Abstract
The biological functions of RNA–protein complexes are, for the most part, poorly defined. Here, we describe experiments that are aimed at understanding the functional significance of alfalfa mosaic virus RNA–coat protein binding, an interaction that parallels the initiation of viral RNA replication. Peptides representing the RNA-binding domain of the viral coat protein are biologically active in initiating replication and bind to a 39-nt 3′-terminal RNA with a stoichiometry of two peptides: 1 RNA. To begin to understand how RNA–peptide interactions induce RNA conformational changes and initiate replication, the AMV RNA fragment was experimentally manipulated by increasing the interhelical spacing, by interrupting the apparent nucleotide symmetry, and by extending the binding site. In general, both asymmetric and symmetric insertions between two proposed hairpins diminished binding, whereas 5′ and 3′ extensions had minimal effects. Exchanging the positions of the binding site hairpins resulted in only a moderate decrease in peptide binding affinity without changing the hydroxyl radical footprint protection pattern. To assess biological relevance in viral RNA replication, the nucleotide changes were transferred into infectious genomic RNA clones. RNA mutations that disrupted coat protein binding also prevented viral RNA replication without diminishing coat protein mRNA (RNA 4) translation. These results, coupled with the highly conserved nature of the AUGC865–868 sequence, suggest that the distance separating the two proposed hairpins is a critical binding determinant. The data may indicate that the 5′ and 3′ hairpins interact with one of the bound peptides to nucleate the observed RNA conformational changes.
Keywords: RNA–protein interactions, virus, peptide, conformational change, replication
INTRODUCTION
RNA–protein interactions play a key role in the regulation of biological processes, including viral RNA replication and encapsidation. One such important interaction involves the RNAs of alfalfa mosaic virus (AMV) and their binding to the viral coat protein. AMV is a positive-sense RNA plant virus with three genomic RNAs (RNAs 1, 2, and 3) and a subgenomic RNA (RNA 4), each of which is separately encapsidated (Jaspars 1985, 1999; Bol 1999). Genomic RNAs 1 and 2 encode proteins with replicase functions (Quadt et al. 1991). The viral movement protein required for cell-to-cell virus movement is translated from RNA 3, whereas the subgenomic RNA 4, which is transcribed from the negative-sense strand of RNA 3, encodes the viral coat protein (Jaspars 1985). In addition to its role in assembly, the viral coat protein is required to initiate RNA replication through a process referred to as genome activation (Bol et al. 1971; Jaspars 1985). AMV and the ilarviruses are unusual among their close relatives because they require coat protein to initiate RNA replication, and because the 3′ termini of the viral RNAs lack the canonical CCA terminus that is characteristic of transfer RNA-like ends (Giege 1996). We propose that coat protein binding organizes the 3′ ends of the AMV and ilarvirus RNAs for functions in the replication process (G.A. Rocheleau, J.E. Petrillo, B. Kelley-Clarke, S. Laforest, G.W. Martin III, and L. Gehrke, in prep.).
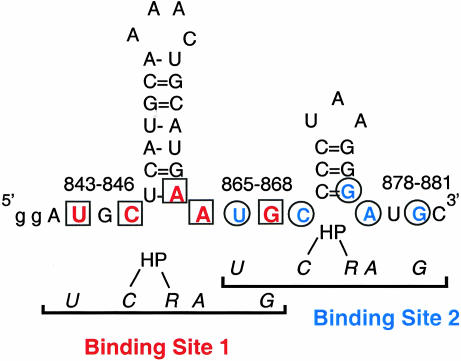
A specific, high-affinity 39-nucleotide AMV coat protein binding site has been identified in the 3′-terminal nucleotides of the AMV RNAs (nucleotides 843–881 in RNA 4; i.e., AMV843–881) through a combination of hydroxyl radical footprinting results, mobility bandshift data, and in vitro genetic selection (Houser-Scott et al. 1994, 1997; Ansel-McKinney and Gehrke 1998). The predicted secondary structure of the 3′-terminal 39 nt (Koper-Zwartoff and Bol 1980; Houwing and Jaspars 1982; Quigley et al. 1984) suggests two hairpins flanked by single-stranded tetranucleotide AUGC sequences (Fig. 1 ▶). The AUGC sequences are highly conserved among AMV and ilarvirus RNAs (Houser-Scott et al. 1994), and in vitro selection experiments reveal that AUGC865–868 is clearly preferred for coat protein binding (Houser-Scott et al. 1997; G.A. Rocheleau, J.E. Petrillo, B. Kelley-Clarke, S. Laforest, G.W. Martin III, and L. Gehrke, in prep.). Chemical modification and ethylation interference data indicate that coat protein interacts with both single-stranded AUGC repeats and the paired nucleotides at the base of the helices, but not with the loop nucleotides (Ansel-McKinney and Gehrke 1998). Nucleotide substitutions or deletions within the AUGC sequences dramatically reduce or eliminate coat protein binding (Houser-Scott et al. 1994; Reusken and Bol 1996; Ansel-McKinney and Gehrke 1998), as do substitutions that are predicted to disrupt the secondary structure of the lower stem loop regions (Reusken and Bol 1996). The pattern of proposed coat protein contacts seems to be arranged symmetrically as two overlapping UGC-hairpin-RAUGC motifs (in which “R” is a purine) that are consistent with the 2:1 peptide to RNA binding stoichiometry (Fig. 1 ▶; Ansel-McKinney and Gehrke 1998). How the 39-nt coat protein binding site physically accommodates two coat protein peptides or two RNA-binding domains from the native coat protein dimer (Kruseman et al. 1971) is not known.
FIGURE 1.
Nucleotides that are important for the coat protein–AMV RNA interaction are arranged in two UGC-hairpin-RAUGC motifs. The data presented in this article are modeled after our earlier publication (Ansel-McKinney and Gehrke 1998), in which possible protein contacts, shown in color, were identified by chemical modification interference analysis. The two UGC-hairpin-RAUGC motifs form binding sites 1 and 2, which overlap at nucleotides 865–868. HP indicates hairpin. The numbers (843–846, etc.) indicate the AUGC sequences at 843–846, 865–868, and 877–881.
The experiments described here support a model stating that both symmetry and interhelical spacing are important determinants of viral coat protein binding and, hence, replication. The role of the spatial arrangement of nucleotide determinants both within and between the two putative binding sites was examined here by constructing variant AMV843–881 RNAs containing single and multiple nucleotide insertions. The majority of insertions both within and between the two sites severely disrupted peptide binding and reduced viral RNA replication in vivo. In contrast, variant AMV843–881 RNAs with multiple nucleotide extensions at the 3′ or 5′ end of the 39-nt fragment had little effect on coat protein binding, whereas a mutant RNA in which the 5′ and 3′ hairpins were exchanged showed only moderately diminished binding affinity. Occupancy of only one of the two protected sites was never observed in hydroxyl radical footprinting experiments, suggesting that two peptides or a native coat protein dimer must bind the RNA to generate a stable conformation. The observation that increased interhelical spacing disrupts RNA–peptide interactions implies that one or both of the bound peptides may bridge the single-stranded AUGC sequence separating the minimal binding site hairpins. The functional significance of the AMV RNA–coat protein interaction and accompanying RNA conformational changes may be to present the 3′ terminus of the RNA as a favorable binding site for the viral replicase.
RESULTS
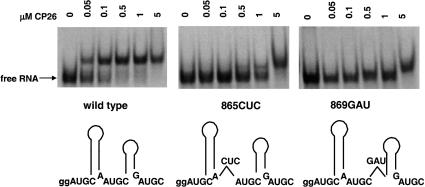
To test the significance of symmetry and interhelical spacing, three-nucleotide insertions were engineered into the UGC-hairpin-RAUGC coat protein binding motifs (Fig. 2 ▶, 865CUC and 869GAU). Although coat protein peptide CP26 bound the wild-type AMV 39-nt RNA with an apparent Kd of 50–100 nM, the corresponding Kd for the two variant RNAs is estimated to be >1 μM (Fig. 2 ▶), suggesting a 10- to 20-fold decrease in binding affinity. Enzymatic structure mapping data demonstrated that the secondary structure of the variant RNAs was not altered significantly compared with the wild type (data not shown). Moreover, in competitive binding experiments, 2.5 μM unlabeled RNAs 865CUC and 869GAU showed only weak competition for labeled wild-type AMV 39-nt RNA, whereas unlabeled wild-type AMV 39-nt RNA fully displaced labeled wild-type AMV 39-nt RNA from the bound form at ≥0.5 μM (data not shown). One interpretation of the evidence presented in Figure 2 ▶ is that disrupting the symmetry of the coat protein binding site significantly reduces coat protein peptide binding. However, an alternate explanation is that nucleotide insertions in the AUGC865–868 region, where the two proposed binding sites overlap (Fig. 1 ▶), is detrimental to high-affinity peptide binding because the hairpins were separated.
FIGURE 2.
Trinucleotide insertions diminish the affinity of the AMV coat protein peptide–RNA interaction. The positions of the nucleotide insertions are indicated schematically at the bottom of the figure, and the electrophoretic mobility shift pattern of the RNA–peptide complex is shown above.
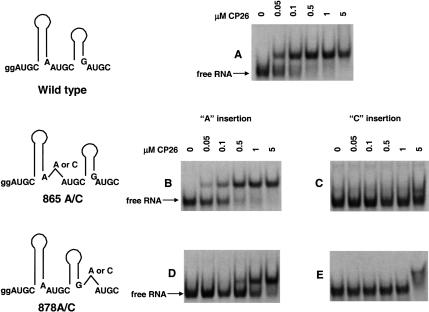
Additional experiments were needed to begin to distinguish between these possibilities. We next asked if single nucleotide insertions at positions 865 and 878 would have a similar effect, this time designing the modifications so that the effects of symmetry and spacing could be distinguished in one case. Adenosine or cytosine insertions at position 865 (865A/C) disrupted the putative symmetry of the binding site 1 (Fig. 1 ▶) and also increased the interhelical distance in the AUGC865–868 overlap region (Fig. 3 ▶). The 878A/C insertions disrupted binding site 2 (Fig. 1 ▶), but did not directly affect the AUGC865–868 overlap region or the interhelical spacing. The results presented in Figure 3A ▶ show that the wild-type RNA bound peptide CP26 with an apparent Kd of ~50 nM. The insertion of an adenosine between nucleotides 864 and 865 (865A) diminished the binding affinity by about twofold to ~100 nM (Fig. 3B ▶), whereas an adenosine inserted between nucleotides 877 and 878 (878A) lowered the affinity ~15-fold to ~750 nM (Fig. 3D ▶). Moreover, a cytosine inserted at the 864/865 (865 C) junction diminished binding by >100-fold (Fig. 3C ▶), as did the same insertion at 877/878 (878C; Fig. 3E ▶). As was the case with the three-nucleotide insertion variants, enzymatic structure mapping data demonstrated that the secondary structure of mutants 865C and 878C was not significantly different from that of the wild type. Moreover, in competitive binding assays, 2.5 μM unlabeled 865C and 878C RNA showed only weak displacement of labeled wild-type AMV 39-nt RNA from the bound form (data not shown). The results of these 878A/C experiments show that peptide binding was diminished by interrupting the putative symmetry of proposed binding sites 1 and 2 and by disrupting the interhelical spacing at 865. Clearly, cytosine insertions were much more detrimental to binding than was the adenosine insertion. The adenosine insertion may have had a smaller effect because it was placed between two purines, where (perhaps unlike the cytosine) it was able to partially mimic the wild-type binding environment. The data are consistent with a hypothesis stating that symmetry is a critical binding determinant. However, the insertions at position 865 altered both symmetry and interhelical distance. Therefore, the results do not rule out the possibility that the number of nucleotides separating the two hairpins (Fig. 1 ▶), which is constant in AMV and the ilarviruses (Houser-Scott et al. 1994), is also important.
FIGURE 3.
The effects of single cytosine and adenosine insertions on peptide binding. The positions of the nucleotide insertions are indicated schematically at the left of the figure, and the electrophoretic mobility shift patterns are shown at the right. (A) Wild-type RNA. (B) 865A RNA; adenosine inserted between 864 and 865. (C) 865C RNA; cytosine inserted between nucleotides 864 and 865. (D) 878A RNA; adenosine inserted between nucleotides 877 and 878. (E) 878C RNA; cytosine inserted between nucleotides 877 and 878.
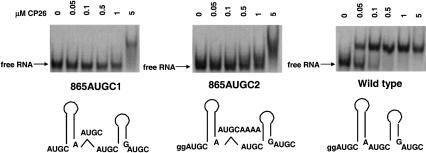
Two additional constructs were tested to analyze the importance of interhelical spacing, using a design that did not disrupt the symmetry of individual binding sites. First, by duplicating AUGC 865–868, two non-overlapping UGC-hairpin-RAUGC motifs were generated (Fig. 4 ▶, 865AUGC1). In a second construct, we added an AUGC but also separated the AUGC repeats with a quartet of adenosines (Fig. 4 ▶, center). Here, the UGC-hairpin-RAUGC motifs are non-overlapping and also separated by an additional four nucleotides. The results of the mobility shift experiments shown in Figure 4 ▶ indicate that the binding affinity of both modified RNAs was decreased by ~20-fold to a Kd of >1 μM. Moreover, at 5 μM, the peptide–RNA interactions became nonspecific (as indicated by the smeared shift pattern). Again, as in control experiments (data not shown), the secondary structure was indistinguishable from wild type by enzymatic structure mapping, and the variant RNAs were poor competitors in competitive binding experiments. The combined data presented in Figures 2 ▶–4 ▶ suggest that both the interhelical spacing and binding site symmetry define high-affinity coat protein peptide binding.
FIGURE 4.
Inserted AUGC nucleotides maintain the symmetry of the proposed binding sites while increasing the interhelical distance. Schematic representations shown at the bottom of the figure show the position of the nucleotide insertions. The corresponding electrophoretic mobility bandshift analysis of each of the labeled RNAs binding to coat protein peptide CP26 is shown above the schematics.
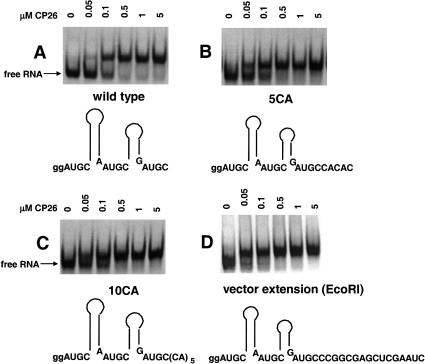
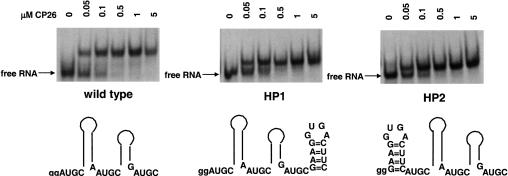
To acquire additional evidence, we next examined the effects of 5′ and 3′ nucleotide extensions on peptide binding. First, we tested the effects of 3′-terminal five- and 10-nt extensions that were designed to be single-stranded. The results (Fig. 5B–D ▶) show that the 5-nt (5CA), 10-nt (10CA), and 16-nt (vector extension) extensions had little effect on peptide binding affinity. Second, we asked how an added hairpin at the 3′ (HP1) or 5′ (HP2) ends of the RNA fragment would affect binding. The results shown in Figure 6 ▶ indicate, again, that neither modification significantly diminished the affinity of the peptide for the viral RNA fragment. These data suggest that the addition of small numbers of nucleotides outside of the binding domain does not affect the RNA–peptide interaction significantly. In contrast, mutations that increased interhelical spacing or disrupted binding site symmetry were detrimental to peptide binding (Figs. 2 ▶–4 ▶).
FIGURE 5.
Analysis of peptide binding using RNAs containing single-stranded 3′ extensions. The figure shows schematic representations of the RNA constructs along with the electrophoretic mobility bandshift data.
FIGURE 6.
Analysis of peptide binding using RNAs containing added 5′ or 3′ hairpins. The figure shows schematic representations of the RNA constructs along with the electrophoretic mobility bandshift data.
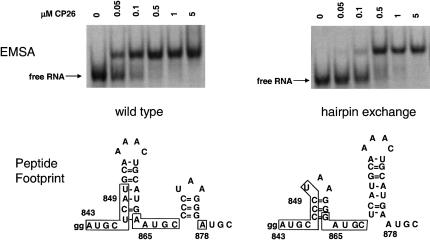
Because of the apparent symmetry formed by the two UGC-hairpin-RAUGC motifs, we predicted that exchanging the relative positions of the two hairpins might not affect peptide binding. Schematic representations of the hairpin exchange RNA and the binding data are shown in Figure 7 ▶, where it can be seen that both the UGC-hairpin-RAUGC sequences and the interhelical spacing are maintained. The mobility shift results suggest that peptides bound the hairpin exchange RNA; however, the affinity was decreased by about fivefold. This decrease in binding affinity is less dramatic than that observed with other variant RNAs (Figs. 2 ▶–4 ▶). Hydroxyl radical footprint methods were used to define the nucleotides that are protected from attack by bound peptide. The boxed nucleotides shown in the lower part of Figure 7 ▶ represent protected nucleotides in the wild-type RNA (left; Ansel-McKinney et al. 1996) and the hairpin exchange RNA (right; gel data not shown). Unexpectedly, the footprint patterns for the wild-type (lower left) and exchange RNA (lower right) fragments are superimposable. The result was unexpected because the lengths of the 5′ hairpins, and both the lengths and sequence composition of the 3′ hairpins are similar among AMV and the related ilarviruses. We therefore anticipated either that the hairpin exchange would preclude peptide binding or that the longer hairpin would be protected in its new, downstream position. Neither of these predictions is consistent with the data. Although the binding affinity for the exchange RNA was diminished about fivefold compared with wild type, the results suggest that two hairpins separated by a single-stranded AUGC represent important determinants for coat protein binding. However, as we demonstrate below, infectious RNA 3 molecules containing the hairpin exchange modification were not active in viral RNA replication.
FIGURE 7.
Peptide binding to the 39-nt hairpin exchange RNA. The two RNAs shown schematically at the bottom were transcribed and tested for peptide binding in a mobility shift assay. The RNA–peptide complexes were also analyzed by hydroxyl radical footprint methods. The nucleotides protected from hydrolysis by bound peptide are enclosed in the boxes.
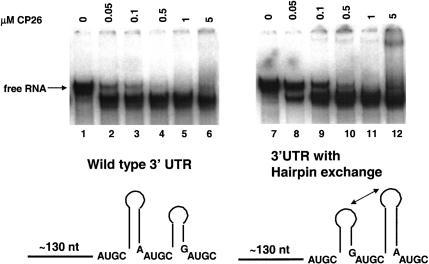
Based on circular dichroism and native gel electrophoresis data (Baer et al. 1994; G.A. Rocheleau, J.E. Petrillo, B. Kelley-Clarke, S. Laforest, G.W. Martin III, and L. Gehrke, in prep.), we have proposed that coat protein binding is accompanied by RNA conformational changes that organize the 3′-terminal structures of the viral RNAs. Evidence for a conformational change can be seen in Figure 8 ▶, where the wild-type 170-nt AMV RNA 4 3′ UTR, upon binding peptide, migrates more rapidly in a non-denaturing gel than does unbound RNA (Fig. 8 ▶, lanes 1–6). We have interpreted this phenomenon as evidence that the RNA structure is being compacted upon peptide binding. Similar results have been reported for the Rev–RRE interaction (Kjems et al. 1992) and for the S15 ribosomal protein–RNA interaction (Batey and Williamson, 1996a, b). We tested the potential effect of the hairpin exchange on the RNA conformational change. The results (Fig. 8 ▶, lanes 7–12) show that in the presence of AMV coat protein peptide CP26, the hairpin exchange RNA migrates rapidly in the native polyacrylamide gel in the same manner as does the wild-type RNA. As was the case with the 39-mer hairpin exchange RNA (Fig. 7 ▶), the 180-nt 3′ UTR hairpin exchange RNA bound peptide with slightly lower affinity (i.e., two- to fivefold) than that of the wild-type RNA. The results are consistent with the hypothesis stating that peptide binding and the RNA conformational change are dependent on the symmetry of determinants found in the UGC-hairpin-RAUGC motifs.
FIGURE 8.
Analysis of RNA conformational changes upon peptide binding to the 170-nt 3′ UTR RNA. (Lanes 1–6) Wild-type 3′ UTR RNA with increasing concentrations of coat protein peptide CP26. (Lanes 7–12) The 170-nt 3′ UTR RNA containing the hairpin exchange.
To determine whether the insertion and deletion mutants affected viral replication, we engineered nucleotide insertions into infectious clones of viral RNA 3. We then mixed the RNA 3 transcripts with wild-type AMV RNAs 1 and 2 (to provide the helicase-methytransferase and RNA-dependent RNA polymerase). This mixture was combined with activator AMV RNA 4 transcripts with truncated 3′ UTR, resulting in a size differential that permitted us to distinguish newly transcribed subgenomic RNA from input activator RNA 4 in Northern blots. Upon translation of the activator RNA 4, coat protein is provided to initiate viral RNA replication in the tobacco cell protoplasts.
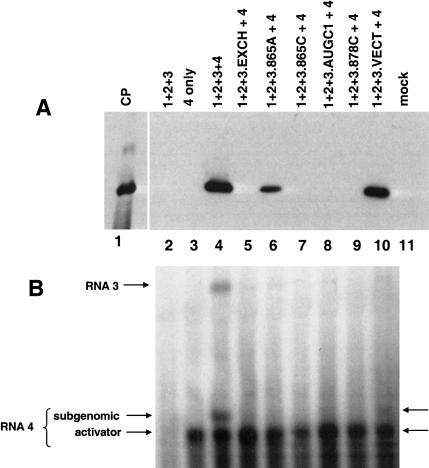
The assay for RNA 3 replication was the production of viral coat protein and subgenomic RNA 4 in transfected tobacco protoplasts (Fig. 9 ▶). In this assay system, viral coat protein and subgenomic RNA 4 are detected only if the viral RNAs are replicating. The genomic RNAs 1–3 are not infectious in the absence of activator coat protein or RNA 4; therefore, no coat protein was detected in this control (lane 2). The results shown in lane 3 demonstrate coat protein translated from the input RNA 4 alone is not detected; that is, coat protein is detected by Western blotting only with viral RNA replication. Coat protein expression and production of subgenomic RNA 4 were observed with a complete inoculum that included viral RNAs 1–4 (Fig. 9A,B ▶, lane 4).
FIGURE 9.
In vivo viral RNA replication assay. Western blot (A) and Northern blot (B) analyses showing the accumulation of AMV coat protein and AMV RNAs 3 and 4 in transfected NT-1 tobacco protoplasts. Details of the replication assay may be found in Materials and Methods. (Lane 1) Viral coat protein standard; (lane 2) cells were transfected with genomic RNA 1–3 only; (lane 3) cells were transfected with viral subgenomic RNA 4 only; (lane 4) complete inoculum where cells were transfected with genomic RNAs 1–3 plus subgenomic RNA 4; (lane 5) cells were transfected with wild-type viral RNAs 1, 2, and 4 and also with genomic RNA 3 carrying the 3′ hairpin exchange construct shown in Fig. 7 ▶; (lane 6) cells were transfected with wild-type viral RNAs 1, 2, and 4 and also with genomic RNA 3 carrying the 865A insertion shown in Figure 3B ▶; (lane 7) cells were transfected with wild-type viral RNAs 1, 2, and 4 along with genomic RNA 3 carrying the 865C mutation shown in Figure 3C ▶; (lane 8) cells were transfected with wild-type viral RNAs 1, 2, and 4 along with genomic RNA 3 carrying the AUGC1 insertion shown in Figure 4 ▶; (lane 9) cells were transfected with wild-type viral RNAs 1, 2, and 4 along with genomic RNA 3 carrying the 878C insertion shown in Figure 3E ▶; (lane 10) cells were transfected with wild-type viral RNAs 1, 2, and 4 along with genomic RNA 3 carrying the 16-nt vector RNA 3′ extension shown in Figure 5D ▶; and (lane 11) cells were mock-transfected; only the Western blot is presented for this sample. (A) An anti-AMV coat protein antibody was used to develop the blot, and the migration of a virion coat protein standard is shown in lane 1. (B) A probe corresponding to the coding region of the viral coat protein was used; therefore, both viral RNAs 3 and 4 (which are coterminal) can be detected (see lane 4). The position of the truncated activator RNA 4 included to initiate replication is indicated (“activator”), as is the position of the newly transcribed subgenomic RNA 4 (“subgenomic”) that was generated during viral RNA replication. In these assays, detection of coat protein by Western blot was a more sensitive indicator of viral RNA replication than was RNA analysis by Northern blot. Viral RNA replication was clearly demonstrated by the accumulation of coat protein (A, lanes 4,6,10), the amount of corresponding subgenomic RNA expressed during replication was low in the samples corresponding to lanes 6 and 10. A faint subgenomic RNA band is present in lane 10, whereas the amount of subgenomic RNA analyzed in lane 6 fell below detection levels.
The results of testing the RNA 3 variant containing the hairpin exchange modification are shown in lanes 5, where neither coat protein nor subgenomic RNA 4 was detected. These results strongly suggest that the hairpin exchange RNA 3 was not replicated. Several of the insertion variants (Figs. 3 ▶–5 ▶) were also tested in the context of infectious RNA 3 constructs to assess their effects on viral RNA replication (Fig. 9A,B ▶, lanes 6–10). Overall, the coat protein–RNA binding data mirrored the detection of viral coat protein and subgenomic RNA in the replication assays. When the 865A mutation was engineered into RNA 3 and transfected along with genomic RNAs 1 and 2 and activator RNA 4, coat protein was detected in the Western blot (Fig. 9 ▶, lane 6A), albeit at a lower level than that observed using the wild-type RNA 3 (Fig. 9 ▶, lane 4A). The corresponding subgenomic RNA 4 level fell below detection in the Northern blot (Fig. 9B ▶, lane 6). Coupled with the binding data (Figure 3 ▶), the results suggest that the 865A mutation diminished, but did not preclude, coat protein binding or viral RNA replication.
Neither coat protein nor subgenomic RNA expression was detected when the 865C, AUGC1, and 878C variants in RNA 3 were tested (Fig. 9 ▶, lanes 7–9). These results correlated with the absence of coat protein binding in vitro to the 39-mer minimal binding site RNA (Figs. 3 ▶, 4 ▶). Results described earlier in this work showed that 3′-terminal extensions of up to 16 nt did not have a significant effect upon coat protein binding in vitro (Fig. 5 ▶). The infectious RNA 3 construct that was extended by 16 nt (Fig. 5D ▶) showed activity in viral RNA replication, but the levels of accumulated coat protein and subgenomic RNA were less than that found with the wild-type RNAs (Fig. 9A,B ▶, cf. lanes 4 and 10). In this case, the strong coat protein binding (Fig. 5D ▶) was not directly reflected in the replication potential. These data suggest that the 3′-terminal extension did not impede coat protein binding significantly but may have affected a subsequent step(s) in the replication reactions.
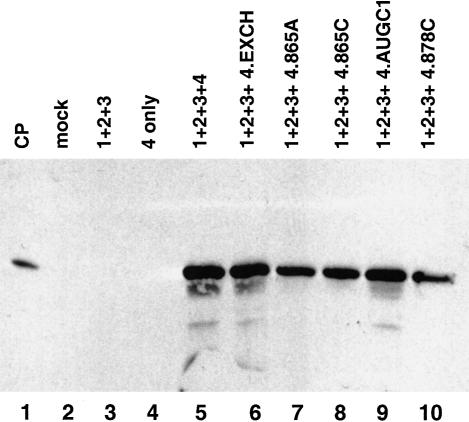
One additional control was done to facilitate interpretation of the mutagenesis and biological activity data. Neeleman et al. (2001) reported that coat protein binding to the viral RNAs enhances their translational efficiency. To help distinguish the effects of the mutations on replication versus translation, we generated subgenomic activator RNA 4 constructs that carried the mutations and used them in the in vivo replication assays (Fig. 10 ▶). In these experiments, genomic RNAs 1–3 were wild-type, but the activator RNA 4 transcripts included to initiate replication carried a number of the mutations described previously in this work. The rationale for the experiment stated that if the nucleotide insertions described in this work diminished RNA translation, then diminished expression of activator coat protein would reduce or prevent viral RNA replication. Although the 865C insertion mutation in RNA 3 did not support viral RNA replication (Fig. 9A,B ▶, lane 7), the same mutation in the context of activator RNA 4 supported significant levels of coat protein expression (Fig. 10 ▶, lane 8). In fact, all of the mutations tested were active in the context of subgenomic RNA 4. These results strongly suggest that the data presented in Figure 9 ▶ reflect effects at the level of viral RNA replication rather than translation.
FIGURE 10.
Activation of viral RNA replication using variant subgenomic RNA 4 constructs. The figure shows a Western blot for viral coat protein expression. (Lane 1) virion coat protein standard; (lane 2) mock-infected cells; (lane 3) genomic RNAs 1–3 only; (lane 4) complete inoculum containing genomic RNAs 1–3 plus subgenomic RNA 4; and (lanes 6–10) wild-type genomic RNAs 1–3 were transfected with the following variant subgenomic RNA 4 constructs: RNA 4 carrying the hairpin exchange (lane 6), RNA 4 carrying the 865A insertion (lane 7), RNA 4 carrying the 865C insertion (lane 8), RNA 4 carrying the AUGC1 insertion (lane 9), and RNA 4 carrying the 878C insertion (lane 10).
DISCUSSION
The AMV coat protein–RNA interaction is significant both as a model system for studying RNA–protein interactions and as a system for analyzing the functional significance of ribonucleoprotein complexes in virus replication. Although coat protein is required to initiate the early stages of AMV and ilarvirus RNA replication (Bol et al. 1971), the mechanism is not understood. The experiments presented here test elements of a coat protein binding model (Ansel-McKinney and Gehrke 1998) while developing predictions of the higher order structure of the complex.
Why is the viral coat protein needed to initiate viral RNA replication? The requirement for coat protein is coupled with a distinguishing structural feature of the alfalmovirus and ilarvirus genera; that is, the RNAs terminate in the sequence AUGC and therefore lack the canonical 3′-terminal CCA sequence that is found on many other bromovirus RNAs. In the related bromoviruses, a tRNA-like structure serves at least two functions. First, it is a substrate for charging by aminoacyl tRNA synthetases, and evidence suggests that the tRNA-like terminus is a key element in replication of these viral RNAs (Dreher and Hall 1988; Kao and Sun 1996). Second, the CCA end serves a telomere function because it contains the sequence elements recognized by the nucleotidyltransferase enzyme that repairs and maintains the correct 3′-terminal nucleotide sequence (Rao et al. 1989). Theorists have proposed that the tRNA-like structures found on many plant viral RNAs are vestiges of an RNA world where they “tagged” the 3′ termini of RNAs to be replicated (Weiner and Maizels 1987; Maizels and Weiner 1994). Therefore, tRNA-like structures found on many plant viral RNAs have a dual function in providing a recognition signal for the viral RNA-dependent RNA polymerase and in preventing loss of 3′-terminal nucleotides during multiple replication rounds.
AMV and the ilarviruses detour from this paradigm by nature of the absence of the canonical 3′-terminal CCA terminus and, second, because of the unusual requirement for coat protein to initiate viral RNA replication. Unlike other bromoviruses, AMV is a very poor substrate for both aminoacyl synthetases (Hall 1979) and nucleotidyltransferase (Olsthoorn et al. 1999). Conversely, AMV RNA replication correlates directly with coat protein binding. In other words, mutations in either the viral RNA or coat protein that reduce the binding interaction also disrupt viral RNA synthesis (van der Vossen et al. 1994; Yusibov and Loesch-Fries 1995 Yusibov and Loesch-Fries 1998; Tenllado and Bol 2000; this work). The literature offers multiple explanations for the action of the coat protein in viral RNA replication, several of which are contradictory. It has been claimed that coat protein is required (Neeleman and Bol 1999) or is not required (van der Vossen et al. 1994) for negative-strand RNA synthesis. Houwing and Jaspars (1978) proposed that coat protein binding induces an RNA conformational change that permits recognition by the RNA-dependent RNA polymerase for minus-strand synthesis, whereas Olsthoorn et al. (1999) recently concluded that coat protein binding converts the AMV RNA from a compact form to an extended form that blocks minus-strand RNA transcription. It has also been proposed that coat protein binding liberates plus-stranded progeny from a double-stranded nonreplicative form and thereby stimulates production of more plus-stranded RNA (Houwing and Jaspars 1993), and that coat protein protects the 3′ termini from degradation (Neeleman and Bol 1999). Houwing and Jaspars (2000) later concluded that protection from degradation was not a key coat protein function.
We have focused on a biochemical analysis of determinants present in both the RNA and protein that facilitate RNA–protein interactions, and also on correlating mutational effects with functional activity in viral RNA replication. The results of the experiments presented here strongly suggest that that strict interhelical spacing is critical for coat protein binding; moreover, the results are consistent with the hypothesis that symmetry may also be an important determinant. By inserting AUGC sequences, symmetry was maintained, but the overlap region at AUGC865–868 was disrupted. The result was that coat protein binding was diminished to a large extent; that is, by >20-fold. Conclusions from these results are that symmetry alone in the context of individual binding sites is insufficient to support coat protein binding and that the number of nucleotides separating the hairpins is significant. The AUGC865–868 is very highly conserved among AMV and ilarviruses (Houser-Scott et al. 1994), and in vitro selection data strongly support its role in coat protein binding (Houser-Scott et al. 1997; G.A. Rocheleau, J.E. Petrillo, L. Guogas, and L. Gehrke, in prep.). We do not, however, understand the structural details of the conserved AUGC nucleotides (Houser-Scott et al. 1994) or the essential arginine 17 (Ansel-McKinney et al. 1996) in the complex.
Nucleotide insertions in the coat protein binding domain generally disrupted coat protein binding. However, the 5′ or 3′ extension variant RNAs (Figs. 5 ▶, 6 ▶) and the hairpin exchange variant RNA (Figs. 7 ▶, 8 ▶) bound peptide with apparent dissociation constants at or within a five- to 10-fold range of wild-type levels (Figs. 5 ▶–7 ▶). Neither of these types of mutations is predicted to disrupt the overlapping UGC-hairpin-RAUGC symmetry motifs (Fig. 1 ▶). The replication experiments suggested that the addition of 16 3′-terminal nucleotides did not disrupt replication (Fig. 9 ▶, lane 10). Neeleman et al. (2001) reported previously that AMV RNAs with 3′ poly(A) extensions were active in replication. An open question is, however, if the replicase is copying the extra nucleotides or initiating at the correct site on the native viral RNA.
The hairpin exchange RNA (Fig. 7 ▶) binds coat protein, and, in the context of the 170-nt 3′ UTR, undergoes the conformational change observed with wild-type AMV RNA (Figure 8 ▶). RNA 3 containing the hairpin exchange is, however, inactive in replication (Fig. 9 ▶, lane 5). Results from other laboratories suggest that altered replicase binding may explain the failure to replicate. The 3′-terminal hairpin 869–877 (Fig. 1 ▶) is proposed to have a UAA triloop. Haasnoot et al. (2000) showed that a hairpin capped by a UAA triloop is important for subgenomic RNA synthesis in brome mosaic virus, and mutating the UAA diminished replication. Following this argument, the predicted 3′-terminal AAAC tetraloop of the exchange mutant RNA (Fig. 7 ▶) might be expected to preclude replication. The hairpin exchange modifications did not affect the proposed subgenomic promoter region (Haasnoot et al. 2000), arguing against the possibility that subgenomic RNA transcription was directly impacted. Although the hairpin exchange RNA undergoes a conformational change upon binding peptide (Fig. 8B ▶), there may be localized structural disruptions that are not revealed in the bandshift and that prevent the correct presentation of the 3′ terminus to the viral replicase. Other features of the hairpin exchange, such as the hairpin length and nucleotide sequence could also have affected replicase binding or the initiation of replication. Little is currently known about how the viral replicase recognizes the 3′ terminus of AMV and ilarvirus RNAs.
In summary, we propose that the viral coat protein recognizes 3′-terminal sequence and structure-specific binding determinants, including the AUGC865–868 and the spatial arrangement of the hairpins. After the initial intermolecular recognition, the unstructured N-terminal RNA binding domain may fold into an α-helix or bent helix structure, whereas concomitant RNA conformational changes (G.A. Rocheleau, J.E. Petrillo, B. Kelley-Clarke, S. Laforest, G.W. Martin III, and L. Gehrke, in prep.) compact the RNA and create an interhelical coat protein binding pocket. The results presented here suggest that one or both of the bound peptides may interact with AUGC865–868 and form bridging RNA contacts between the two hairpins, thereby explaining the protection of residue A878 in hydroxyl radical footprint experiments (Ansel-McKinney et al. 1996). The function of the RNA conformational change would be to convert the viral RNA into a substrate that is competent for viral RNA replication activities.
MATERIALS AND METHODS
RNA in vitro transcription and end-labeling
Unlabeled RNAs were transcribed in vitro from synthetic DNA templates (Milligan et al. 1987) by using the MEGAshortscript T7 transcription kit (Ambion). To generate labeled transcripts, 20 μL reactions contained rATP, rCTP, and rGTP at 5.6 mM; rUTP at 0.6 mM; and 50 μCi (α-32P) UTP (PerkinElmer, 3000 Ci/mmole). Labeled and unlabeled RNAs were gel-purified by electrophoresis into a 15% polyacrylamide-urea gel, visualized by ultraviolet light shadowing or autoradiography, and eluted overnight at 4°C (Calnan et al. 1991). The solution containing the eluted RNA was phenol-chloroform–extracted, chloroform-extracted, and ethanol-precipitated. The purified RNA was then resuspended in water and quantified by absorption spectroscopy. For 5′ end-labeling, 100 picomoles transcribed RNAs were dephosphorylated by incubation with 20 units of calf intestinal alkaline phosphatase (CIP; New England Biolabs) for 2 h at 37°C. Twenty picomoles of dephosphorylated RNA were 5′ end-labeled by incubation with 20 units of T4 polynucleotide kinase (NEB) and 13 pmole 3000Ci/mmole or 6000Ci/mmole (γ-32P) ATP for 2 h at 37°C. End-labeled RNA was separated from unincorporated nucleotides by electrophoresis into a 15% polyacrylamide-urea gel, localized by autoradiography, and eluted as described above.
A variant RNA construct was engineered to test the effects of exchanging the positions of the two 3′-terminal hairpins in the infectious RNA 3 clone. The exchange clone was generated by annealing two DNA oligonucleotides at an 18-bp overlap that served as a double-stranded primer for extension using T4 DNA polymerase. One DNA oligomer represented the mRNA-sense polarity of nucleotides 1859–1957 (GenBank accession no. K02703) of the AMV RNA 3 3′ UTR. The second DNA oligomer was of antisense polarity and represented nucleotides 2037–1939. When annealed, a double-stranded primer region was created at nucleotides 1939–1957. After annealing at 65°C, the termini were extended by incubation DNA for 1 h at 37°C with 3 units T4 DNA polymerase (New England Biolabs) and 5mM dNTPs in 1× T4 DNA polymerase buffer (New England Biolabs). The extension products were subsequently separated by electrophoresis into a 2% agarose gel, and the correctly sized product was extracted and purified. The double-stranded fragment contained a 5′ BstX1 restriction site and a 3′ Xma1 site for use in subcloning. The purified DNA was digested with BstX1 and Xma1 and ligated into a plasmid containing a cDNA copy of RNA 3 that had been cleaved with the same enzymes, creating the hairpin exchange mutant.
Peptides
AMV peptide CP26 was synthesized with an acetylated N terminus as described (Baer et al. 1994). Peptides were purified by high-performance liquid chromatography, and the concentration of peptide solutions was determined by amino acid analysis (Ansel-McKinney et al. 1996). AMV peptide CP26 contains the N-terminal 26 residues of the AMV coat protein. The sequence is N-acetyl-SSSQKKAGGKAGKPTKRSQNYAALRK.
Electrophoretic mobility bandshift analysis
Electrophoretic mobility bandshift analysis (EMSA) conditions were adapted from Weinberger et al. (1986) as described previously (Baer et al. 1994; Ansel-McKinney et al. 1996). The RNA concentration in the EMSA reactions was 20 nM, and the peptide concentrations used in the binding reactions are indicated in each figure. The nucleotide insertions analyzed in this work caused significant changes in peptide binding affinity in most cases. The relative binding affinities were estimated by a visual examination of the bandshift patterns.
Hydroxyl radical footprinting
Hydroxyl radical protection assays are described in detail elsewhere (Ansel-McKinney et al. 1996; Ansel-McKinney and Gehrke 1997). The cleavage reactions were performed for 1 h at room temperature in 2 mM Fe(II), 4mM EDTA, and 10 mM DTT. Reactions were quenched by addition of thiourea to a final concentration of 9.1 mM; next, the solution was heated to 68°C for 3 min and analyzed by electrophoresis into a 20% polyacrylamide/urea gel.
Infectious RNA transcripts
Constructs encoding infectious clones of AMV RNAs 1 and 2 were generated by reverse transcription and rapid amplification of cDNA ends (RACE) using total viral genomic RNA as template. The RACE products were ligated into pGEM-T vector (Promega). Positive clones were amplified by using primers that contained the bacteriophage T7 promoter sequence for in vitro transcription in addition to a nucleotide change in the 5′ leader sequence that optimized the transcriptional yield of the infectious clone RNAs (Neeleman and Bol 1999). The DNA template used to generate the AMV RNA 3 clone by PCR amplification was provided by L. Sue Loesch-Fries (Purdue University). Again, the upstream primer included a nucleotide change reported by Neeleman and Bol (1999) that increases transcriptional yield while maintaining infectivity. The RNA 4 clone was provided by L. Sue Loesch-Fries (Loesch-Fries et al. 1985). The P14A and Y21A mutations were introduced into the RNA 3 and RNA 4 clones by using PCR mutagenesis (QuikChange, Stratagene).
Viral RNA replication assays
Nucleotide substitutions were introduced into the infectious clone DNA by using the Quick-Change Site-Directed Mutagenesis kit (Stratagene). In vitro-transcribed, capped AMV RNAs were transfected by electroporation into tobacco protoplasts prepared from Nicotiana tabacum NT-1 cells grown in liquid culture (Watanabe et al. 1987). To maintain the culture, cells were inoculated into maintenance media (1× Murashige and Skoog salt mixture, 88 mM sucrose, 0.6 mM myo-Inositol, 1 mg/L thiamine-HCl, and 0.2 mg/L 2,4-dichloro-phenoxyacetic acid at pH 5.8). The cultured cells were incubated at 28°C with shaking for 3 d, harvested by gentle sedimentation, and washed twice in 0.4 M mannitol and 20 mM Mes (pH 5.8). To remove the cell walls, the cells were resuspended in 0.4 M mannitol, 20 mM Mes (pH 5.8), 1% cellulase (Calbiochem), and 0.1% pectolyase (Sigma); transferred to Petri dishes; and incubated for 1 h in the dark with gentle shaking (60 rpm). After sedimenting the cells by centrifugation at 100g for 2 min, they were washed twice by resuspension in 0.4 M mannitol and 20 mM Mes buffer (pH 5.8) and counted. After a final sedimentation step, the protoplasts were resuspended in cold 0.4 M mannitol and 20 mM Mes (pH 5.8), at a concentration of 5 × 106 cells/mL. For the replication assay, ~200 μL of the protoplast suspension (2 × 106 cells) were mixed with 600 μL of electroporation buffer (0.14 M NaCl, 3 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, 0.4 M D-mannitol at pH 6.5) containing 12 μg of a mixture of in vitro transcribed AMV RNAs at molar ratios of 1:1:1:2 (4.4 μg RNA 1, 3.1 μg RNA 2, 2.4 μg RNA 3, 2.1 μg RNA 4). The protoplasts were electroporated at 300 volts, 325 μF (BioRad Gene Pulser) and then allowed to recover for 30 min on ice. The cells were diluted into 5 mL protoplast media (1× Murashige and Skoog salt mixture, 88 mM sucrose, 0.6 mM myo-inositol, 1 mg/L thiamine-HCl, 0.2 mg/L 2,4-dichloro-phenoxyacetic acid, and 0.4 M mannitol at pH 5.8) and incubated without agitation at 28°C.
Replication was activated by transfecting a truncated activator RNA 4 along with genomic RNAs 1 through 3. This truncated activator RNA encodes the full-length coat protein; however, the 3′ untranslated region was shortened from 170 nt to ~60 nt to distinguish activator RNA from newly transcribed subgenomic RNAs in Northern blots. This approach provided unequivocal evidence of viral RNA replication. Viral RNA replication levels, as measured by coat protein expression, were similar when full-length RNA 4 or the truncated activator RNA 4 was used (to be described in detail elsewhere; G.W. Martin and L. Gehrke, in prep.).
For protein analysis, protoplasts were pelleted 24–48 h after transfection, resuspended in 1× SDS gel loading buffer (Maniatis et al. 1982), and boiled for 5 min. Cellular debris was removed by centrifugation, and an aliquot of the supernatant was analyzed by electrophoresis into a 12% polyacrylamide protein gel. AMV coat protein was detected by Western blotting with a specific polyclonal rabbit anti-AMV coat protein antiserum.
Acknowledgments
We thank A.L.N. Rao, Cynthia Hemenway, and Neeta Pillai for advising us during the establishment of the protoplast-based virus replication assay. We also thank other members of the Gehrke laboratory and members of Jane-Jane Chen’s laboratory for critical discussions. This work was supported by NIH award GM42504.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5154104.
REFERENCES
- Ansel-McKinney, P. and Gehrke, L. 1997. Footprinting RNA–protein complexes with hydroxyl radicals. In Analysis of mRNA formation and function (ed. J.D. Richter), pp. 285–303. Academic Press, New York.
- ———. 1998. RNA determinants of a specific RNA–coat protein peptide interaction in alfalfa mosaic virus: Conservation of homologous features in ilarvirus RNAs. J. Mol. Biol. 278: 767–785. [DOI] [PubMed] [Google Scholar]
- Ansel-McKinney, P., Scott, S.W., Swanson, M., Ge, X., and Gehrke, L. 1996. A plant viral coat protein RNA-binding consensus sequence contains a crucial arginine. EMBO J. 15: 5077–5084. [PMC free article] [PubMed] [Google Scholar]
- Baer, M., Houser, F., Loesch-Fries, L.S., and Gehrke, L. 1994. Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 13: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T. and Williamson, J.R. 1996a. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA: I. Defining the minimal RNA site. J. Mol. Biol. 261: 536–549. [DOI] [PubMed] [Google Scholar]
- ———. 1996b. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA: II. Specificity determinants of RNA–protein recognition. J. Mol. Biol. 261: 550–567. [DOI] [PubMed] [Google Scholar]
- Bol, J.F. 1999. Alfalfa mosaic virus and ilarviruses: Involvement of coat protein in multiple steps of the replication cycle. J. Gen. Virol. 82: 947–951. [DOI] [PubMed] [Google Scholar]
- Bol, J.F., Van Vloten-Doting, L., and Jaspars, E.M.J. 1971. A functional equivalence of top component a RNA and coat protein in the initiation of infection by alfalfa mosaic virus. Virology 46: 73–85. [DOI] [PubMed] [Google Scholar]
- Calnan, B.J., Biancalana, S., Hudson, D., and Frankel, A.D. 1991. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA protein recognition. Genes & Dev. 5: 201–210. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W. and Hall, T.C. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 201: 31–40. [DOI] [PubMed] [Google Scholar]
- Giege, R. 1996. Interplay of tRNA-like structures from plant viral RNAs with partners of the translation and replication machineries. Proc. Natl. Acad. Sci. 93: 12078–12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot, P.C., Brederode, F.T., Olsthoorn, R.C., and Bol, J.F. 2000. A conserved hairpin structure in alfamovirus and bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.C. 1979. Transfer RNA-like structures in viral genomes. In International review of cytology (eds. G.H. Bourne and J.R. Danielli), pp. 1–26. Academic Press, New York. [DOI] [PubMed]
- Houser-Scott, F., Baer, M.L., Liem Jr., K.F., Cai, J.-M., and Gehrke, L. 1994. Nucleotide sequence and structural determinants of specific binding of coat protein or coat protein peptides to the 3′ untranslated region of alfalfa mosaic virus RNA 4. J. Virol. 68: 2194–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser-Scott, F., Ansel-McKinney, P.A., Cai, J.M., and Gehrke, L. 1997. In vitro genetic selection analysis of alfalfa mosaic virus coat protein binding to 3′-terminal AUGC repeats. J. Virol. 71: 2310–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing, C.J. and Jaspars, E.M.J. 1978. Coat protein binds to the 3′-terminal part of RNA 4 of alfalfa mosaic virus. Biochemistry 17: 2927–2933. [DOI] [PubMed] [Google Scholar]
- ———. 1982. Protein binding sites in nucleation complexes of alfalfa mosaic virus RNA 4. Biochemistry 21: 3408–3414. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Coat protein stimulates replication complexes of alfalfa mosaic virus to produce virion RNAs in vitro. Biochimie 75: 617–622. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Activation of the alfalfa mosaic virus genome by viral coat protein in non-transgenic plants and protoplasts: The protection model biochemically tested. Arch. Virol. 145: 13–35. [DOI] [PubMed] [Google Scholar]
- Jaspars, E.M.J. 1985. Interaction of alfalfa mosaic virus nucleic acid and protein. In Molecular plant virology (ed. J.W. Davies), pp. 155–221. CRC Press, New York.
- Jaspars, E.M. 1999. Genome activation in alfamo- and ilarviruses. Arch. Virol. 144: 843–863. [DOI] [PubMed] [Google Scholar]
- Kao, C.C. and Sun, J.H. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: Use of oligoribonucleotide primers. J. Virol. 70: 6826–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems, J., Calnan, B.J., Frankel, A.D., and Sharp, P.A. 1992. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 11: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koper-Zwartoff, E.C. and Bol, J.F. 1980. Nucleotide sequence of the putative recognition site for coat protein in the RNAs of alfalfa mosaic virus and tobacco streak virus. Nucleic Acids Res. 8: 3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruseman, J., Kraal, B., Jaspars, E., Bol, J., Brederode, F., and Veldstra, H. 1971. Molecular weight of the coat protein of alfalfa mosaic virus. Biochemistry 10: 447–455. [DOI] [PubMed] [Google Scholar]
- Loesch-Fries, L.S., Jarvis, N.P., Krahn, K.J., Nelson, S.E., and Hall, T.C. 1985. Expression of alfalfa mosaic virus RNA 4 cDNA transcripts in vitro and in vivo. Virology 146: 177–187. [DOI] [PubMed] [Google Scholar]
- Maizels, N. and Weiner, A.M. 1994. Phylogeny from function: Evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. 91: 6729–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeleman, L. and Bol, J.F. 1999. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology 254: 324–333. [DOI] [PubMed] [Google Scholar]
- Neeleman, L., Olsthoorn, R.C., Linthorst, H.J., and Bol, J.F. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. 98: 14286–14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn, R.C., Mertens, S., Brederode, F.T., and Bol, J.F. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 18: 4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadt, R., Rosdorff, H.J.M., Hunt, T.W., and Jaspars, E.M.J. 1991. Analysis of the protein composition of alfalfa mosaic virus RNA-dependent RNA polymerase. Virology 182: 309–315. [DOI] [PubMed] [Google Scholar]
- Quigley, G.J., Gehrke, L., Roth, D.A., and Auron, P.E. 1984. Computer-aided nucleic acid secondary structure modeling incorporating enzymatic digestion data. Nucleic Acids Res. 12: 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A.L.N., Dreher, T.W., Marsh, L.E., and Hall, T.C. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. 86: 5335–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken, C.B.E.M. and Bol, J.F. 1996. Structural elements of 3′-terminal coat protein binding site in alfalfa mosaic virus RNAs. Nucleic Acids Res. 24: 2660–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado, F. and Bol, J.F. 2000. Genetic dissection of the multiple functions of alfalfa mosaic virus coat protein in viral RNA replication, encapsidation, and movement. Virology 268: 29–40. [DOI] [PubMed] [Google Scholar]
- van der Vossen, E.A.G., Neeleman, L., and Bol, J.F. 1994. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology 202: 891–903. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., Meshi, T., and Okada, Y. 1987. Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett. 219: 65–69. [Google Scholar]
- Weinberger, J., Baltimore, D., and Sharp, P. 1986. Distinct factors bind to apparently homologous sequences in the immunoglobulin heavy-chain enhancer. Nature 322: 846–848. [DOI] [PubMed] [Google Scholar]
- Weiner, A.M. and Maizels, N. 1987. tRNA-like structures tag the 3′ ends of genomic RNA molecules for replication: Implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. 84: 7383–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov, V. and Loesch-Fries, L.S. 1995. N-terminal basic amino acids of alfalfa mosaic virus coat protein involved in the initiation of infection. Virology 208: 405–407. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Functional significance of three basic N-terminal amino acids of alfalfa mosaic virus coat protein. Virology 242: 1–5. [DOI] [PubMed] [Google Scholar]