Abstract
Most cellular processes requiring RNA structure rearrangement necessitate the action of Asp-Glu-Ala-Asp (DEAD) proteins. Members of the family, named originally for the conserved DEAD amino acid sequence, are thought to disrupt RNA structure and facilitate its rearrangement by unwinding short stretches of duplex RNA. BstDEAD is a novel 436 amino acid representative of the DEAD protein family from Bacillus stearothermophilus that contains all eight conserved motifs found in DEAD proteins and is homologous with other members of the family. Here, we describe the 1.85 Å resolution structure of the N-terminal domain (residues 1–211) of BstDEAD (BstDEAD-NT). Similar to the corresponding domains of related helicases, BstDEAD-NT adopts a parallel α/β structure with RecA-like topology. In general, the conserved motifs superimpose on closely related DEAD proteins and on more distantly related helicases such as RecA. This affirms the current belief that the core helicase domains, responsible for mechanistic activity, are structurally similar in DEAD proteins. In contrast, however, the so-called Walker A P-loop, which binds the β- and γ-phosphates of ATP, adopts a rarely seen “closed” conformation that would sterically block ATP binding. The closed conformation may be indicative of a general regulatory feature among DEAD proteins (and RNA helicases) that differs from that used by DNA helicases. BstDEAD also contains a unique extension of ~60 residues at the C terminus that is highly basic, suggesting that it might bind nucleic acids and, in so doing, confer specificity to the helicase activity of the core region.
Keywords: RNA, helicase, DEAD protein, RecA, ATPase
INTRODUCTION
Almost every cellular process that involves RNA at some point requires the activity of DExD/H proteins. Such processes include RNA splicing (Staley and Guthrie 1998; Mohr et al. 2002) and ribosome biogenesis (Venema and Tollervey 1995). DExD/H proteins are members of helicase Superfamily II (SFII), within which the DEAD, DEAH, and DExH subfamilies are classified by variations in the so-called Walker B motif (Gorbalenya and Koonin 1993; de la Cruz et al. 1999; Hall and Matson 1999). The widely held belief is that DExD/H proteins are responsible for the mechanistic activity of duplex RNA unwinding, which facilitates the rearrangement of RNA structure. DExD/H proteins have also been implicated in disrupting protein–RNA interactions as a result of duplex RNA unwinding (Jankowsky et al. 2001). Members of the DEAD family were originally named for the presence of the conserved Asp-Glu-Ala-Asp (DEAD) amino acid sequence (Linder et al. 1989). Most DEAD proteins are thought to possess ATP-dependent RNA helicase activity, although this has been shown directly for only a few members of the family (Rogers et al. 1999).
Including the namesake DEAD sequence and the well-characterized Walker A motif found in proteins with ATPase activity (Walker et al. 1982), DEAD proteins are identified by homology to a family with members that contain eight highly conserved signature sequences, originally elucidated by Gorbalenya and Koonin (1993). These sequences are characteristic of DNA and RNA helicases (Gorbalenya and Koonin 1993). Recent work by Tanner et al. (2003) has identified yet another motif, conserved exclusively among DEAD proteins, known as the Q-motif, referring to an invariant Gln residue in the N-terminal region. Despite the fact that the number of DEAD proteins identified by sequence homology is large, the biological functions of most of these proteins remain unknown. Furthermore, high-resolution structures remain limited to two DEAD proteins, yeast eIF4A and mjDEAD from Methanococcus janaschii (Caruthers et al. 2000; Story et al. 2001), and the HCV helicase, a DExH protein from hepatitis C (Kim et al. 1998).
The minimal DEAD protein core is composed of two independent domains connected by a flexible linker. The core, which typically includes 350–370 amino acids, constitutes the framework within which all the conserved motifs necessary for ATPase and helicase activity are contained. However, many DEAD proteins possess variable N- or C-terminal extensions, which are thought to grant unique roles to individual proteins (for review, see Tanner and Linder 2001). Recent work by Kossen et al. (2002) describes how binding specificity for region of 23S rRNA is conferred by the C-terminal extension of Bacillus subtilis YxiN. Although only a small number of DEAD proteins have demonstrated significant RNA binding specificity, this provides the most compelling evidence that residues responsible for specific RNA recognition and binding may be contained within an extension. The aforementioned structures of eIF4A and mjDEAD represent the minimal DEAD protein core lacking any extensions.
We here describe the 1.85 Å resolution crystal structure of the N-terminal domain of BstDEAD, a novel DEAD protein from Bacillus stearothermophilus. Although the precise biological function of BstDEAD remains unknown, a preliminary biochemical analysis indicates that BstDEAD is a functional ATPase in the presence of nucleic acids. Furthermore, BstDEAD possesses a highly basic C-terminal extension that may provide the specificity for recognition of a biological RNA target.
RESULTS AND DISCUSSION
Structure of the N-terminal domain (BstDEAD-NT)
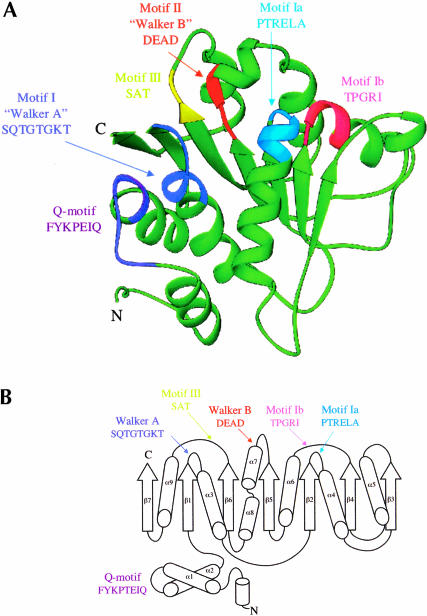
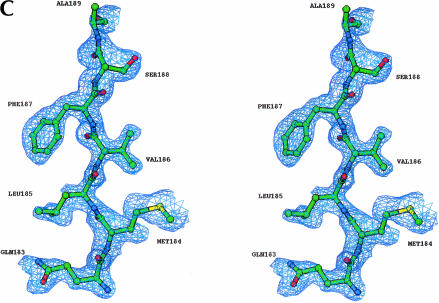
The N-terminal domain has a parallel α/β structure with RecA-like topology (Fig. 1A,B ▶). The RecA-like architecture is common in helicase proteins, and every reported helicase structure to date contains at least one RecA-like domain. The RecA-like core consists of five parallel β-strands surrounded by five α-helices (Story et al. 1992; Caruthers and McKay, 2002). The BstDEAD-NT core is somewhat larger and includes seven parallel β-strands that are surrounded by seven α-helices (Fig. 1A,B ▶). The two additional α-helices (labeled α1 and α2 in Figs. 1B ▶, 2 ▶) flank the core at the N terminus. Although the two N-terminal helices and the C-terminal β-strand appear to be extraneous to the RecA-like core, they are present in all known DEAD proteins. Experimental electron density, including a portion of the final refined model, is shown in Figure 1C ▶. As seen in Figure 1A ▶, the loop connecting the two N-terminal helices contains the Q-motif, a recently identified stretch of highly conserved amino acid residues unique to the DEAD protein family (Tanner et al. 2003). In the crystal structures of eIF4A and mjDEAD (Caruthers et al. 2000; Story et al. 2001), as in the putative structure of full-length BstDEAD, the β7 strand leads into the flexible polypeptide linker that connects the two core domains. In addition to the Q-motif, the N-terminal domain contains the conserved helicase motifs I, II, and III that are responsible for binding and hydrolysis of ATP plus motifs Ia and Ib that play poorly defined roles in substrate recognition. The positions of these motifs, highlighted in Figure 1A,B ▶, are situated primarily in loop regions, at the end of β-strands or capping the ends of α-helices.
FIGURE 1.
Structure of the BstDEAD N-terminal domain. (A) Ribbon representation of BstDEAD-NT consisting of residues 4–211. The color coding used to distinguish the conserved motifs follows the method of Tanner and Linder (2001) and is maintained through the other figures as follows: Q-motif, purple; motif I (Walker A), blue; motif Ia, light blue; motif Ib, pink; motif II (Walker B or DEAD), red; and motif III (SAT), yellow. The motifs found in the N-terminal domain include all that is necessary for ATP recognition and hydrolysis. This figure, as well as Figures 1C ▶, 3 ▶, 5 ▶, and 6A–C ▶, was made with RIBBONS (Carson 1997). (B) Topological organization of the BstDEAD N-terminal domain. β-Strands are depicted as arrows; α-helices, as cylinders. The elements that are not included in the RecA-like core are α1, α2, α5, β3, and β7. Locations of the conserved motifs are shown and colored as in A. (C) Stereo diagram showing representative experimentally phased electron density using MAD data from BstDEAD-NT calculated to 1.85Å and contoured at 1 σ. The amino acids are from the refined BstDEAD-NT model and include part of the SAT motif (motif III).
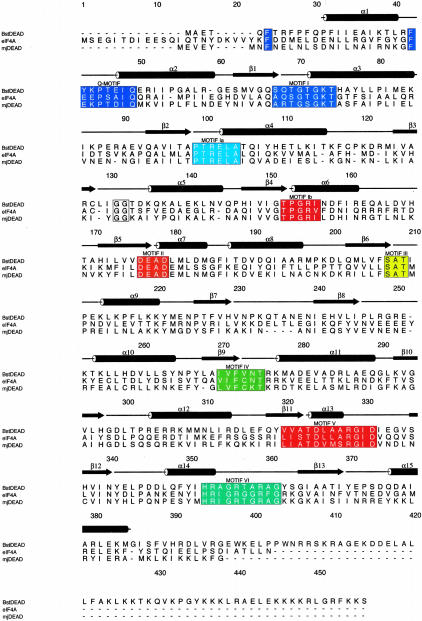
FIGURE 2.
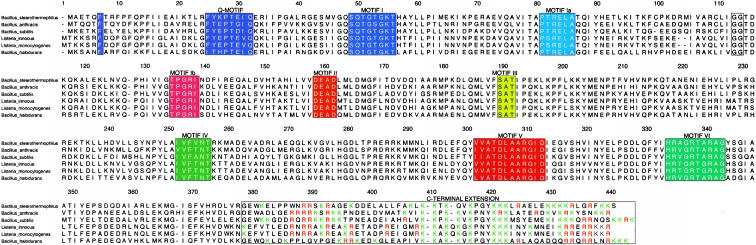
Sequence comparison of Bacillus stearothermophilus BstDEAD, yeast eIF4A, and Methanococcus janaschii mjDEAD. BstDEAD contains all eight conserved helicase motifs in the classification of Gorbalenya and Koonin (1993). These are shown in boxes and are colored as in Figure 1 ▶. Secondary structure topology is based on DALI (Holm and Sander 1993) structural relatedness search using BstDEAD-NT model and preliminary full-length BstDEAD model, eIF4A (PDB ID no. 1FUU), and mjDEAD (PDB ID no. 1HV8). Boundaries of secondary structure elements are approximate due to variations in loop regions. Sequence alignment calculated by using CLUSTALW (Thompson et al. 1994) and generated by using ALSCRIPT (Barton 1993).
Relationship to other proteins
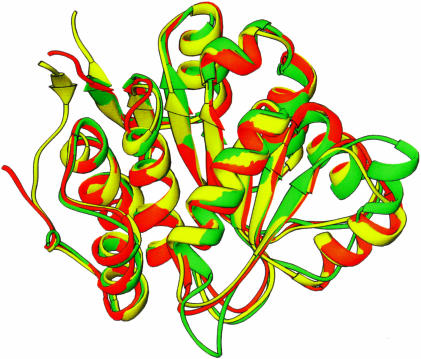
As shown in Figure 2 ▶, BstDEAD shares a moderate degree of sequence similarity with yeast eIF4A and Methanococcus janaschii mjDEAD, with 27% and 30% overall sequence identity, respectively. As the conserved motifs necessary for ATP binding and hydrolysis are contained within the N-terminal domain, the structure of BstDEAD-NT is expected to be similar to the corresponding ATPase domains of other DEAD proteins. A structural relatedness search performed with DALI (Holm and Sander 1993) identified similarity with a large number of proteins possessing ATPase domains, the most closely related of which are the ATPase (N-terminal) domains of eIF4A (Caruthers et al. 2000) and mjDEAD (Story et al. 2001), with 199 Cα positions that superimpose with a root mean square difference (RMSD) of 1.3Å and 1.4Å, respectively. An overlay of these structures is shown in Figure 3 ▶, with the orientation of BstDEAD-NT the same as in Figure 1A ▶. Excepting variations in the loop regions, the overall fold, including the positions of the signature motifs, is conserved. As well, this structural conservation extends to more distantly related proteins in Superfamily I (SFI) and the DExH subfamily of SFII helicases, including PcrA (Subramanya et al. 1996), RecA (Story et al. 1992), and HCV helicase (Kim et al. 1998), in which the Cα backbone superimposes with 2.8Å, 3.8Å, and 3.1Å RMSD for 152, 119, and 128 positions, respectively.
FIGURE 3.
Superimposition of BstDEAD-NT on the N-terminal domains of other DEAD proteins. Ribbon representation of BstDEAD-NT, shown in green, superimposed on the N-terminal domain structures of eIF4A (PDB ID no. 1QVA) in yellow and mjDEAD (PDB ID no. 1HV8) in orange. The orientation of the BstDEAD N-terminal domain is the same as in Figure 1A ▶.
A search of the protein sequence database reveals that BstDEAD is homologous with a small number of uncharacterized bacterial DEAD proteins. Those that are the most similar are putative DEAD proteins from Bacillus anthracis and B. subtilis, with 62% and 61% overall sequence identity, respectively. Sequence homology within the core is expected, as DEAD proteins are identified and classified based on the presence and variability of the conserved motifs. Outside of the common two-domain core, extensions of various lengths on either termini may specify a unique biochemical role, such as RNA binding. Of those proteins most similar to BstDEAD, all possess a C-terminal extension of similar length and sequence (Fig. 4 ▶). In BstDEAD, this region is highly basic (25 of the terminal 70 amino acids are positively charged), suggesting that it may bind nucleic acids. An alignment indicates that the C-terminal extension of BstDEAD shares 39% and 42% sequence identity with the C-terminal extensions of the putative DEAD proteins from B. anthracis and B. subtilis, respectively (Fig. 4 ▶). Notably, within this extension are clusters of conserved arginine and/or lysine residues. Although the biological activity of BstDEAD is not currently known, we postulate that the C-terminal extension might specify a unique biochemical role through interactions with an RNA substrate.
FIGURE 4.
Sequence comparison between BstDEAD and the five most closely related proteins. The conserved motifs in the N-terminal domain are highlighted in colored boxes as in Figure 1 ▶. The conserved motifs found in the C-terminal domain are colored as follows: motif IV, green; motif V, magenta; and motif VI, aqua. The approximate boundary of the C-terminal extension is boxed, and basic residues within this region are colored red (Arg) and green (Lys). The sequence alignment was done with CLUSTALW (Thompson et al. 1994), and the figure was generated by using ALSCRIPT (Barton 1993). Swiss-Prot/TrEMBL accession codes are as follows: Bacillus anthracis, Q81LV0; Bacillus subtilis, P54475; Listeria innocua, Q92BQ9; Listeria monocytogenes, Q8Y755; and Bacillus halodurans, Q9KD34.
The biochemical activity of BstDEAD was investigated by using both thin layer chromatography (TLC) and spectroscopic ATPase assays. Preliminary results indicate that BstDEAD has a characteristically low basal level of ATPase activity in the absence of nucleic acids, and this activity is greatly stimulated in the presence of RNA (data not shown). In an effort to identify a specific RNA target for BstDEAD, we measured the rates of ATP hydrolysis in the presence of numerous RNA cofactors. Although the rates did vary slightly, the differences were not sufficient to suggest an RNA target that specifically interacts with BstDEAD.
Although the two-domain core contains all necessary motifs for ATPase and helicase activity, only a few DEAD proteins have demonstrated RNA binding specificity (Fuller-Pace et al. 1993; Kossen and Uhlenbeck 1999). B. subtilis YxiN protein is targeted to a particular region of 23S rRNA solely by residues within its C-terminal extension (Kossen et al. 2002). Thus, the C-terminal extension seen in BstDEAD and the related DEAD proteins may provide the residues necessary for specific recognition of a yet-to-be identified RNA substrate. The C-terminal extension could define a novel subfamily of DEAD proteins all sharing homology within this region. This might provide clues to elucidating the precise biological roles of these proteins.
Conserved motifs
The conserved residues in motif I, commonly referred to as the Walker A motif, were originally identified by Walker et al. (1982) by comparing the sequences of numerous proteins having NTPase activity. The consensus Walker A sequence (GxxxGKT) comprises the phosphate binding loop, or so-called P-loop, forming part of a cleft that interacts with the β- and γ-phosphates of ATP. In the crystal structure of PcrA, the conserved lysine in the P-loop occupies the position of the bound magnesium ion, but when bound to AMPPNP, this residue undergoes a conformational shift that vacates the magnesium binding site and allows it to interact with the β-phosphate (Velankar et al. 1999). The conserved threonine residue in the P-loop also contributes to the ATP binding cleft through interactions with the bound magnesium ion.
Motif II, often called the Walker B or DEAD motif, contains the conserved DEAD amino acids for which members of this family are named. The strictly conserved D and E residues interact with the β- and γ-phosphates of ATP through a coordinated magnesium ion (Fry et al. 1986). The Glu residue is also thought to be involved in the water-mediated hydrolysis of ATP. The Ala and Asp residues are somewhat less conserved, and variations in these positions are the basis for classification of other helicase subfamilies.
The SAT motif (motif III) is thought to be involved in linking the binding and hydrolysis of ATP with helicase activity. Mutations in this region maintain both ATPase activity and RNA binding but cause dissociation between ATP hydrolysis and duplex unwinding activity (Pause and Sonenberg 1992). The role of the SAT motif in linking ATP hydrolysis and duplex unwinding may be a result of interactions made with the last D of the DEAD motif. A recent report by Tanner et al. (2003) identified another highly conserved stretch of residues in the N terminus known as the Q-motif. This newly identified motif appears to be unique to members of the DEAD protein family and is suggested to be involved in regulation of ATP binding by modulating the state of the P loop (see below; Tanner et al. 2003). The remaining signature sequences found in the N-terminal domain, motifs Ia and Ib, likely are involved in binding RNA substrates, although the precise roles of these motifs are not well defined.
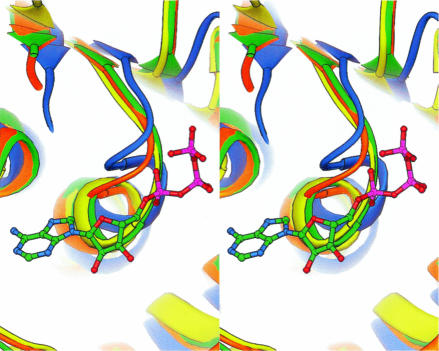
When the structure of BstDEAD-NT is superimposed on the structures of eIF4A (Caruthers et al. 2000) and mjDEAD (Story et al. 2001), the overall fold is conserved as well as the positions of the conserved motifs (Fig. 3 ▶). The P-loop of BstDEAD, however, is atypical. It adopts a closed conformation, similar to that first described in the N-terminal domain structure of eIF4A (Johnson and McKay 1999) but not seen in any other RNA or DNA helicase, with or without ATP. Although no biological relevance has thus far been ascribed to the unique closed conformation, it appears as though the closed P-loop of BstDEAD would sterically inhibit ATP binding. In Figure 5 ▶, the closed P-loop conformations of BstDEAD and eIF4a (Johnson and McKay 1999) are shown compared with the open conformations of mjDEAD (Story et al. 2001) and PcrA (Velankar et al. 1999) with a bound ATP analog. The closed P-loop conformation of eIF4A results from swiveling around Gly70 and Ala65 (Johnson and McKay 1999). Gly53 in BstDEAD is analogous to Gly70 in eIF4A as the “pivot” around which the P-loop rotates. In the crystal structure of eIF4A with a bound nucleotide (Benz et al. 1999), it was observed that, in addition to having an open P-loop conformation, the conserved residues of the Q-motif interact with the conserved residues in the P-loop. In the BstDEAD structure, the corresponding residues in the P-loop are situated too far away to make contact with the Q-motif (Fig. 6C ▶). Thus, it is suggested that the Q-motif, through interactions with the P-loop, may be acting as a switch to modify binding and hydrolysis of ATP.
FIGURE 5.
Stereo view of the Walker A phosphate binding loops (P-loop) from the superimposition of BstDEAD-NT (green) with eIF4A (yellow, PDB ID no. 1QVA), mjDEAD (orange, PDB ID no. 1HV8), and PcrA (blue, PDB ID no. 3PJR) shown with bound ATP analog. The P-loops of BstDEAD and eIF4A are seen in the “closed” conformation that would preclude ATP binding. Structure comparison was performed by using DALI (Holm and Sander 1993), and the figure was generated with RIBBONS (Carson 1997).
FIGURE 6.
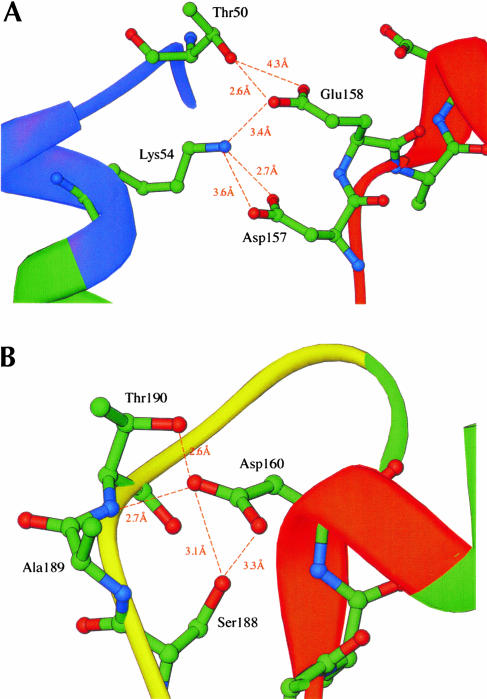
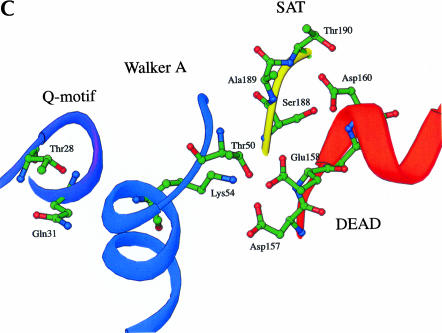
Conserved residues and interactions forming the ATP binding pocket. The ribbon loops and strands are colored as in Figure 1A,B. Distances between residues are shown as dashed lines and are given in Å. (A) Interactions between residues in the closed P-loop and DEAD motif. (B) Interactions between the DEAD motif and the SAT motif. (C) Overall view of the ATP binding pocket formed by the Q-motif, Walker A motif, DEAD motif, and SAT motif.
Residues in the Walker A P-loop are central to the formation of the ATP binding pocket. In addition to interactions with the Q-motif described above, coordinated interactions with the DEAD motif are important for formation of the ATP binding cleft. The conserved residues in the Q-motif, P-loop, and DEAD motif of BstDEAD-NT are positioned almost identically to those in the structure of eIF4A (Johnson and McKay 1999). With the exception of the closed P-loop conformation, the conserved motifs in BstDEAD also superimpose on those of mjDEAD (Story et al. 2001) and PcrA (Subramanya et al. 1996). In these structures, the strictly conserved Walker A lysine (Lys54 in BstDEAD) side-chain is positioned to make interactions with the Asp and Glu (Asp157 and Glu158 in BstDEAD) of the DEAD motif (Fig. 6A ▶). As described above for PcrA, the lysine side-chain occupies the position of the coordinated magnesium ion in the absence of ATP (Velankar et al. 1999). In BstDEAD, the lysine side-chain likely is involved in electrostatic and hydrogen bond interactions, as it is positioned between 2.7 Å and 3.6 Å away from the side-chains of Asp157 and Glu158 (Fig. 6A ▶). Glu158 in BstDEAD is also positioned within range (~2.6 Å) to interact with Thr50 in the P-loop. Johnson and McKay (1999) suggested that a similar hydrogen bond interaction between Ser67 and Glu170 in eIF4A might stabilize the closed conformation of the P loop. Thus, these interactions could effectively stabilize the closed conformation of the P loop as another means of regulating ATP binding and hydrolysis.
The final D in the DEAD motif does not appear to directly interact with the phosphates of ATP. However, through interactions with the SAT motif, this Asp residue (Asp160 in BstDEAD) might be the link between ATP hydrolysis and duplex unwinding. In the structure of BstDEAD-NT, the side-chain of Asp160 is positioned within 2.6 Å and 3.1 Å of the hydroxyl side-chains of Thr190 and Ser188, respectively (Fig. 6B ▶). Furthermore, the side-chain of Asp160 is situated (within 2.7 Å) such that it might participate in hydrogen bond interactions with the main-chain. The potential for similar hydrogen bond interactions is also observed in the structures of eIF4A and mjDEAD (Johnson and McKay 1999; Story et al. 2001). Thus, through interactions with the DEAD motif, an indirect link can be established between the coupling of ATP hydrolysis and duplex unwinding via the SAT motif.
The P-loop as a conformational switch
In the context of the very limited amount of structural information available on DEAD proteins, the description of the BstDEAD N-terminal domain does provide some new insights. Based on structure comparisons with previously documented DNA and RNA helicases, it seems clear that the core domains adopt very similar three-dimensional conformations and that the conserved signature sequences within these domains are positioned nearly identically. However, even slight differences in the positions of these conserved motifs may signify characteristics that are unique to the individual helicase families. The closed Walker A P-loop conformation, first observed in the structure of the eIF4A N-terminal domain, may represent a general feature of DEAD proteins in regulating ATP binding (Johnson and McKay 1999). However, the position of the P-loop in the mjDEAD structure was seen in the open conformation, such as that observed in all reported DNA helicase structures (Story et al. 2001). Now, our structure of the BstDEAD N-terminal domain showing the P-loop in the closed conformation corroborates that seen in eIF4A, and this suggests that the closed conformation may in fact represent a general regulatory feature of DEAD proteins. This finding, along with the recent identification of the conserved Q-motif among members of the DEAD protein family and its suggested role in modulating the state of the P-loop, suggests that the helicase activity of DEAD proteins may be regulated by a conformational “on/off” switch represented by the “open/closed” state of the P-loop (Tanner et al. 2003). As all DNA helicases, with or without bound ATP, show the P-loop in the open conformation, the current structure, along with that of eIF4A, suggests that DEAD proteins (RNA helicases) may regulate ATP binding in a manner different from that of DNA helicases.
MATERIAL AND METHODS
Cloning and purification
The gene for the full-length DEAD protein BstDEAD was originally identified as an uncharacterized open reading frame in B. stearothermophilus. The 1308-basepair gene, which codes for a 436 amino acid protein, was amplified by using PCR from genomic B. stearothermophilus DNA (Carmel and Matthews 2003). By using PCR, the putative N-terminal domain-coding region, corresponding to amino acids 1–211, was cloned from the full-length gene and inserted into pET22b vector (Novagen) under control of the T7 promoter. Selenomethionine (SeMet)-substituted BstDEAD-NT protein with a C-terminal hexahistidine tag was overexpressed in Escherichia coli BL21(DE3) gold cells (Stratagene) following the method for SeMet incorporation described by Gassner et al. (1999). Harvested cells were resuspended in 20 mM Tris (pH 7.9), 1 M KCl, 10 mM imidazole, 10% glycerol, and 0.1% triton and lysed by sonication. Cell debris was removed by centrifugation at 17,000 rpm, and the resulting supernatant containing soluble BstDEAD-NT protein was loaded on a Ni-NTA agarose (QIAGEN) column. Purified BstDEAD-NT protein was eluted by using a 10- to 200-mM imidazole gradient followed by dialysis into 20 mM Hepes (pH 7.5), 125 mM NaCl, 1 mM DTT, and 1 mM EDTA. Further purification involved heating to 60° for 30 min, followed by cooling on ice for 30 min and centrifugation at 17,000 rpm to denature and remove any protein contaminants. A 4 L prep yielded ~75 mg of purified protein, which was concentrated to ~57 mg/mL prior to crystallization.
Crystallography
Crystals of SeMet-substituted BstDEAD-NT were grown by using hanging drops by mixing protein solution 1 : 1 with, and equilibrating over, a reservoir solution containing 100 mM CAPS (pH 10.5) and 30% (w/v) PEG 1550. Large single crystals grew to a size of ~0.7 × 0.3 × 0.2 mm in ~1 wk at room temperature. Crystals were then transferred briefly into a cryoprotectant solution of paratone and flash frozen in liquid nitrogen at 100 K. The crystals have space group P64 with unit cell dimensions a = b = 113.46 Å, c = 65.34 Å. Three wavelength anomalous data, collected on beamline 1–5 at the Stanford Synchrotron Radiation Laboratory (SSRL), were based on the Se K edge (Table 1 ▶). Diffraction data, measured to 1.85 Å, were processed by using MOSFLM (CCP4 1994). The data were merged and scaled by using SCALA and TRUNCATE (CCP4 1994). The putative selenium sites were located with the program SOLVE (Terwilliger and Berendzen 1999). The heavy atom parameters were refined by using SHARP (de la Fortelle and Bricogne 1997), and the resultant electron density map (Fig. 1C ▶) was used to build the preliminary protein model. The asymmetric unit contains two BstDEAD-NT molecules (VM = 2.42 Å3/D with 49% solvent), with molecule A modeled from residue 4–211 and molecule B modeled from residue 2–211 using O (Jones et al. 1991). Electron density for the N-terminal two residues of molecule A was not seen, nor was density for the C-terminal His-tag of either molecule. The model was refined by using TNT (Tronrud 1997), these results being summarized in Tables 1 ▶ and 2 ▶. A Ramachandran analysis, performed by using PROCHECK (Laskowski et al. 1993), indicates that 92.0% of the nonproline and nonglycine residues lie in the most favored regions of the plot with 0% found in the disallowed regions.
TABLE 1.
Crystallographic data collection and phasing statistics for SeMet BstDEAD-NT
| BstDEAD (SeMet) MAD | |||
| Data set | λ1 (peak) | λ2 (infl) | λ3 (high) |
| Source | SSRL 1-5 | SSRL 1-5 | SSRL 1-5 |
| Temperature (K) | 100 | 100 | 100 |
| Wavelength (Å) | 0.9789 | 0.9792 | 0.9252 |
| Resolution (Å) | 25.0–1.85 | 25.0–1.85 | 25.0–1.85 |
| Measured reflections | 334,071 | 334,232 | 335,416 |
| Unique reflections | 40,587 | 40,586 | 40,958 |
| Rsym (%) | 7.1 (41.0) | 7.1 (39.3) | 6.8 (32.6) |
| Completeness (%) | |||
| All data | 98.8 (98.8) | 98.8 (98.8) | 99.8 (100) |
| Anomalous data | 98.5 (85.4) | 98.5 (85.3) | 99.6 (99.6) |
| Figure of merit (centric/acentric) | 0.544/0.719 | ||
Statistics for the outermost shell of data (1.9–1.85 Å) are given in parentheses.
TABLE 2.
Crystallographic model and refinement statistics
| Resolution (Å) | 1.85 |
| Space group | P64 |
| Unit cell dimensions (Å) | a = b = 113.4, c = 65.3 |
| Rfactor (%) | 20.1 |
| Rfree (%) | 28.6 |
| Residues modeled | 417 |
| Waters modeled | 237 |
| RMSD bond lengths (Å) | 0.011 |
| RMSD bond angles (°) | 2.3 |
Coordinates have been deposited in the Protein Data Bank (PDB; access code 1Q0U).
Acknowledgments
We thank Drs. Andrew Hausrath, Blaine Mooers, Michael Quillin, and Martin Sagermann for their assistance and expertise with crystallographic aspects. Special thanks are due to Emanuelle Delagoutte and Peter von Hippel for helpful comments and suggestions, and to Zvi Pasman for assistance with biochemical experiments. This work was supported in part by a grant to B.W.M. from the National Institutes of Health (GM20066). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5134304.
REFERENCES
- Barton, G.J. 1993. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6: 37–40. [DOI] [PubMed] [Google Scholar]
- Benz, J., Trachsel, H., and Baumann, U. 1999. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae: The prototype of the DEAD box protein family. Structure Fold. Des. 7: 671–679. [DOI] [PubMed] [Google Scholar]
- Carmel, A.B. and Matthews, B.W. 2003. Purification, crystallization and preliminary X-ray analysis of the novel DEAD protein BstDEAD from Bacillus stearothermophilus. Acta Cryst. D 59: 1869–1870. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1997. Ribbons. Methods Enzymol. 277: 493–505. [PubMed] [Google Scholar]
- Caruthers, J.M. and McKay, D.B. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12: 123–133. [DOI] [PubMed] [Google Scholar]
- Caruthers, J.M., Johnson, E.R., and McKay, D.B. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. 97: 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Cryst. D 50: 760–763. [DOI] [PubMed] [Google Scholar]
- de la Cruz, J., Kressler, D., and Linder, P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24: 192–198. [DOI] [PubMed] [Google Scholar]
- de la Fortelle, E. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276: 472–494. [DOI] [PubMed] [Google Scholar]
- Fry, D.C., Kuby, S.A., and Mildvan, A.S. 1986. ATP-binding site of adenylate kinase: Mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc. Natl. Acad. Sci. 83: 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace, F.V., Nicol, S.M., Reid, A.D., and Lane, D.P. 1993. DbpA: A DEAD box protein specifically activated by 23S rRNA. EMBO J. 12: 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassner, N.C., Baase, W.A., Hausrath, A.C., and Matthews, B.W. 1999. Substitution with selenomethionine can enhance the stability of methionine-rich proteins. J. Mol. Biol. 294: 17–20. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A.E. and Koonin, E.V. 1993. Helicases: Amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3: 419–429. [Google Scholar]
- Hall, M.C. and Matson, S.W. 1999. Helicase motifs: The engine that powers DNA unwinding. Mol. Microbiol. 34: 867–877. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Jankowsky, E., Gross, C.H., Shuman, S., and Pyle, A.M. 2001. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science 291: 121–125. [DOI] [PubMed] [Google Scholar]
- Johnson, E.R. and McKay, D.B. 1999. Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. RNA 5: 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Cryst. D 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kim, J.L., Morgenstern, K.A., Griffith, J.P., Dwyer, M.D., Thomson, J.A., Murcko, M.A., Lin, C., and Caron, P.R. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: The crystal structure provides insights into the mode of unwinding. Structure 6: 89–100. [DOI] [PubMed] [Google Scholar]
- Kossen, K. and Uhlenbeck, O.C. 1999. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: Delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 27: 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossen, K., Karginov, F.V., and Uhlenbeck, O.C. 2002. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 324: 625–636. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291. [Google Scholar]
- Linder, P., Lasko, P.F., Ashburner, M., Leroy, P., Nielsen, P.J., Nishi, K., Schnier, J., and Slonimski, P.P. 1989. Birth of the D-E-A-D box. Nature 337: 121–122. [DOI] [PubMed] [Google Scholar]
- Mohr, S., Stryker, J.M., and Lambowitz, A.M. 2002. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109: 769–779. [DOI] [PubMed] [Google Scholar]
- Pause, A. and Sonenberg, N. 1992. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF4A. EMBO J. 11: 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers Jr., G.W., Richter, N.J., and Merrick, W.C. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274: 12236–12244. [DOI] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Story, R.M., Weber, I.T., and Steitz, T.A. 1992. The structure of the E. coli recA protein monomer and polymer. Nature 355: 318–325. [DOI] [PubMed] [Google Scholar]
- Story, R.M., Li, H., and Abelson, J.N. 2001. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl. Acad. Sci. 98: 1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya, H.S., Bird, L.E., Brannigan, J.A., and Wigley, D.B. 1996. Crystal structure of a DExx box DNA helicase. Nature 384: 379–383. [DOI] [PubMed] [Google Scholar]
- Tanner, N.K. and Linder, P. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8: 251–262. [DOI] [PubMed] [Google Scholar]
- Tanner, N.K., Cordin, O., Banroques, J., Doere, M., and Linder, P. 2003. The Q motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11: 127– 138. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Cryst. D 55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronrud, D.E. 1997. TNT refinement package. Methods Enzymol. 277: 306–319. [DOI] [PubMed] [Google Scholar]
- Velankar, S.S., Soultanas, P., Dillingham, M.S., Subramanya, H.S., and Wigley, D.B. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97: 75–84. [DOI] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1995. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11: 1629–1650. [DOI] [PubMed] [Google Scholar]
- Walker, J.E., Saraste, M., Runswick, M.J., and Gay, N.J. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]