated before the synthesis of adenosine derivatives. Modification with a 6-aminohexyl group at either the N6 or 8 of AMP was introduced, and its effect was examined for incorporation into the 5′ end of RNA. Figure 1 ▶ shows how modification at the two different sites (N6 and 8) of adenine affects transcription initiation, using a previously defined transcription system containing 35 nucleotides (nt) in DNA template and [α-32P]ATP as the label (Huang 2003). All transcription solutions contained 0.25 mM ATP and 1 mM each of GTP, CTP, and UTP, as well as 4 mM of an AMP derivative as indicated. Under the normal transcription conditions (Fig. 1 ▶, lane 1), the system produces the full-size RNA (N band) of 35 nt, along with the N+1 and N+2 bands of RNA (adding 1 or 2 nontemplated nucleotides at the 3′ end). In the presence of N6-(6-aminohexyl) adenosine 5′-monophosphate (N6-HDA-AMP; lane 3), the three major RNA bands (four bands total) shift upwards (slower) by approximately 1 nt. The shift is caused by the incorporation of N6-HDA-AMP into the 5′ end of RNA. Similar to a coenzyme-initiated transcription (Huang 2003), transcription in the presence of N6-HDA-AMP synthesizes two types of RNA: the 5′ end modified RNA (N6-HDA-AMP-RNA) and normal pppRNA. Each type contains the N, N+1, and N+2 bands of RNA. Due to the upward 1-nt shift by N6-HDA-AMP-RNA, four RNA bands are produced (lane 3). They represent (from bottom to top) N band pppRNA (very weak), N+1 pppRNA and N band N6-HDA-AMP-RNA, N+2 pppRNA and N+1 band N6-HDA-AMP-RNA, and N+2 band N6-HDA-AMP-RNA. On the other hand, 8-(6-aminohexyl) aminoadenosine 5′-monophosphate (8-HDA-AMP) does not have any detectable effects on transcription (lane 5), indicating that 8-HDA-AMP can neither initiate nor inhibit transcription. Total RNA yields remain constant within the error of the experiments (typically ±20%) in the presence or absence of either modified adenosine derivative.
FIGURE 1.
Effects of N6- or 8-modification of adenosine nucleotides on transcription initiation under the T7 φ2.5 promoter. RNA was prepared in the absence (lane 1) or presence of 4 mM of either N6-HDA-AMP (lane 3) or 8-HDA-AMP (lane 5). Transcription solutions contained 0.25 mM ATP and 1 mM each of GTP, CTP, and UTP. In addition, ~0.1 μM of [α-32P]ATP was included to label RNA products for visualization and quantitation. Samples were run to single-nucleotide resolution on an 8% denaturing polyacrylamide gel. Normal transcription produces three RNA bands (lane 1). There are N, N+1, and N+2 bands of RNA. After transcription, half of each RNA preparation was reacted with FAM-SE (lanes 2,4,6, 100 mM FAM-SE in 0.5 M NaHCO3, 30 min at room temperature). Amino- and fluorescein-RNA yields are indicated (lanes 1–6). Compared with normal pppRNA (lane 1), the amino-RNA (lane 3) and FAM-labeled RNA (lane 4) migrate with slower rates, shifting upwards by roughly 1 and 2.4 nt, respectively.
To confirm N6-HDA-AMP incorporation into RNA, all three RNA samples were treated with 5(6)-carboxyfluorescein N-hydroxysuccinimide ester (FAM-SE) and loaded onto the same gel for electrophoresis (Fig. 1 ▶, lanes 2,4,6). If the amino derivative of AMP (either N6-HDA-AMP or 8-HDA-AMP) can act as a transcription initiator, the resulting RNA would bear a free amino group at the 5′ end, which can be labeled by FAM-SE and would lead to further migration retardation in the gel. As can be seen from Figure 1 ▶, reaction of N6-HDA-AMP-initiated RNA with FAM-SE produces RNA products with slower migration in the gel by 1–2 nt relative to N6-HDA-AMP-RNA (cf. lanes 3 and 4) and by 2–3 nt relative to normal RNA (cf. lanes 1 and 4). After individual RNA band quantitation by phosphorimaging, the transcription initiation efficiency of N6-HDA-AMP can be calculated by comparing the relative band intensity of lanes 3 and 4 with lane 1. Excellent transcription initiation efficiency (76%–80%) can be achieved with N6-HDA-AMP. However, treatment of FAM-SE with RNA transcribed under either the normal conditions (lane 1) or in the presence of 8-HDA-AMP (lane 5) does not result in slower-migrating RNA products (lanes 2,6), further demonstrating the inability of 8-HDA-AMP to initiate transcription. Therefore, the N6 position of adenine may be modified, and the resulting adenosine derivatives maintain transcription initiation capacity. On the other hand, an unmodified 8-position of adenine appears to be essential for adenosine nucleotides to act as transcription initiators.
Synthesis of amino derivatives of adenosine
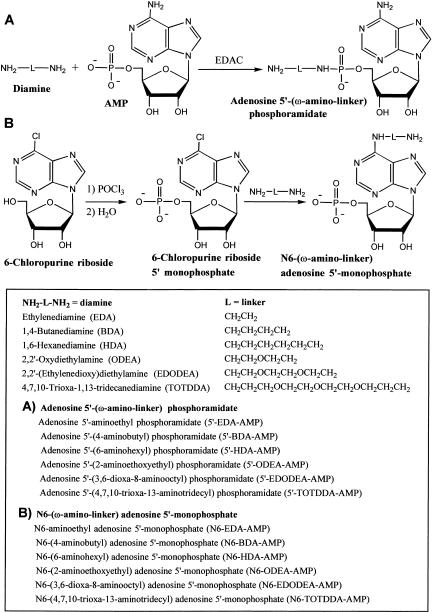
From the above experiment (Fig. 1 ▶) and our previous results (Huang 2003), either 5′- or N6-modification on adenosine is acceptable for transcription initiation under the T7 class II promoter. To demonstrate the utility of modification tolerance at these two sites, it is desirable to label RNA with fluorophores and biotin at the 5′ end. We synthesized two series (each consisting of six different linkers) of 5′- and 6-amino derivatives of adenosine nucleotides (Fig. 2 ▶) to optimize the transcription initiation efficiency and facilitate broad applications of this transcription system. The linkers in these derivatives differ in both size and hydrophobicity. Synthesis of 5′-amino derivatives of adenosine phosphoramidate followed the standard direct phosphate-amine coupling procedure by the water-soluble carbodiimide, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDAC; Chu et al. 1983). Coupling efficiencies as determined by HPLC are excellent, ranging from 70% to 95% after a 2-h reaction at room temperature. Because all of the adenosine phosphoramidate products are more hydrophobic than the other components in the reaction mixture (AMP, diamines, urea derivatives), product purification can be achieved by simply loading the reaction mixture onto a reverse-phase HPLC column (equilibrated in 100% water), followed by elution with 30% MeOH.
FIGURE 2.
Synthetic scheme for amino derivatives of adenosine. (A) Synthesis of 5′ amino derivatives of adenosine phosphoramidate by EDAC-assisted direct coupling of diamines with AMP. (B) Preparation of N6-amino derivatives of AMP from 6-chloropurine riboside. Chlorophosphorylation of 6-chloropurine riboside followed by hydrolysis yields 6-chloropurine riboside 5′-monophosphate. Displacement of the 6-chloride by diamines produces the desired products. The linkers vary in length (3–14 bonds) and hydrophobicity. Abbreviations for diamines and their adenosine derivatives are in parentheses.
For the synthesis of the N6-derivative series, 6-chloropurine riboside 5′-monophosphate was first prepared by phosphorylation of the 5′ hydroxyl of 6-chloropurine riboside by phosphorus oxychloride. Hydrolysis produced the intermediate 6-chloropurine riboside 5′-monophosphate. Direct chloride displacement of 6-chloropurine riboside 5′-monophosphate by diamines (Brodelius et al. 1974) led to high yields of N6-amino derivatives of AMP (60%–90%). Product purification was based on differing hydrophobicities, and was easily achieved by reverse-phase HPLC.
Synthesis of 5′- and N6-fluorescein and biotin derivatives of adenosine
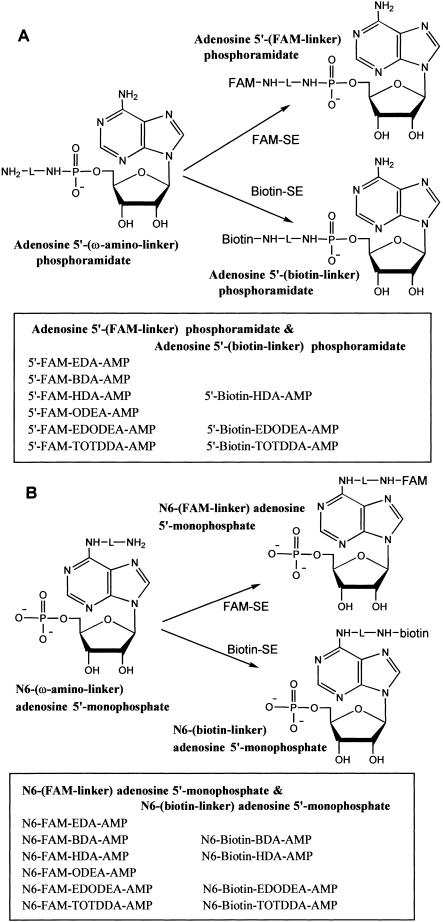
Fluorescein coupling to all above synthesized amino-modified adenosine derivatives was achieved by reaction with FAM-SE (1:1 ratio), shown in Figure 3A ▶ (5′-fluorescein coupling) and Figure 3B ▶ (for N6-fluorescein coupling). HPLC analysis of the fluorescein coupling reactions gave 70%–90% yields of fluorescein products. Seven biotinyl compounds (Fig. 3 ▶) of the 5′- and N6-amino derivatives of AMP were synthesized by coupling with biotin N-hydroxysuccinimide ester (biotin-SE) with a 2:1 ratio of amine:biotin-SE. Near quantitative conversion of amino derivatives to their corresponding biotin derivatives was achieved. Purification of the fluorescein- and biotin-linked adenosine nucleotides was performed by reverse-phase HPLC. To confirm correct product formation, two fluorescein-coupled samples (5′-FAM-HDA-AMP and N6-FAM-BDA-AMP) from each series were analyzed by mass spectrometry (MS): 5′-FAM-HDA-AMP, C37H38N7O12P, calc. MW 803.7, found MH+ 804.9; N6-FAM-BDA-AMP, C35H33N6O13P, calc. MW 776.6, found MH+ 777.8.
FIGURE 3.
Synthesis of fluorescein and biotin derivatives of AMP by reaction of amino derivatives of AMP with FAM-SE and biotin-SE. (A) Preparation of adenosine 5′-fluorescein- and 5′-biotin-phosphoramidates, and (B) synthesis of N6-fluorescein and N6-biotin derivatives of AMP.
Incorporation of 5′- and N6-amino derivatives into RNA by in vitro transcription
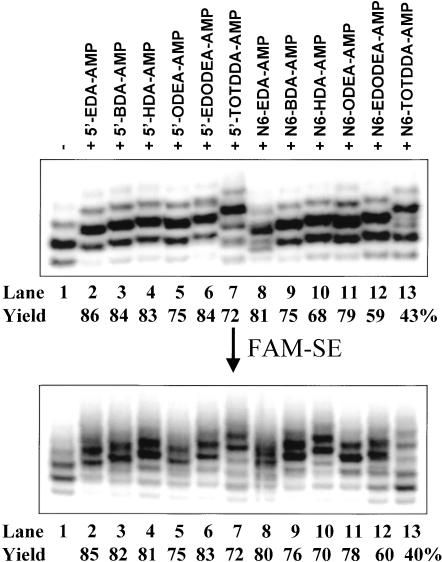
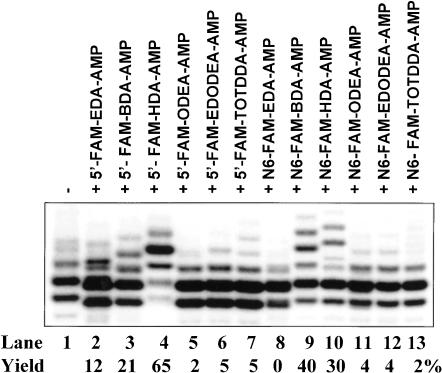
All of the above synthesized amino derivatives of adenosine nucleotide were tested for their ability to initiate transcription under the T7 φ2.5 promoter. As shown in Figure 4 ▶ (top panel), these 12 amino derivatives (containing either 5′- or N6-modification) can all act as efficient transcription initiators, with yields ranging from 43% ± 5% to 86% ± 5% (concentration ratio of 0.25:4 mM of ATP:amino derivative in transcription solution). Neither the linker length nor hydrophobicity appears to significantly affect the transcription initiation efficiency, although hydrophilic and the longest TOTDDA (14 bonds) linker seems to lower the initiation efficiency. In addition, modification at either the 5′ or N6 site has little effect on transcription initiation (with the exception of N6-TOTDDA modification). To confirm the presence of a free amino group and incorporation yields, the RNA samples shown in the top panel of Figure 4 ▶ were reacted with FAM-SE for fluorescein derivatization via the free amino group. As can be seen from the bottom panel of Figure 4 ▶, all of the samples (lanes 2–13) can be fluorescein-modified except the first lane (unmodified RNA). The results from the two gels are in excellent agreement (within the experimental error of ± 5%) to show that (1) all of the 12 amino derivatives of adenosine nucleotide can act as transcription initiators under the T7 φ2.5 promoter, producing free amino-bearing RNAs at the 5′ end; and (2) they are all good-to-excellent transcription initiators, yielding 43%–86% of 5′ amino-modified RNA.
FIGURE 4.
Transcription initiation with 5′- and N6-amino derivatives of adenosine nucleotides under the T7 φ2.5 promoter (top) and their posttranscriptional derivatization with fluorescein (bottom). RNA samples were run to single-nucleotide resolution on an 8% denaturing PAGE and visualized by phosphorimaging. Top panel: Transcription in the presence (4 mM) of one of the 12 amino derivatives produces high yields of 5′ end amino-modified RNAs, with migration rates ranging from slightly below that of the N+1 band to that of the N+2 band of unmodified RNA, depending on the linker size. Bottom panel: Coupling of fluorescein with 5′ end amino-modified RNAs further slows down the RNA migration in the gel by approximately 1 nt (i.e., with migration rates varying from slightly below that of the N+2 band to that of the N+3 band of unmodified RNA). Both amino-RNA yields and fluorescein-labeled RNA yields are indicated under the corresponding gels. The yields of fluorescein-linked RNA agree well with the results of amino-RNA in the top panel.
Incorporation of 5′- and N6-fluorescein and biotin derivatives into RNA
After demonstrating high initiation efficiency of 5′- and N6-amino-modified adenosine derivatives, we investigated fluorescein- and biotin-linked adenosine nucleotides for their incorporation into the first nucleotide of RNA transcripts. If successful, this approach would provide a one-step procedure for specific site (+1) labeling of RNA by fluorophores and biotin. Figure 5 ▶ shows transcription initiation by various fluorescein-linked adenosine nucleotides. Although all of the 12 amino derivatives of adenosine nucleotides are efficient transcription initiators (Fig. 4 ▶), further derivatization of these amino-modified adenosine nucleotides with fluorescein substantially changes their ability to initiate transcription. Both the linker size and hydrophobicity can have significant effects. If the linker is too short (three bonds with a CH2CH2 unit), initiation efficiency is very low (Fig. 5 ▶, lanes 2,8). Long linkers (EDODEA and TOTDDA, nine and 14 bonds, respectively) also lead to poor transcription initiation (lanes 6,7,12,13). However, good to excellent transcription initiators (lanes 3,4,9,10) are produced from middle-sized linkers (BDA and HDA, five and seven bonds). On the other hand, ODEA as a linker (six bonds) results in very poor transcription initiation (lanes 5,11). Therefore, for fluorescein-linked adenosine nucleotides, both the linker size and hydrophobicity play an important role in determining initiation efficiency. Different modification sites (5′ versus N6) on adenosine nucleotide also have some effect. The three best fluorescein-containing initiators are 5′-FAM-HDA-AMP, N6-FAM-BDA-AMP, and N6-FAM-HDA-AMP; the other fluorescein derivatives of AMP are fair to poor transcription initiators. In general, modification at the 5′ site has less negative effect on transcription initiation compared to modification at the N6 site. Transcription initiation by fluorescein-AMP derivatives was confirmed by nuclease P1-digestion of the resulting RNA (gel-purified) followed by MS analysis. From RNAs prepared with N6-FAM-BDA-AMP (MW 776.6) and 5′-FAM-HDA-AMP (MW 803.7) as the initiators, MH+ peaks of 777.8 and 804.9, respectively, were found.
FIGURE 5.
Fluorescein-linked adenosine nucleotide-initiated transcription. RNA products were resolved by 8% denaturing PAGE and quantitated by phosphorimaging. Transcription in the absence (lane 1) or presence (2 mM) of fluorescein derivatives (5-isomers) of adenosine nucleotides (lanes 2–13). Transcription initiation efficiency varies from 0% to 65% (indicated under the gel, depending on the linker size and hydrophobicity) under the transcription conditions.
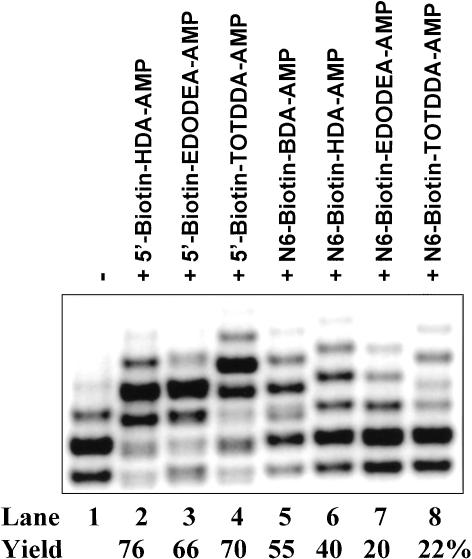
Further demonstration of functional adenosine derivative-initiated transcription was performed with biotin derivatives. Based on the results shown in Figure 5 ▶ and considering potential applications of biotinylated RNA, only seven biotin derivatives of adenosine nucleotides were synthesized. These biotin derivatives may have either high transcription initiation efficiency (based on the results of BDA and HDA derivatives in Fig. 5 ▶) or favorable properties such as long linker and hydrophilicity (EDODEA and TOTDDA). As shown in Figure 6 ▶, biotin-modified adenosine nucleotides are in general better transcription initiators than their corresponding fluorescein derivatives. All seven biotinylated derivatives are able to initiate transcription with good to excellent efficiency (from 20%–76%). Unlike fluorescein derivatives, the acceptance for long and hydrophilic linkers (EDODEA and TOTDDA) ranges from moderate to excellent (Fig. 6 ▶, lanes 3,4,7,8). As in the case of fluorescein derivatives, biotin-modified adenosine nucleotides show more tolerance towards modification at the 5′ site than the N6 site.
FIGURE 6.
Biotin labeling of RNA through 5′- and N6-biotin-modified adenosine nucleotides under the T7 class II promoter. Transcription was performed with either 0 mM (lane 1) or 4 mM biotin derivatives (lanes 2–8). Different biotinylated RNA products were resolved by 8% denaturing PAGE. Biotinylated RNA products comigrated with the N+2 and N+3 bands of unmodified RNA (depending on the linker size).
DISCUSSION
Commonly used in vitro transcription systems all use guanosine and its derivatives for initiation, producing RNA with guanosine and its derivatives at the 5′ end (+1), although there are a few exceptions of other nucleotides such as adenosine and cytidine derivatives to initiate transcription with certain sequences (Nam et al. 1986; Helm et al. 1999). We recently developed a different in vitro transcription system derived from the T7 φ2.5 promoter (Dunn and Studier 1983), from which transcription is initiated by adenosine and its 5′-modified derivatives (Huang et al. 2000; Huang 2003). There is no apparent sequence restriction for the A-initiated transcription system. Our experiments (data not shown) have demonstrated similar transcription efficiencies between a T7 class III promoter (G-initiation) and the φ2.5 promoter (A-initiation).
Many applications require site-specific modifications of RNA. For example, fluorophore labeling of RNA at predefined sites is essential for structural and functional investigation (Walter and Burke 2000; Klostermeier and Millar 2001b; Sastry 2001; Walter 2001; Walter et al. 2001). Specific 5′-end modifications of RNA may be achieved through transcription initiation with 5′-modified guanosine derivatives (Burgin and Pace 1990; Seelig and Jaschke 1999; Sastry 2001; Zhang et al. 2001a,b). However, a few limitations exist for the guanosine-initiated transcription system. (1) Only 5′-modified guanosine derivatives can be used as transcription initiators, rendering the 5′ end of resulting RNA unavailable for other functionalities, such as further derivatization. Although 6-thioguanosine 5′-monophosphate (6sGMP) has been used to initiate transcription, resulting in photo-crosslinkable 6-thioG-linked RNA (Christian et al. 1998; Harris and Christian 1999), it is doubtful that 6sGMP can be used for efficient labeling of RNA at the specific +1 site. (2) The promoter of the guanosine-initiated transcription systems usually requires two or more consecutive guanosines at the first few nucleotide positions of RNA for efficient transcription (13 out of 15 G-initiating promoters from T7 phage produce ≥ two consecutive guanosines at the 5′ end of RNA; Dunn and Studier 1983). Repeated abortive initiation may produce substantial concentrations of oligomers of G. Initiation by both GTP and its oligomers can result in significant 5′ heterogeneity of RNA (Nam et al. 1986; Pleiss et al. 1998). Such 5′ RNA heterogeneity may hinder its applications that require high purity of RNA, such as in structural investigation by NMR and X-ray crystallography. (3) Guanosine-initiated transcription systems restrict the first nucleotide of the RNA to be a guanosine or its derivative, although desired RNA sequences may have other nucleotides (such as A) at their 5′ ends. (4) Compared to other nucleoside derivatives, guanosine derivatives tend to have relatively low solubility in aqueous solutions.
Certain advantages of the A-initiated transcription system become apparent. First, the promoter contains AG as the first two nucleotides for RNA sequences (Huang et al. 2000; Huang 2003). The mechanism of causing 5′ RNA heterogeneity by repeated abortive initiation and re-initiation (Nam et al. 1986; Pleiss et al. 1998) does not exist. As a result, the A-initiated transcription system can produce RNA with a high degree of 5′ RNA homogeneity, which has been confirmed by DNazyme trimming of the 3′ portion of transcribed RNA, leading to one band of RNA at single-nucleotide resolution by PAGE (Huang 2003). A direct comparison between a T7 class III promoter and the φ2.5 promoter has been made from transcription followed by DNazyme cutting, resulting in a major N band RNA plus 5%–8% N+1 and N+2 bands from the class III promoter and a single band RNA from the φ2.5 promoter (data not shown). Second, in combination with guanosine-initiated transcription systems, choices are provided to synthesize RNA with either an A or a G as the first nucleotide. Third, adenosine derivatives usually have much better solubility than guanosine derivatives in aqueous solutions. Adenosine derivatives are not capable of forming G-quartet structures that can lead to aggregation. Therefore, many potential functional groups may be linked to adenosine to make a variety of derivatives.
Based on the results of the present investigation and above discussion, the T7 φ2.5 promoter provides a simple but versatile mechanism for preparing a variety of 5′ end modified RNA, either by a two-step procedure (transcription followed by posttranscriptional modification) or a single-step transcription. The modification site can be at the 5′, N6, or both 5′ and N6 of adenosine and its nucleotides. An assortment of linkers with varying length and hydrophobicity can be introduced between a functional group and adenosine or its nucleotides. Different chemistries, such as carboxyl-amine coupling and haloacetamide-, halide-, or maleimide-thiol coupling, are available for functional group derivatization of adenosine, its nucleotides, and RNA. In particular, N6-modified AMP derivatives constitute a new type of transcription initiators, from which site-specific RNA labeling can be achieved by transcription and RNA ligation. For example, doubly fluorescent-labeled RNA may be prepared by two steps: synthesizing two RNA fragments, each containing either the donor or the acceptor [Donor-L-A-RNAfrag1, pA(-L-Donor)-RNAfrag1, and pA(-L-Acceptor)-RNAfrag2], and then joining the two RNA fragments by RNA/DNA ligase to produce full RNA with both donor and acceptor anchored at specific sites (i.e., Donor-L-A-RNAfrag1-pA(-L-Acceptor)-RNAfrag2, or pA(-L-Donor)-RNAfrag1-pA(-L-Acceptor)-RNAfrag2). For small RNA fragments, site-specific fluorophore labeling can be achieved by chemical synthesis (Earnshaw and Gait 1998; Scaringe et al. 1998). However, for most functional RNAs (from ribozymes to ribosomal RNA), chemical synthesis becomes impractical (due to high costs and low yields). The transcription–ligation method offers a general procedure to label RNA at predefined sites. The resulting RNA may be used to study RNA folding, kinetics, and three-dimensional structure by fluorescence resonance energy transfer (FRET; Tuschl et al. 1994; Clegg 1995; Klostermeier and Millar 2001a).
MATERIALS AND METHODS
Reagents
ATP, UTP, GTP, and CTP were from Boehringer Mannheim. T7 RNA polymerase was purchased from Epicentre Technologies. Taq DNA polymerase, DTT, dATP, dTTP, dGTP, and dCTP were obtained from Promega Life Science Technologies. DNA oligonucleotides were from Integrated DNA Technologies.
The following chemicals were from Sigma-Aldrich-Fluka: ethylenediamine (EDA), Sigma E4379; 1,6-hexanediamine (HDA), Sigma H2381; biotin N-hydroxysuccinimide ester (biotin-SE), Sigma H1759; 5(6)-carboxyfluorescein N-hydroxysuccinimide ester [5(6)-FAM-SE], Sigma C1609; N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDAC), Sigma E7750; 6-chloropurine riboside, Sigma C8276; 8-(6-aminohexyl) aminoadenosine 5′-monophosphate lithium salt (8-HDA-AMP), Sigma A3771; 1,4-butanediamine (BDA), Fluka 32791; 2,2′-oxydiethylamine dihydrochloride (ODEA), Fluka 75961; 2,2′-(ethylenedioxy)diethylamine (EDODEA), Fluka 03739; 4,7,10-trioxa-1,13-tridecanediamine (TOTDDA), Fluka 92892; phosphorus oxychloride, Fluka 79580; triethyl phosphate, Aldrich T6110-7.
Synthesis of 5′-amino derivatives of adenosine phosphoramidate
Direct coupling (Chu et al. 1983) of diamines with AMP by the water-soluble carbodiimide, EDAC, was used to synthesize a series of six 5′-amino derivatives of adenosine phosphoramidate. The reactions and structures are shown in Figure 2A ▶. Briefly, 0.25 μmole of AMP and 5.0 μmole of one of the six diamines (EDA, BDA, HDA, ODEA, EDODEA, and TOTDDA) were dissolved in 1.0 mL water. The solution pH was adjusted to 6.0–6.5 with 6 N HCl. Solid EDAC (hydrochloride, 1.5 μmol) was then added to the AMP-diamine mixture, and the reaction was allowed to proceed for 2 h at room temperature with constant stirring. Product yields were determined by reverse-phase HPLC analysis under the following eluting conditions (column, Alltech Expedite C18, 10 × 4.6 mm and flow rate 1 mL/min): 5% MeOH/95% 20 mM phosphate (pH 7.0) for AMP and EDA reaction; 10% MeOH/90% 20 mM phosphate (pH 7.0) for the reactions of AMP with BDA, HDA, ODEA, and EDODEA; 15% MeOH/85% 20 mM phosphate for the AMP and TOTDDA reaction.
A semipreparative HPLC column (Waters Delta Pak C18, 300 × 7.8 mm) was used to purify all six 5′-amino derivatives of adenosine phosphoramidate. For each product purification, the column was first equilibrated in 100% water. All of the reaction mixture (1–2 mL) was loaded onto the column, followed by washing with approximately 30 mL of water. The pure product was then eluted with 30% MeOH. Collected product solutions were concentrated under vacuum to a final volume of ~0.5 mL. Concentrations of purified 5′-amino derivatives of adenosine phosphoramidate were determined by their absorbance at 260 nm, using a molar extinction coefficient of 15,000 M−1.cm−1.
Synthesis of N6-amino derivatives of adenosine 5′-monophosphate
A series of six N6-amino derivatives of AMP were synthesized by diamine-displacement of 6-chloropurine riboside 5′-monophosphate, which was prepared by phosphorylation of 6-chloropurine riboside with phosphorus oxychloride (Brodelius et al. 1974): To 7.7 mL of triethylphosphate, 3 mmole of 6-chloropurine riboside was suspended at 0°C. Small fractions of phosphorus oxychloride (0.56 mL, 6 mmol) were added over a period of 30 min with constant stirring. The suspension was kept stirring at 0°C for an additional 1.5 h, when the white suspension became clear solution. Water (2 mL) was added to hydrolyze the 5′-phosphoryl chloride to 5′-monophosphate. The solution was then neutralized to pH 5 by NaOH. HPLC analysis (Alltech Expedite C18, 10 × 4.6 mm; 10% MeOH/90% 20 mM phosphate; flow rate, 0.5 mL/min) indicated near complete conversion of 6-chloropurine riboside to 6-chloropurine riboside 5′-monophosphate. The reaction mixture was used directly for the synthesis of the following N6 amino derivatives of AMP.
To 0.1 mmole of the above 6-chloropurine riboside 5′-monophosphate solution, 1 mmole of diamine (EDA, BDA, HDA, ODEA, EDODEA, or TOTDDA in their free amine state) was added. After stirring at room temperature for 30 min, the solution was acidified to pH 3.5 by HCl. HPLC analysis (Alltech Expedite C18, 10 × 4.6 mm; 100%–70% ammonium formate, pH 3.2, 0%–30% MeOH; flow rate, 0.5 mL/min) indicated 60%–90% conversion of 6-chloropurine riboside 5′-monophosphate to N6-amino-derivatives of AMP.
Products were purified by semipreparative HPLC under the following conditions: Waters Delta Pak C18, 300 × 7.8 mm; flow rate, 2 mL/min. The reaction mixture was loaded onto the column pre-equilibrated with 20 mM ammonium formate (pH 3.2). Products were eluted with 30%–50% MeOH. Collected product solutions were concentrated under vacuum and re-injected back onto the semipreparative column that was equilibrated in 100% water. After washing with 20–30 mL of water, the product was eluted with 50% MeOH. After concentration of collected product fractions, concentrations were determined by absorbance at 267 nm (ɛ267 = 18,500 M−1.cm−1) (Brodelius et al. 1974).
Synthesis of 5′- and N6-fluorescein derivatives of AMP
Fluorescein-tagged nucleotides of the above 5′- and N6-amino derivatives of adenosine phosphoramidate and AMP were prepared by reacting the 5′- and N6-amino derivatives with 5(6)-carboxyfluorescein N-hydroxysuccinimide ester (FAM-SE) according to the following procedure: to 5 μL of 0.4 M 5′-amino derivatives of adenosine phosphoramidate or N6-amino derivatives of AMP, 5 μL of 1 M NaHCO3 (pH 8.0) and 10 μL of 0.2 M 5(6)-FAM-SE solution (freshly made in DMSO) were added. The reaction mixture was incubated for 30 min at room temperature. HPLC analysis (Alltech Expedite C18, 10 × 4.6 mm; 30%–40% MeOH and 70%–60% 20 mM KH2PO4, pH 4.6) showed 70%–90% fluorescein conversion to a pair of isomers (5 and 6) of fluorescein-tagged nucleotides.
Purification was performed by HPLC under the following conditions: Reaction solution was loaded in 25%–40% MeOH and 75%–60% KH2PO4. After 15–30 min, the MeOH concentration was increased by 5%. Two fractions containing 5-fluorescein and 5-fluorescein nucleotides were collected and concentrated under vacuum. Their concentrations were determined by absorbance at 492 nm in 20 mM phosphate buffer (pH 7.0), using ɛ492 = 80,000 M−1.cm−1.
Synthesis of 5′- and N6-biotin derivatives of AMP
Seven biotinyl compounds of 5′- and N6-amino derivatives of adenosine phosphoramidate and AMP were prepared by reaction with biotin N-hydroxysuccinimide ester (biotin-SE). To a mixture of 20 μL of 0.4 M amino derivative of AMP or adenosine phosphoramidate and 10 μL of 1 M NaHCO3, 20 μL of freshly made 0.2 M biotin-SE solution (in DMSO) was added. The reaction mixture was allowed to incubate for 30 min at room temperature. HPLC analysis (Alltech Expedite C18, 10 × 4.6 mm; 30% MeOH/70% 20 mM KH2PO4, pH 4.6) showed 40%–60% conversion of amino derivatives of AMP and adenosine phosphoramidate to biotin-tagged nucleotides.
Product purification was achieved by HPLC under the following conditions: all of the reaction mixture was loaded onto a Waters Delta Pak C18, 150 × 3.9 mm, equilibrated in 100% water. After 5–10 min, the mobile phase was changed to 15%–20% MeOH. One major fraction containing the product biotin nucleotide was eluted. Collected biotin nucleotide solutions were concentrated, and concentrations were determined by absorbance at 260 nm, assuming ɛ260 = 15,000 M−1.cm−1.
Incorporation of 5′- and N6-amino, fluorescein, and biotin derivatives into RNA by in vitro transcription
For all RNA transcription experiments, the promoter sequence was derived from the T7 φ2.5 promoter (Dunn and Studier 1983; Huang et al. 2000; Huang 2003). The RNA sequence was a 35-mer used in previous transcription studies (Huang 2003): AGGGA AGCGGGCAUGCGGCCAGCCAUAGCCGAUCA. Transcription was carried out under the following conditions (final concentrations): 1 mM each of UTP, GTP, and CTP, 0.25 mM ATP, 40 mM Tris (pH 8.0), 6 mM MgCl2, 2 mM spermidine, 0.01% Triton X-100, 5 mM DTT, 0.2 μM DNA templates, and 5 units/μL T7 RNA polymerase. In addition, different concentrations of a transcription initiator (5′-amino derivatives of adenosine phosphoramidate, N6-amino derivatives of AMP, or their fluorescein and biotin derivatives) were added. The radiolabel, α-32P-ATP (NEN Life Science), was also included in transcription solutions to internally label RNA transcripts. All transcription reactions were incubated for 2 h at 37°C before product analysis by 8% denaturing PAGE.
Posttranscriptional fluorescein labeling of 5′-amino-RNA
Fluorescein labeling of 5′-NH2-RNA (prepared above as internally 32P-labeled RNA) was achieved by reaction with FAM-SE in a total volume of 2 μL containing 0.5 M NaHCO3 and 0.1 M FAM-SE (freshly prepared in DMSO). Reactions were incubated for 30 min at room temperature. A gel-loading dye solution (8 μL) was then added, and 1 μL of samples was loaded onto an 8% denaturing gel for analysis.
RNA quantitation
After polyacrylamide gel electrophoresis, the intensity of individual RNA bands was quantitated through phosphorimaging (Molecular Imager, Bio-Rad Laboratories). Different RNA bands were analyzed through profile integration after horizontal background subtraction. The yields of derivatized RNA (5′-amino, fluorescein, and biotin) were calculated according to a previously established procedure (Huang 2003). The relative intensity of pppRNA bands (N, N+1, and N+2 bands) in the absence of an AMP derivative (lane 1 in Figs. 1 ▶, 4 ▶, 5 ▶, 6 ▶) was first calculated. The relative intensity of all bands from a derivatized RNA lane was then quantitated. The change in relative intensity among different RNA bands was used to calculate derivatized RNA product yields.
Acknowledgments
This work was supported by NSF grant MCB9974487 and NASA grant NAG5-10668. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5106403.
REFERENCES
- Brodelius, P., Larsson, P.O., and Mosbach, K. 1974. The synthesis of three AMP-analogues: N6–(6-aminohexyl)-adenosine 5′-monophosphate, N6–(6-aminohexyl)-adenosine 2′,5′-bisphosphate, and N6-(6-aminohexyl)-adenosine 3′,5′-bisphosphate and their application as general ligands in biospecific affinity chromatography. Eur. J. Biochem. 47: 81–89. [DOI] [PubMed] [Google Scholar]
- Burgin, A.B. and Pace, N.R. 1990. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 9: 4111–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, E.L., McPheeters, D.S., and Harris, M.E. 1998. Identification of individual nucleotides in the bacterial ribonuclease P ribozyme adjacent to the pre-tRNA cleavage site by short-range photo-cross-linking. Biochemistry 37: 17618–17628. [DOI] [PubMed] [Google Scholar]
- Chu, B.C., Wahl, G.M., and Orgel, L.E. 1983. Derivatization of unprotected polynucleotides. Nucleic Acids Res. 11: 6513–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg, R.M. 1995. Fluorescence resonance energy transfer. Curr. Opin. Biotechnol. 6: 103–110. [DOI] [PubMed] [Google Scholar]
- Dunn, J.J. and Studier, F.W. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166: 477–535. [DOI] [PubMed] [Google Scholar]
- Earnshaw, J. and Gait, M.J. 1998. Modified oligoribonucleotides as site-specific probes of RNA structure and function. Biopolymers 48: 39–55. [Google Scholar]
- Harris, M.E. and Christian, E.L. 1999. Use of circular permutation and end modification to position photoaffinity probes for analysis of RNA structure. Methods 18: 51–59. [DOI] [PubMed] [Google Scholar]
- Helm, M., Brule, H., Giege, R., and Florentz, C. 1999. More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA 5: 618–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F. 2003. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 31: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F., Bugg, C.W., and Yarus, M. 2000. RNA-Catalyzed CoA, NAD, and FAD synthesis from phosphopantetheine, NMN, and FMN. Biochemistry 39: 15548–15555. [DOI] [PubMed] [Google Scholar]
- Klostermeier, D. and Millar, D.P. 2001a. RNA conformation and folding studied with fluorescence resonance energy transfer. Methods 23: 240–254. [DOI] [PubMed] [Google Scholar]
- Klostermeier, D. and Millar, D.P. 2001b. Time-resolved fluorescence resonance energy transfer: A versatile tool for the analysis of nucleic acids. Biopolymers 61: 159–179. [DOI] [PubMed] [Google Scholar]
- Krieg, P.A. and Melton, D.A. 1987. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 155: 397–415. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, H.G., Loechel, S., and Fried, H.M. 1986. Plasmids allowing transcription of cloned DNA by Salmonella typhimurium phage SP6 RNA polymerase to produce RNAs with authentic 5′-terminal sequences. Gene 46: 57–64. [DOI] [PubMed] [Google Scholar]
- Pleiss, J.A., Derrick, M.L., and Uhlenbeck, O.C. 1998. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 4: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya, I.D. and Gurevich, V.V. 1994. In vitro transcription: Preparative RNA yields in analytical scale reactions. Anal. Biochem. 220: 420–423. [DOI] [PubMed] [Google Scholar]
- Sastry, S. 2001. A fluorescence-based assay for transcription using a novel fluorescent GTP analogue. Biophys. Chem. 91: 191–208. [DOI] [PubMed] [Google Scholar]
- Scaringe, S.A., Wincott, F.E., and Caruthers, M.H. 1998. Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J. Am. Chem. Soc. 120: 11820–11821. [Google Scholar]
- Seelig, B. and Jaschke, A. 1999. Ternary conjugates of guanosine monophosphate as initiator nucleotides for the enzymatic synthesis of 5′-modified RNAs. Bioconjug. Chem. 10: 371–378. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Gohlke, C., Jovin, T.M., Westhof, E., and Eckstein, F. 1994. A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science 266: 785–789. [DOI] [PubMed] [Google Scholar]
- Walter, N.G. 2001. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods 25: 19–30. [DOI] [PubMed] [Google Scholar]
- Walter, N.G. and Burke, J.M. 2000. Fluorescence assays to study structure, dynamics, and function of RNA and RNA-ligand complexes. Methods Enzymol. 317: 409–440. [DOI] [PubMed] [Google Scholar]
- Walter, N.G., Harris, D.A., Pereira, M.J., and Rueda, D. 2001. In the fluorescent spotlight: Global and local conformational changes of small catalytic RNAs. Biopolymers 61: 224–242. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Cui, Z., and Sun, L. 2001a. Synthesis of 5′-deoxy-5′-thioguanosine-5′-monophosphorothioate and its incorporation into RNA 5′-termini. Org. Lett. 3: 275–278. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Sun, L., Cui, Z., Gottlieb, R.L., and Zhang, B. 2001b. 5′-sulfhydryl-modified RNA: Initiator synthesis, in vitro transcription, and enzymatic incorporation. Bioconjug. Chem. 12: 939–948. [DOI] [PubMed] [Google Scholar]