Abstract
The anticodon loop of tRNA contains a number of conserved or semiconserved nucleotides. In most tRNAs, a highly modified purine is found at position 37 immediately 3′ to the anticodon. Here, we examined the role of the base at position 37 for tRNAPhe binding to the A site of Escherichia coli ribosomes. Affinities and rate constants of A-site binding of native yeast peptidyl-tRNAPhe with hypermodified G (wybutine), or of unmodified peptidyl-tRNAPhe transcripts with G, A, C, or U, at position 37 were measured. The data indicate that purines stabilize binding due to stronger stacking and additional interactions with the ribosome mediated by Mg2+ ions. Paromomycin, an antibiotic that binds to 16S rRNA in the decoding center, greatly stabilized tRNAs in the A site and abolished the Mg2+-dependence of binding. Comparison of binding enthalpies and entropies suggests that hypermodification of the base at position 37 does not affect stacking in the codon–anticodon complex, but rather decreases the entropic penalty for A-site binding. Substitution of purines with pyrimidines at position 37 increases the rates of tRNA binding to and dissociation from the A site. The data suggest that initial binding of tRNA to the A site is followed by a rate-limiting rearrangement of the anticodon loop or the ribosome decoding center that is favored by purines at position 37 and involves stronger stacking, additional Mg2+ binding, and interactions with 16S rRNA.
Keywords: translation, ribosome decoding, tRNA selection, tRNA conformational change, tRNA modifications
INTRODUCTION
The interaction between codon and anticodon plays a central role in mRNA decoding on the ribosome. The canonical structure of the anticodon loop is essential for interactions with both A and P sites (Yusupov et al. 2001) and is evolutionary conserved. The anticodon loop is defined by the presence of a number of conserved and semiconserved nucleotides that form an extended structural signature (Auffinger and Westhof 2001). At the 5′ end of the loop, a pyrimidine base is found at position 32, followed by an invariant U at position 33. Various modified nucleotides are found at position 34, which base-pairs to position 3 of the codon, whereas nt 35 and 36, which interact with positions 2 and 1 of the codon, respectively, exhibit only a limited number of modifications. Bases at positions 37 and 38 are mostly purines, which at position 37 are frequently hypermodified. Sequence analysis of >3000 tRNA genes from different organisms shows that A is found at position 37 in ~80% and G in 20% of all tRNAs (Sprinzl et al. 1998). In ~26% of tRNAs an unmodified A is found; the most common modifications at position 37 are t6A and m1G. The type of modification varies with the coding specificity. In general, tRNAs that have only G and C bases in their anticodons never have a modified purine adjacent to the 3′ side of the anticodon, whereas all tRNAs with at least two A or U bases have a hypermodified purine at that position. For example, almost all phenylalanine-specific tRNAs (anticodon GAA) have such modifications, most often to wybutine (Fig. 1 ▶), peroxywybutine, or 2-methylthio-N6-isopentenyladenine.
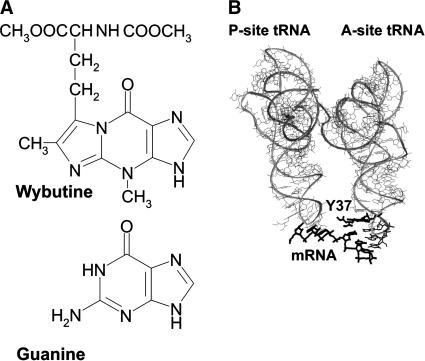
FIGURE 1.
(A) Structure of the Y base (commonly used symbol for wybutine, systematic symbol yW; The RNA Modification Database, http://medlib.med.utah.edu/RNAmods) compared to G. (B) Arrangement of the Y base (Y37) in the complex of yeast tRNAPhe with U6-mRNA in the A site of Thermus thermophilus ribosomes (Yusupov et al. 2001); P-site tRNA is also shown. Y37 and U6-mRNA are highlighted.
Studies in model systems indicate that base modifications at position 37 stabilize tRNA•mRNA interactions by improving intrastrand stacking within the anticodon loop and interstrand stacking between codon and anticodon bases (Grosjean et al. 1998). Using a model system of two tRNAs with complementary anticodons, it was shown that the tRNA•tRNA complex is six orders of magnitude more stable than expected for a comparable three base-pair double helix between trinucleotides (Grosjean et al. 1976, 1978). Three structural features were identified as sources for the enhancement of stability: the loop constraint, the general influence of dangling ends, and the particular effect of modified nucleotides, the latter contributing up to an order of magnitude to stability of the complex. The formation of complexes containing tRNAs with a modified nucleotide at position 37 was accompanied by a larger enthalpy change, compared to those without the modification or without Y base, suggesting an effect of the modification on stacking (Grosjean et al. 1976).
Similar conclusions were reached when the role of modified nucleotides at position 37 for tRNA binding to the P site of the ribosome was studied (Katunin et al. 1994). Removal of the Y base from yeast tRNAPhe or its replacement by unmodified A reduced the enthalpy of binding by 23 kcal/mole and 15 kcal/mole, respectively, suggesting the important role of the nucleotide and its modification in stacking within the anticodon loop. Furthermore, removal of the Y base changed the number of Mg2+ ions involved in P-site binding (Katunin et al. 1994), indicating a loss of Mg2+ coordination. Interestingly, much smaller affinity effects of nt 37 modification were found when the binding of anticodon-stem-loop (ASL) fragments of yeast tRNAPhe to 30S subunits was studied (Ashraf et al. 2000), suggesting a contribution of the 50S subunit in positioning the P-site tRNA. The structural reasons for an important role of the nt 37 modification at the P site are not clear. Footprinting studies indicated that the modification was responsible for the enhanced chemical reactivity of C1400 in 16S rRNA (Moazed and Noller 1986; Ashraf et al. 2000). Because Y37 is located too far from C1400 to affect the modification pattern directly (Yusupov et al. 2001), the effect is most likely due to a change in the structure of the anticodon loop that results in a slightly different position of the anticodon relative to C1400.
Probably the most important functional effects of tRNA modifications, including that of nt 37, are on the accuracy of codon reading in the A site. The nucleotide 3′-adjacent to the anticodon was shown to influence the level of both frameshifting (Curran 1998; Urbonavicius et al. 2001) and misreading (Bouadloun et al. 1986; Petrullo and Elseviers 1986; Wilson and Roe 1989; Diaz and Ehrenberg 1991; Li et al. 1997). Although it is conceivable that the correct three-dimensional structure of the anticodon loop and the stabilization of the codon–anticodon complex are important to prevent both types of errors, the molecular basis of the effect is not known. The effect of removing all nucleotide modifications by using an in vitro transcript of tRNAPhe was to moderately alter the rates of elemental steps of A-site binding (Harrington et al. 1993). Some of these effects, for instance, a three- to fourfold increase of the rate constants of tRNA dissociation from the ribosome could be attributed mostly to changes in the anticodon loop. Importantly, the solution structure of yeast tRNAPhe in the presence of Mg2+ ions was not affected by nucleoside modifications (Hall et al. 1989). This suggests that the changes in tRNA interaction with the A site result from different dynamic properties of modified and unmodified anticodon loops and the respective codon–anticodon complexes or different recognition of these complexes by the ribosome. However, information on thermodynamic or kinetic properties of such complexes in the A site of the ribosome is not available.
The present study was undertaken to examine the mechanism by which nt 37 and its modification influence A-site binding of tRNA. We compared fully modified wild-type (wt) yeast tRNAPhe(Y37) and unmodified tRNAPhe transcripts with purine (Y37G, Y37A) or pyrimidine (Y37U, Y37C) bases at position 37, and a derivative of yeast tRNAPhe where the Y base was excised (−Y), with respect to affinities and kinetics of binding to the A site. Thermodynamic parameters of A-site binding, ΔH° and ΔS°, were measured, which provide information regarding the contribution of stacking and loop flexibility. The role of Mg2+ ions in the interaction was assessed from the Mg2+ dependencies of the affinity constants. To measure these values, a model system must be used where tRNA binding to the ribosome can be studied at equilibrium. For this purpose, several experimental systems can be considered. Either full-length tRNA may be used, in the form of aminoacyl-, peptidyl-, or deacylated tRNA, or ASL. The functional activity of tRNA transcripts, both full-length and ASL, is incomplete and may vary significantly, and it is not possible to separate active and inactive species. Full-length aminoacylated tRNA or tRNA transcripts can be purified from nonaminoacylated material by HPLC (see Materials and Methods), but the respective A-site complexes are too stable to be amenable for Kd measurements over a broad range of Mg2+ and temperature conditions (Semenkov et al. 2000). Thus, we have chosen the corresponding peptidyl-tRNAs (pept-tRNA)—that is, fMetPhe-tRNAs formed on the ribosome—for the present study. The stability of A-site binding of fMetPhe-tRNAPhe, which presumably occupies the A/P state (Moazed and Noller 1989), is much lower than that of Phe-tRNAPhe in the A/A state and allows it to follow the binding and dissociation equilibrium over a wide range of conditions (Semenkov et al. 2000). The affinity difference is attributed to changes in contacts of the 3′ end of tRNA with the peptidyl transferase center, whereas tRNA interactions with the ribosome decoding site do not appear to be affected (Moazed and Noller 1989). A-site binding of various pept-tRNAs with different bases at position 37 was studied by nitrocellulose filtration. In addition to equilibrium-binding constants, rate constants of pept-tRNA binding to and dissociation from the A site could be measured. On the basis of comparisons of thermodynamic parameters of A-site binding, Mg2+ interaction, and kinetic behavior of pept-tRNAs, the role of the base at position 37 in the modulation of mRNA decoding in the A site could be assessed.
RESULTS
Base substitutions at position 37 affect the stability of pept-tRNA binding in the A site
The preparation of unmodified transcripts of yeast tRNAPhe, aminoacylation, and purification of aminoacyl-tRNA were performed as described in Materials and Methods. The purified preparations used for the measurements contained 1400–1700 pmoles of [14C]Phe-tRNAPhe per A260 unit (80%–97% purity assuming 1750 pmoles/A260 for tRNAPhe). To obtain quantitative information about the interaction of tRNAs with the A site, the following experimental system was used (Semenkov et al. 2000). The 70S initiation complexes were prepared using f[3H]Met-tRNAfMet, initiation factors, and mRNA with a UUU codon following the AUG initiation codon. Then, aminoacylated tRNAPhe transcripts or wt Phe-tRNAPhe were bound to the A site in the presence of EF-Tu•GTP. After peptide bond formation, ribosome complexes contained deacylated tRNAfMet in the P site and f[3H]Met[14C]Phe-tRNAPhe (pept-tRNA) in the A site, pept-tRNA having different nucleotides at position 37. The complexes were purified from unbound tRNAs and translation factors by ultracentrifugation. Under appropriate conditions, the resulting complexes dissociated reversibly, depending on the stability of the pept-tRNA binding in the A site (Semenkov et al. 2000). From the rates of dissociation and the extent of pept-tRNA binding after the new equilibrium is established, the equilibrium dissociation constant, Kd, and the rate constants of tRNA dissociation from and association to the A site, koff and kon, can be calculated.
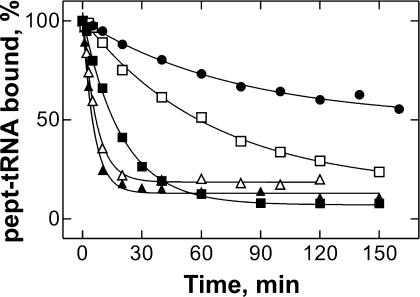
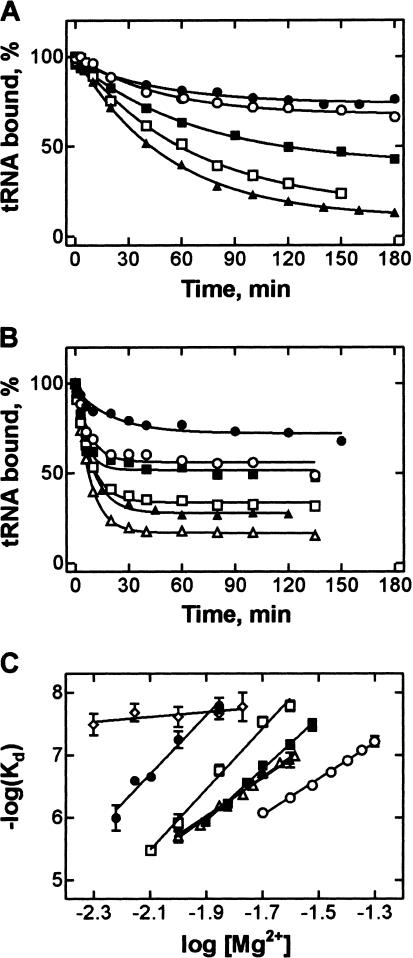
When the ribosomal complexes with wt pept-tRNAPhe were incubated at 37°C and 10 mM Mg2+, <50% of tRNA dissociated from the A site within 2.5 h (Fig. 2 ▶). In contrast, the dissociation of all unmodified pept-tRNA transcripts was much faster, and there were significant differences in the A-site stability, depending on the nature of the base at position 37, with half-lifes of complexes ranging from 1 h (Y37A) to a few minutes (Y37U, Y37C). This demonstrates that A-site binding is strongly affected by the nature of nt 37 in the anticodon loop. In the following, the thermodynamic basis for these effects is studied.
FIGURE 2.
Effect of base replacements at position 37 on the stability of A-site binding of pept-tRNA. Dissociation from the A site was measured at 37°C in buffer A (10 mM MgCl2). Wild-type pept-tRNA (Y37) (•), pept-tRNA(Y37G) (▪), Y37A (□), Y37U (▴), Y37C (▵).
Thermodynamic parameters of binding
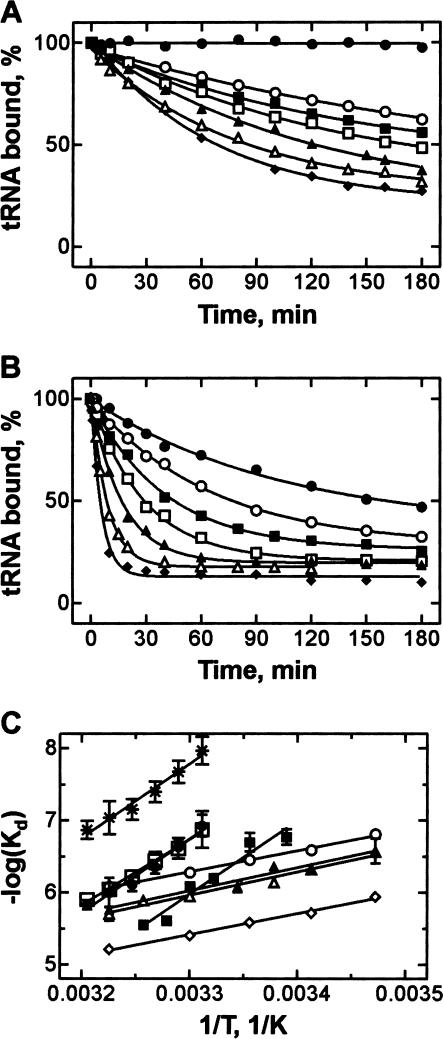
The structure of the tRNA anticodon is stabilized by stacking between nt 34–39 of the anticodon loop, which may be further enhanced by a modification of nt 37 (Saenger 1984). Removal of the modification or replacement of purine at position 37 by pyrimidine is likely to weaken the stacking interactions and to destabilize the codon–anticodon duplex. The loss of stacking is expected to decrease the enthalpy of interaction. This was verified by measuring thermodynamic parameters of A-site binding for pept-tRNAs with substitutions at position 37. The temperature dependence of Kd values was measured at 10 mM Mg2+, except for wt pept-tRNAPhe, which was measured at 6 mM Mg2+, and pept-tRNAPhe(-Y), which was measured at 20 mM Mg2+, because at 10 mM Mg2+ the stability of these tRNAs in the A site were too high or too low, respectively, to measure reliably the level of pept-tRNA binding at equilibrium. However, given the linear dependence of log(Kd) on log[Mg2+] (see below and Kirillov and Semenkov 1982), the Kd values for wt pept-tRNAPhe and pept-tRNAPhe(−Y) at 10 mM Mg2+ could be obtained by extrapolation.
The stability of pept-tRNA in the A site decreased with increasing temperature, both with wt pept-tRNAPhe (Fig. 3A ▶) and pept-tRNA transcripts (shown for Y37U, Fig. 3B ▶). Pept-tRNA transcripts with a purine at position 37 were bound to the A site at least 10 times more weakly than wt pept-tRNA. However, plotting log(Kd) as a function of 1/T (van’t Hoff plot) showed that the slopes of these linear plots are similar for wt pept-tRNA and pept-tRNA transcripts with G or A at position 37 (Fig. 3C ▶) indicating similar values of ΔH° (Table 1 ▶). In contrast, both pyrimidine substitutions, Y37U and Y37C, or removal of the Y base changed the slope of the van’t Hoff plot significantly. The ΔH° values were ~30 kcal/mole higher for tRNAs with pyrimidine or without a base at position 37 than those for purine-containing tRNAs. These data suggest that the replacement of purine by pyrimidine at position 37 resulted in substantial weakening of stacking interactions in the codon–anticodon duplex, whereas the lack of G37 modification had practically no effect on stacking.
FIGURE 3.
Temperature dependence of the Kd of A-site binding. (A) Time courses of dissociation of wt pept-tRNA(Y37) at 6 mM MgCl2. From top to bottom: 0°C, 29°C, 31°C, 33°C, 35°C, 37°C, and 39°C. (B) Time courses of dissociation of pept-tRNA(Y37U) at 10 mM MgCl2. From top to bottom: 15°C, 20°C, 23°C, 26°C, 30°C, 34°C, 37°C. (C) Temperature dependence of the Kd values of wt pept-tRNAPhe(Y37) at 6 mM MgCl2 (•) and extrapolated to 10 mM MgCl2 from the linear Mg2+ dependence of Kd (Fig. 5 ▶) (★); pept-tRNA(−Y) at 20 mM MgCl2 (○) and extrapolated to 10 mM MgCl2 (⋄); pept-tRNA(Y37G)(▪), Y37A (□), Y37U (▴), Y37C (▵) at 10 mM MgCl2.
TABLE 1.
Thermodynamic parameters of A-site binding of pept-tRNAa
| pept-tRNA | ΔH° (kcal/mole) | ΔG° (kcal/mole) | ΔS° (cal/mole/°K) | TΔS° (kcal/mole) |
| Y37 (10 mM Mg2+)b | −47 ± 4 | −10.0 ± 0.3 | −120 ± 11 | −37 ± 4 |
| Y37 (6 mM Mg2+) | −47 ± 4 | −8.5 ± 0.3 | −125 ± 11 | −39 ± 4 |
| Y37G | −47 ± 6 | −6.9 ± 0.3 | −128 ± 19 | −40 ± 6 |
| Y37A | −42 ± 2 | −8.6 ± 0.1 | −107 ± 6 | −33 ± 2 |
| Y37U | −15 ± 2 | −8.2 ± 0.1 | −21 ± 6 | −7 ± 2 |
| Y37C | −15 ± 1 | −8.1 ± 0.1 | −22 ± 4 | −7 ± 1 |
| −Y (20 mM Mg2+) | −14 ± 1 | −8.6 ± 0.1 | −15 ± 2 | −5 ± 1 |
| −Y (10 mM Mg2+)b | −14 ± 1 | −7.4 ± 0.1 | −19 ± 2 | −6 ± 1 |
aΔG° values were calculated from Kd values measured at 310°K according to the equation ΔG° = RTlnKd; ΔH° and ΔS° values were determined from the slope and the Y-axis intercept of the log(Kd) versus 1/T plot, respectively (Fig. 3C ▶).
bExtrapolated on the basis of the linear Mg2+ dependence of the respective Kd values (Fig. 5 ▶).
Values of ΔS° calculated from the Y-axis intercept (Fig. 3C ▶) were negative, indicating an unfavorable entropic contribution to codon–anticodon duplex formation (Table 1 ▶). The contribution was particularly large, between −40 and −33 kcal/mole (TΔS° at 310°K) for purine-containing pept-tRNAs, and was reduced to −7 kcal/mole with replacement of purines by pyrimidines, or to −6 kcal/mole by removal of the Y-base.
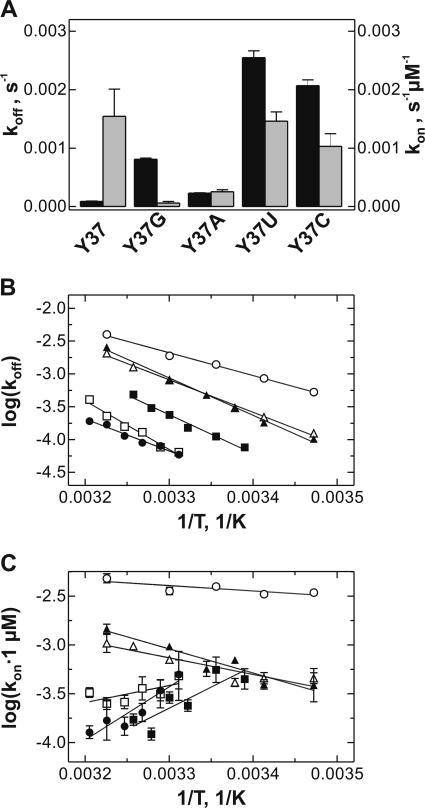
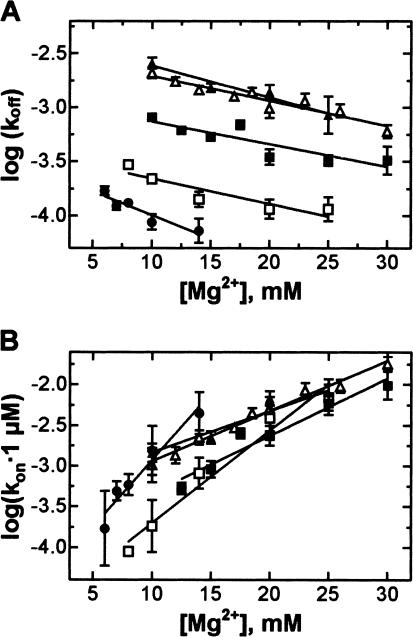
The rate constants of pept-tRNA dissociation from, and association with, the A site, koff and kon, were also affected by the nature of base 37. At 10 mM Mg2+, pept-tRNAs with pyrimidine at position 37 bound faster to the A site, and dissociated faster, than those containing purine (Fig. 4A ▶). Analysis of the temperature dependence of the dissociation rate constants showed that the slopes of the Arrhenius plots were very similar for all tRNAs, indicating that activation energies and enthalpies, Ea and ΔH≠, of the dissociation reaction were similar and that different rates were mainly due to different entropies (Fig. 4B ▶). With pyrimidine-containing pept-tRNAs, the association rate constants, kon, also increased with temperature (Fig. 4C ▶). Surprisingly, the kon values decreased, rather than increased, with temperature for wt pept-tRNA or purine-containing pept-tRNA transcripts. The true bimolecular association rate constant is expected to increase with temperature because it depends on the rate of diffusion. However, if the overall binding reaction is comprised of two steps—that is, of a rapid bimolecular association reaction followed by a slower rate-limiting rearrangement—then the observed temperature dependence may pertain to this latter conformational change. The decrease of the reaction rate with temperature then would indicate that the rearrangement step has an unfavorable reaction enthalpy and is driven by a favorable entropic term.
FIGURE 4.
Temperature dependence of the rate constants of A-site binding. (A) Rate constants with different pept-tRNA constructs measured at 10 mM Mg2+ and 37°C. Black bars, koff; gray bars, kon. (B) Temperature dependence of the dissociation rate constants, koff, of wt pept-tRNA(Y37) at 6 mM Mg2+ (•) or pept-tRNA(−Y) at 20 mM Mg2+(○) and pept-tRNA(Y37G) (▪), Y37A (□), Y37U (▴), Y37C (▵) at 10 mM MgCl2. (C) Temperature dependence of the association rate constants, kon, calculated for 1 μM total concentration of the ligands. Symbols as in B.
Role of Mg2+ ions
The Mg2+ is known to play an important role in tRNA binding to the ribosome. Previous analyses showed that five Mg2+ ions take part in the stabilization of pept-tRNA from Escherichia coli in the A site of 70S ribosomes (Kirillov and Semenkov 1982; Semenkov et al. 2000). The nature of nt 37 modulated the Mg2+ dependence of binding (Fig. 5A,B ▶). To attain the same Kd values, higher Mg2+ concentrations were required for pept-tRNAs with pyrimidines or without a base at position 37 than with purines. From the slopes of the linear log(Kd) versus log[Mg2+] plots (Fig. 5C ▶), the number of Mg2+ ions involved in the interaction was calculated. This amounted to five Mg2+ ions for wt pept-tRNA and pept-tRNA(Y37A) or four for tRNAPhe(Y37G). The replacement of base 37 with pyrimidines or removal of Y at nt 37 resulted in a reduction of the number to three, indicating the loss of Mg2+-mediated interactions between the pept-tRNAs and the ribosome. These data suggest that purines at position 37 are involved in recruiting functionally important Mg2+ ions.
FIGURE 5.
Mg2+ dependence of the Kd of A-site binding. (A) Time courses of dissociation of pept-tRNA(Y37A) at 37°C. From top to bottom: 25, 20, 14, 10, 8 mM Mg2+. (B) Time courses of dissociation of pept-tRNA(Y37C) at 37°C. From top to bottom: 30, 26, 23, 18, 14, 12 mM Mg2+. (C) log(Kd) versus log[Mg2+] plot for wt pept-tRNA(Y37) (•), pept-tRNAs(Y37G) (▪), Y37A (□), Y37U (▴), Y37C (▵), −Y (○), and Y37C in the presence of paromomycin (⋄). The slopes of the plots give the numbers of Mg2+ ions involved in the interaction between pept-tRNA and the A site.
Crystal structures identified three Mg2+ ions located in the decoding center, two of which were found close to A1492 and A1493 in helix 44 at the decoding center of 16S rRNA. The latter two Mg2+ ions can be specifically replaced by the antibiotic paromomycin (Ogle et al. 2001; Vicens and Westhof 2001). We expected that substitution by paromomycin could help us to identify the positions of those Mg2+ ions that are affected by base exchanges at position 37 of tRNA, and therefore, measured the affinity constants of pept-tRNA binding to the A site in the presence of paromomycin at different Mg2+ concentrations. Paromomycin strongly stabilized the A-site binding of pept-tRNAs, both purine- and pyrimidine-containing. In the presence of paromomycin, the affinity of wt pept-tRNA was 1 nM, whereas that of Y37C was ~25 nM, compared to 80 nM and 2000 nM, respectively, in the absence of the antibiotic at 10 mM Mg2+. Interestingly, in the presence of paromomycin, binding of pept-tRNA was not only tight, but also independent of Mg2+ (Fig. 5C ▶) suggesting that (1) a Mg2+-mediated conformational change in helix 44 is—at least in part—responsible for the Mg2+ dependence of tRNA binding, and (2) paromomycin had a more profound effect than would be expected from just the occupancy of Mg2+ binding sites in helix 44.
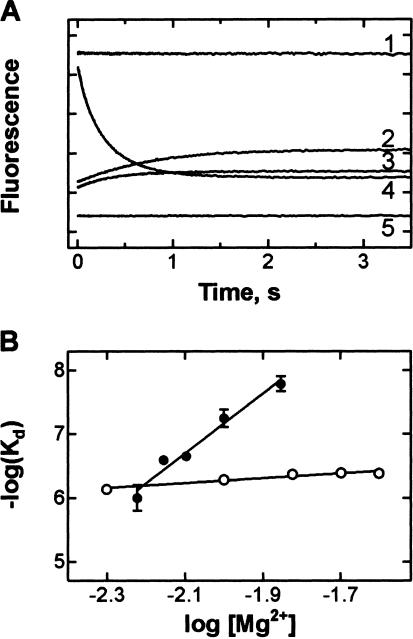
Several Mg2+ ions are bound to the tRNA molecule (Jovine et al. 2000; Shi and Moore 2000). Some of them are bound tightly and are required for the formation of the tRNA tertiary structure, but some may modulate the local conformation at their binding sites in the tRNA molecule and, thereby, influence the formation of the codon–anticodon complex. If the tRNA-bound Mg2+ ions, rather than those bound to the ribosome or to the ribosome•tRNA•mRNA complex, lead to the strong Mg2+ dependence of the Kd described, a similar Mg2+ dependence of Kd should be observed for codon–anticodon interactions in the absence or presence of the ribosome. To test this possibility, we studied the interaction between two tRNAs with complementary anticodons, tRNAPhe (yeast, 3′-AAG-5′) and tRNAGlu2 (E. coli, anticodon 5′-mnm5s2UUC-3′) in solution (Grosjean et al. 1976, 1978). Complex formation was measured by stopped-flow monitoring the fluorescence decrease of Y37 in tRNAPhe (Fig. 6A ▶). The E. coli tRNAPhe lacking Y37 competed with tRNAPhe(Y37) for binding to tRNAGlu2, hence the increase of Y37 fluorescence due to dissociation of yeast tRNAPhe from the complex with the addition of E. coli tRNAPhe. Likewise, dissociation of the tRNAPhe(Y37)•tRNAGlu2 complex with dilution resulted in an increase of Y37 fluorescence. Thus, the formation of the tRNAPhe(Y37)•tRNAGlu2 complex was reversible and Kd values can be determined using the changes in Y37 fluorescence.
FIGURE 6.
Mg2+ dependence of the anticodon–anticodon duplex formation between tRNAPhe and tRNAGlu2. (A) Time courses of complex formation between tRNAPhe (yeast, anticodon 3′-AAG-5′) and tRNAGlu2 (E. coli, anticodon 5′-mnm5s2UUC-3′) measured by stopped-flow monitoring the fluorescence of Y37 in tRNAPhe (transient 4) and the dissociation of the complex with addition of tRNAPhe (E. coli, anticodon 3′-AAG-5′) (transient 2) or with twofold buffer dilution (transient 3). Transients 1 and 5, controls measured by mixing yeast tRNAPhe with noncomplementary E. coli tRNAGly (anticodon 5′-CCG-3′) or nonfluorescent E. coli tRNAPhe with tRNAGlu2, respectively. (B) Mg2+ dependence of the Kd of the tRNAPhe(Y37)•tRNAGlu2 complex formation (open circles) or of A-site binding (cf. Fig. 5C ▶; closed circles).
To determine Kd values, the apparent rates of tRNAPhe(Y37)•tRNAGlu2 complex formation and its dissociation in the presence of excess E. coli tRNAPhe were measured. From the competition experiment, the dissociation rate constant can be calculated directly by exponential fitting. The apparent rate constant of complex formation is given by kon([A]+[B])+koff, where kon and koff are the association and dissociation rate constants, respectively, and [A] and [B] are equilibrium concentrations of tRNAPhe and tRNAGlu2. Using the koff value determined from the competition experiments and knowing the added concentrations of both tRNAs, the association rate constant can be determined (see Materials and Methods). At 10 mM Mg2+ and 10.5°C, kon = 1.2 μM−1s−1, koff = 0.64 s−1, and Kd = 0.5 μM, similar to the values reported earlier (Grosjean et al. 1976, 1978). Varying the Mg2+ concentration from 5 mM to 25 mM did not change the Kd of the tRNAPhe•tRNAGlu2 complex formation appreciably (Fig. 6B ▶); the slope of the log(Kd) versus log([Mg2+]) dependence was small, ~0.3. This suggests that the formation of the codon–anticodon complex in solution does not require Mg2+ ions other than those tightly bound to tRNA and involved in maintaining the tRNA tertiary structure; the latter ions cannot be monitored by the present experimental approach.
Generally, association rate constants of pept-tRNA binding to the ribosome increased, whereas dissociation rate constants of the complex decreased, with increasing Mg2+ concentration (Fig. 7 ▶). At all Mg2+ concentrations, pept-tRNA with pyrimidine 37 bound to, and dissociated from, the A site more rapidly that the purine-containing pept-tRNA. Dissociation rate constants, koff, increased in the order Y37<A<G<C/U. Within the precision of the present measurements, the slopes of the Mg2+ dependencies of koff were similar (Fig. 7A ▶), suggesting that a similar number of Mg2+-mediated contacts were broken with dissociation of different pept-tRNAs. Association rate constants showed a more complex pattern (Fig. 7B ▶). At lower Mg2+ concentrations, pept-tRNAs with purine at position 37 were ~10 times slower in A-site binding than those with pyrimidine. The difference became smaller at elevated Mg2+ concentrations, indicating that the A-site association depended more strongly on Mg2+ for tRNA with purine at position 37 than with pyrimidine. A strong dependence of kon on Mg2+ was found for wt pept-tRNA; however, at all Mg2+ concentrations, wt pept-tRNA bound 10 times more rapidly to the A site than unmodified purine-containing pept-tRNA transcripts. Thus, Mg2+ modulates the kinetics of tRNA interaction with the A site in a fashion that depends on tRNA modifications in general and on the nature of the base at position 37 in particular.
FIGURE 7.
Mg2+ dependence of association and dissociation rate constants of A-site binding. (A) Dissociation rate constants, koff, measured with wt pept-tRNA(Y37) (•) and with pept-tRNA(Y37G) (▪), Y37A (□), Y37U (▴), Y37C (▵) at 37°C. (B) Association rate constants, kon calculated for 1 μM total concentration of ligands. Symbols as in A.
DISCUSSION
Thermodynamic stabilization of the codon–anticodon duplex in the A site
The energetic contribution of stacking to the free energy of pept-tRNA interaction with the A site is very high, −47 kcal/mole. In contrast, when pyrimidine is present at position 37, the enthalpy of interaction drops to −15 kcal/mole, which is close to the value expected for a double helix with three base pairs and two stacking interactions (Saenger 1984). Thus, the presence of a purine base (but not its hypermodification; discussed later) at position 37 in the anticodon loop strongly stabilizes the codon–anticodon complex by increasing the stacking energy. Furthermore, the enthalpy of binding of tRNAPhe to the A site (−47 kcal/mole, this paper) and the P site (−39 kcal/mole; Katunin et al. 1994) are similar, but significantly larger than reported for the interaction of two tRNAs in solution (−25 kcal/mole; Grosjean et al. 1976). This suggests that the ribosome contributes to the favorable enthalpic term, either directly by providing additional interactions, or indirectly by stabilizing a particular conformation of the codon–anticodon duplex.
The large favorable negative enthalpy of codon–anticodon interaction is partially compensated by the unfavorable entropy of binding (Table 1 ▶). Interestingly, in none of the cases studied here was the measured value of ΔS° on the ribosome comparable to that predicted for the interaction of the trinucleotide UUC with yeast tRNAPhe (Borer et al. 1974) or measured for the complex of two tRNAs with complementary anticodons (Grosjean et al. 1976), which was between −36 and −59 cal/mole/°K. When stacking is discontinued by the presence of pyrimidine at position 37 or removal of wybutine, the value of ΔS° was less unfavorable than expected, about −20 cal/mole/°K, both in the A site (Table 1 ▶) and the P site (Katunin et al. 1994). In contrast, when extensive stacking was established, a much larger negative entropy change was found, between −130 and −100 cal/mole/°K, in both A and P sites. This suggests that the ribosome modulates the entropic cost of the codon–anticodon interaction, and that the unfavorable entropy of the formation of continuous stacking interactions is probably due to the loss of conformational freedom of the anticodon loop.
Importance of modified nucleotides in yeast tRNAPhe
Hypermodified nucleotides in the anticodon loop are known to be important for the interaction with the ribosome and are required for the recognition of some tRNAs by the ribosome (Yokoyama and Nishimura 1995; Li et al. 1997; Ashraf et al. 2000; Yarian et al. 2000, 2002). The principal function of modified nucleosides seems to ensure the correct conformation of the three anticodon residues in the context of a seven-nucleotide loop to allow for correct reading of the codon (Yokoyama and Nishimura 1995). The structural influence of modified nucleotides can be quite significant, particularly when the modifications are required to stabilize a canonical U-turn structure of the loop, as shown for tRNALys (Agris 1996; Stuart et al. 2000; Sundaram et al. 2000) or E. coli tRNAPhe (Cabello-Villegas et al. 2002). In other cases, exemplified by tRNAAsp (Perret et al. 1990), the lack of modifications has little effect on the structure of the anticodon loop, whereas the interactions stabilizing the elbow region may be altered. The lack of modified nucleotides in transcripts of yeast tRNAPhe had essentially no effect on its native structure in solution (Hall et al. 1989) and did not impair ribosome binding (Harrington et al. 1993). Our present data are consistent with earlier results (Harrington et al. 1993) regarding the effects of modifications on the kinetic stability of tRNA binding in the A site. In fact, we found that the dissociation rate constant of the fully modified tRNA was only fivefold lower than that of the unmodified transcript, compared to three- to fourfold reported previously (Harrington et al. 1993). Together, modifications contribute 3.1 kcal/mole to the free energy of tRNA binding to the A site, compared to 1.1 kcal/mole at the P site (Katunin et al. 1994). An unexpected result of the present study was that nucleotide modifications did not contribute to the stacking interactions within the codon–anticodon duplex in the A site, unlike in the P site of the ribosome (Katunin et al. 1994) or in tRNA– tRNA duplexes in solution (Grosjean et al. 1976). In the A site, the higher affinity of modified, compared to unmodified, tRNA was due to a less unfavorable entropy of the interaction. This is in marked contrast to the results obtained with tRNA–tRNA duplexes, where entropies were in the range expected for binding of a triplet to the anticodon and the unexpectedly large stability of the complexes was due to an unusual extent of base stacking (Grosjean et al. 1976).
Role of Mg2+
The Mg2+ ion can alter RNA conformation and stabilize tRNA tertiary structure (Laing and Draper 1994; Agris 1996). Analysis of Mg2+ dependencies of the affinity constants suggested that five Mg2+ ions took part in the A-site binding of wt pept-tRNAPhe from yeast (Fig. 5C ▶) or E. coli (Semenkov et al. 2000), or an unmodified pept-tRNA transcript with A at position 37, whereas four ions were involved in the interaction with tRNAPhe(Y37G). Replacement of Y37 with C or U or removal of Y decreased the number of Mg2+ ions involved in binding to three. The location of these Mg2+ ions is not known. Crystal structures identified a number of Mg2+ bound to the tRNAPhe molecule (Jovine et al. 2000; Shi and Moore 2000). Strongly bound Mg2+ ions were found mainly in the D stem-loop, whereas a number of weakly bound Mg2+ were found at several different independent sites, including one in the anticodon loop. Although earlier reports suggested that wybutine 37 was involved in binding of that Mg2+ ion (Saenger 1984), in the high-resolution crystal structure Mg2+ is coordinated by N7 of A38 and O1P of Y37 (Shi and Moore 2000). Upon interaction with the ribosome, the occupancy of the weakly bound Mg2+ binding sites may change due to interactions with the ribosome or conformational rearrangements of tRNA. Notably, the involvement of Mg2+ ions in the interactions seems to be specific for the ribosome complexes, because codon–anticodon interaction between two tRNA molecules with complementary anticodons was independent of Mg2+. The replacement of wybutine by pyrimidine bases may change the orientation of A38 in a way that precludes the efficient coordination of Mg2+ in the anticodon loop and thus loss of Mg2+ binding. This, in turn, could change the dynamic properties of the loop and thus the interaction with the ribosome, resulting in a change of the number of Mg2+ ions involved in the formation of the A-site complex.
In addition to Mg2+ ions bound to tRNA, there are three Mg2+ ions that mediate contacts between the codon–anticodon complex and 16S rRNA: two are bound within helix 44 of 16S rRNA and seem to stabilize the flipped-out conformation of nt 1492 and 1493 in the decoding center of 16S rRNA, and one takes part in binding interactions with the third base pair of the codon–anticodon complex (Carter et al. 2000; Ogle et al. 2001). Whereas the latter seems to be too far to be influenced by the base at position 37 of tRNA, the coordination of Mg2+ ions in helix 44 is likely to be affected when the replacement of wybutine at position 37, particularly by pyrimidine, alters the geometric or dynamic properties of the U1:A36 base pair and its interactions with A1493 helix 44. In fact, the replacement of the two Mg2+ ions in helix 44 by paromomycin abolished the Mg2+ dependence of binding of tRNA, both wt and Y37C, indicating that these Mg2+ ions are involved, most likely indirectly by supporting a particular conformation of the decoding region, in the stabilization of the codon–anticodon complex. Finally, there may be other Mg2+ ions taking part in the interaction between tRNA and the ribosome that were not identified so far in the crystal structures.
Rearrangements in the anticodon loop with binding to the ribosome
Two lines of evidence suggested that, with binding to the A site, purine-containing pept-tRNAs undergo a structural rearrangement, presumably in the anticodon loop, which was not observed with pyrimidine-containing pept-tRNAs. As expected for a bimolecular reaction, the association rate constant increased with temperature for tRNAs with pyrimidine at position 37. In contrast, with purine at position 37, binding of tRNA to the A site was slowed down with increasing the temperature (Fig. 4B ▶)—that is, it behaved abnormally for a reaction that was expected to depend on the rate of diffusion. This suggests that with purine at position 37 an energetically unfavorable, slow conformational rearrangement, rather than the bimolecular association reaction, limited the rate of tRNA binding to the ribosome.
Evidence along the same lines came from the analysis of the Mg2+ dependence of the rate constants (Fig. 7B ▶), which showed that tRNAs with unmodified purine at position 37 were slower in both binding to, and dissociation from, the A site, than those with pyrimidine. A single base exchange in the anticodon loop is unlikely to change the bimolecular rate constant of tRNA association with the ribosome, assuming that the replacement does not change the structure of tRNA appreciably, and that the base in question does not form a direct contact with the ribosome, which is also unlikely. Thus, the results may be explained by assuming a slow Mg2+-dependent rate-limiting rearrangement with tRNA binding to the A site, which is favored by purine at position 37. The same rearrangement may stabilize tRNA on the ribosome, resulting in lower dissociation rates of purine-containing tRNAs.
A number of Mg2+ ions may act cooperatively on tRNA and 16S rRNA to induce or stabilize the rearrangement of tRNA and the decoding region resulting in a stable codon–anticodon complex on the ribosome. However, when the Mg2+ ions in helix 44 of 16S rRNA were replaced by paromomycin, the complex was strongly stabilized and the complex formation was no longer influenced by Mg2+. Paromomycin induces a conformation of the decoding region with A1492 and A1493 of 16S rRNA oriented toward the minor groove of the codon–anticodon duplex, thus freezing the complex on the ribosome (Pape et al. 2000; Ogle et al. 2001; Vicens and Westhof 2001). It is possible that the binding of paromomycin obviates the requirement for Mg2+, because it provides sufficient energy to induce those rearrangements that otherwise require the combined contributions of several Mg2+-dependent interactions. In the absence of the ribosome, the codon–anticodon duplex seems to form in a way that does not require a significant conformational change (Grosjean et al. 1978) and is independent of Mg2+ (this paper). Thus, the mutual adjustment of the decoding site and the codon–anticodon complex is the property of the ribosome that may have evolved to achieve the high fidelity and speed of tRNA selection on the ribosome.
Conclusions
Binding of tRNA to the A site of the ribosome is determined by a number of interactions that are responsible for the specificity of aminoacyl-tRNA selection and maintenance of the correct reading frame. The A-site binding is a multistep process that requires conformational adjustments of both the ribosome and tRNA. In particular, the anticodon region of the tRNA molecule, which is dynamic and adaptable, is important for recognition. The present data suggest an important contribution of the ribosome to the thermodynamics of codon–anticodon complex formation. Codon–anticodon complexes with a purine at position 37 in the anticodon loop have a large favorable interaction enthalpy that is partly neutralized by a large entropic penalty. Stacking is disrupted when pyrimidine is present at position 37, lowering the free enthalpy of binding to values expected for three base pairs in the codon–anticodon triplet and the binding entropy being less unfavorable than expected from model systems. In addition to stacking, tRNA is stabilized in the A site by Mg2+. The number of Mg2+ ions involved in complex formation depends on the nature of nt 37 and amounts to five with tRNAs containing Y or A, compared to four with tRNA (Y37G) or three with pyrimidine-containing tRNAs. The formation of the codon–anticodon duplex apparently takes place in two steps: the bimolecular binding step, which is not affected by the nature of the base present at position 37, and the second step, which stabilizes the binding and requires purine at position 37. Thus, codon recognition in the A site is a dynamic process that is sensitive to divalent cations, temperature, and base modifications, and that is modulated by interactions with the ribosome.
MATERIALS AND METHODS
Materials
Experiments were carried out in buffer A (50 mM Tris•HCl at pH 7.5, 70 mM NH4Cl, 30 mM KCl, and MgCl2 as indicated) at 37°C, if not stated otherwise. Biochemicals were from Roche, radioactive compounds from ICN. Inorganic pyrophosphatase was from Sigma, RNase inhibitor from MBI Fermentas. f[3H]Met-tRNAfMet was prepared as described (Rodnina et al. 1994b). Ribosomes, initiation factors, mRNA, and EF-Tu were prepared as described (Rodnina et al. 1994a; Rodnina and Wintermeyer 1995; Rodnina et al. 1999). tRNAPhe(−Y) was prepared by incubating yeast tRNAPhe in ammonium formate buffer, pH 2.9, for 3.5 h at 37°C (Katunin et al. 1994).
Mutagenesis and preparation of tRNAPhe constructs
Plasmid p67YF0 containing the gene for yeast tRNAPhe under a T7-RNA polymerase promoter was donated by O.C. Uhlenbeck (Northwestern University, Evanston, IL). Mutants of tRNAPhe with G37 substitutions to A, T, or C were generated by megaprimer PCR. Mutations were confirmed by DNA sequencing. tRNAs were produced by T7-RNA polymerase transcription (Sampson and Uhlenbeck 1988). Transcription reactions contained 0.1 mg/mL plasmid DNA, linearized by BstNI, 3 mM NTPs, 5 mM GMP, 5 units/mL inorganic pyrophosphatase, and 0.1 mg/mL T7-RNA polymerase (prepared as described; Davanloo et al. 1984) in buffer B (40 mM Tris-HCl at pH 7.5, 15 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 10 mM DTT). After incubation for 3 h at 37°C, the reaction was stopped by addition of 1/10 volume of 20% potassium acetate at pH 5.0, and tRNA was precipitated with ethanol. The tRNA transcripts were purified under nondenaturing conditions on a MonoQ HR5/5 (Pharmacia) anion exchange column using a linear NaCl (0.3 M–1 M) gradient in buffer C (30 mM Bis-Tris at pH 6.0, and 10 mM MgCl2). A typical yield after transcription and purification procedure was 20–25 A260 units of tRNA from 1 mL of transcription mixture.
Preparative aminoacylation of wt tRNA and tRNA transcripts was carried out in 2-mL reaction volume containing 100 mM Tris-HCl at pH 7.8 (25°C), 3 mM ATP, 1 mM dithiothreitol, 30 A260 units tRNA, 7 mM MgCl2 for native yeast tRNAPhe (or 20 mM MgCl2 for tRNA transcripts), 40 μM [14C]Phe, and 0.1 mg/mL of partially purified S100 fraction from yeast as a source of phenylalanyl-tRNA synthetase. After incubation for 30 min at 37°C, the reaction was stopped by adding 1/10 volume of cold 20% potassium acetate at pH 5.0, protein extracted with phenol, and aminoacylated tRNA precipitated with ethanol. The [14C]Phe-tRNAPhe was purified by HPLC on a Lichrospher WP300 RP18 column (Merck) using a linear 5%–15% ethanol gradient in buffer D (20 mM ammonium acetate at pH 4.5, 10 mM magnesium chloride, 400 mM NaCl). Final purity of [14C]Phe-tRNAPhe, wt, and transcripts, was 1400–1700 pmoles/A260 unit. Samples were shock-frozen in liquid nitrogen and stored in small aliquots at −80°C.
Biochemical assays
To prepare 70S initiation complexes, ribosomes (1 μM) were incubated with a fourfold excess of MFTI-mRNA (Rodnina et al. 1999) in the presence of 1.5 μM initiation factors IF1, IF2, IF3, 1.5 μM f[3H]Met-tRNAfMet, and 1 mM GTP in buffer A (7 mM Mg2+) for 1 h at 37°C. Ternary complex, EF-Tu•GTP•[14C]Phe-tRNAPhe, was prepared by incubating 14 μM EF-Tu with 1 mM GTP, 3 mM phosphoenol pyruvate, 0.1 mg/L pyruvate kinase for 15 min at 37°C, followed by addition of 7 μM [14C]Phe-tRNAPhe. Ternary complex was added to the initiation complex and incubated for 30 sec at 20°C to form the pretranslocation complex carrying fMetPhe-tRNAPhe in the A site. Then, the Mg2+ concentration was adjusted to 20 mM to prevent premature drop-off of fMetPhe-tRNAPhe from the A site and the complex was kept on ice before further purification. The pretranslocation complex with Phe-tRNAPhe(−Y), was formed at 50 mM Mg2+. Pretranslocation complexes were purified by ultracentrifugation through 400 μL 1.1 M sucrose cushion in buffer A (20 mM Mg2+, except pept-tRNA(−Y) where 50 mM Mg2+ was used) for 2 h at 259,000 g in a Sorvall M120GX centrifuge. The amount of [14C]Phe and [3H]Met bound to ribosomes was determined by nitrocellulose filtration by directly applying aliquots of the reaction mixture to the filters (0.45 μm, Sartorius) and subsequent washing with 5 mL of buffer A. Filters were dissolved and radioactivity measured in QS361 scintillation cocktail.
To induce the dissociation of fMetPhe-tRNAPhe from the A site, the Mg2+ concentration was adjusted as indicated in Figure legends, and the amount of pept-tRNA bound to the A site at different incubation times was determined by nitrocellulose filtration. In parallel, the amount of fMetPhe-tRNAPhe translocated to the P site was controlled by puromycin reaction (1 mM puromycin, 10 sec, 20°C) and was found to be less than 7% at the longest incubation times.
Kinetic experiments
Fluorescence stopped-flow measurements were performed as described previously (Pape et al. 1998) using a stopped-flow apparatus from Applied Photophysics. Y37 fluorescence was excited at 325 nm and measured after passing KV 408 filter (Schott). Formation of the complex between two tRNAs was initiated by rapidly mixing equal volumes (60 μL each) of yeast tRNAPhe (2 μM in the experiment of Fig. 7A ▶ or 0.5 μM when the Mg2+-dependence of binding was measured) with E. coli tRNAGlu2 (equimolar to yeast tRNAPhe) in buffer A containing 5, 10, 15, 20, or 25 mM Mg2+ at 10.5°C. For the competition experiments, the tRNAPhe•tRNAGlu2 complex was formed at concentrations indicated above and mixed with E. coli tRNAPhe added either in equimolar amount to the tRNAPhe•tRNAGlu2 complex (Fig. 7A ▶) or in fivefold excess (all other experiments).
Calculations
Dissociation and association rate constants, koff and kon, were calculated from time courses of dissociation by numerical integration using Scientist software (Micromath). The following model was used: A ⇔ B + C, and B ⇒ D, where A denotes ribosomes with fMetPhe-tRNAPhe bound to the A site; B, unbound fMetPhe-tRNAPhe; C, ribosomes with unoccupied A site; D, hydrolyzed fMetPhe-tRNAPhe. koff and kon give Kd = koff/kon; khydr is the rate constant of fMetPhe-tRNAPhe hydrolysis free in solution. The first reaction is reversible (Semenkov et al. 2000); thus Kd values can be estimated from the ratio of A, B, and C at equilibrium. The second reaction is slow but irreversible. The starting concentrations of A and C were measured at the beginning of each experiment (initial concentration of B was zero because purified ribosome complexes were used) and the changes of the concentrations of A, B, and C with time were measured by nitrocellulose filtration. Concentrations of D at different time points were measured by extraction of fMetPhe-A into ethyl acetate at pH 4.5. Combining the time courses of A-site binding and puromycin reaction yielded values of Kd, kon and koff.
Rate constants of interaction between tRNAPhe and tRNAGlu2 were calculated in the following way. The dissociation rate constant, koff, was determined by exponential fitting of the time courses of tRNAPhe(Y37)•tRNAGlu2 complex dissociation induced by the addition of excess E. coli tRNAPhe. The association rate constant was calculated by numerical integration from time courses of tRNAPhe(Y37) binding to tRNAGlu2 using the value of koff determined as described above according to the reaction model A + B ⇔ C, where A, B, and C denote tRNAPhe(Y37), tRNAGlu2, and the complex, respectively.
Acknowledgments
We thank Olke Uhlenbeck for providing the plasmid p67YF0 and constructs for T7 RNA polymerase expression, and Petra Striebeck, Astrid Böhm, Carmen Schillings, and Simone Möbitz for expert technical assistance. The work was supported by the Russian Foundation for Basic Research, the International Bureau of BMBF, the Deutsche Forschungsgemeinschaft, the European Commission, the Alfried Krupp von Bohlen und Halbach-Stiftung, the Fonds der Chemischen Industrie, and the Young Scientists INTAS Fellowship for ALK (YSF 00–192).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5142404.
REFERENCES
- Agris, P.F. 1996. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 53: 79–129. [DOI] [PubMed] [Google Scholar]
- Ashraf, S.S., Guenther, R.H., Ansari, G., Malkiewicz, A., Sochacka, E., and Agris, P.F. 2000. Role of modified nucleosides of yeast tRNAPhe in ribosomal binding. Cell Biochem. Biophys. 33: 241–252. [DOI] [PubMed] [Google Scholar]
- Auffinger, P. and Westhof, E. 2001. An extended structural signature for the tRNA anticodon loop. RNA 7: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer, P.N., Dengler, B., Tinoco Jr., I. and Uhlenbeck, O.C. 1974. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 86: 843–853. [DOI] [PubMed] [Google Scholar]
- Bouadloun, F., Srichaiyo, T., Isaksson, L.A., and Bjork, G.R. 1986. Influence of modification next to the anticodon in tRNA on codon context sensitivity of translational suppression and accuracy. J. Bacteriol. 166: 1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Villegas, J., Winkler, M.E., and Nikonowicz, E.P. 2002. Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe). J. Mol. Biol. 319: 1015–1034. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., and Ramakrishnan, V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Curran, J.F. 1998. Modified nucleosides in translation. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 493–516. ASM Press, Washington, DC.
- Davanloo, P., Rosenberg, A.H., Dunn, J.J., and Studier, F.W. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. 81: 2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, I. and Ehrenberg, M. 1991. ms2i6A deficiency enhances proofreading in translation. J. Mol. Biol. 222: 1161–1171. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Soll, D.G., and Crothers, D.M. 1976. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J. Mol. Biol. 103: 499–519. [DOI] [PubMed] [Google Scholar]
- Grosjean, H.J., de Henau, S., and Crothers, D.M. 1978. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. 75: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, H., Houssier, C., Romby, P., and Marquet, R. 1998. Modulation role of modified nucleotides in RNA loop-loop interactions. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 113–134. ASM Press, Washington, DC.
- Hall, K.B., Sampson, J.R., Uhlenbeck, O.C., and Redfield, A.G. 1989. Structure of an unmodified tRNA molecule. Biochemistry 28: 5794–5801. [DOI] [PubMed] [Google Scholar]
- Harrington, K.M., Nazarenko, I.A., Dix, D.B., Thompson, R.C., and Uhlenbeck, O.C. 1993. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry 32: 7617–7622. [DOI] [PubMed] [Google Scholar]
- Jovine, L., Djordjevic, S., and Rhodes, D. 2000. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J. Mol. Biol. 301: 401–414. [DOI] [PubMed] [Google Scholar]
- Katunin, V., Soboleva, N., Mahkno, V., Sedelnikova, E., Zhenodarova, S., and Kirillov, S. 1994. Effect of the nucleotide-37 on the interaction of tRNA(Phe) with the P site of Escherichia coli ribosomes. Biochimie 76: 51–57. [DOI] [PubMed] [Google Scholar]
- Kirillov, S.V. and Semenkov, Y.P. 1982. Non-exclusion principle of Ac-Phe-tRNAPhe interaction with the donor and acceptor sites of Escherichia coli ribosomes. FEBS Lett. 148: 235–238. [DOI] [PubMed] [Google Scholar]
- Laing, L.G. and Draper, D.E. 1994. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J. Mol. Biol. 237: 560–576. [DOI] [PubMed] [Google Scholar]
- Li, J., Esberg, B., Curran, J.F., and Bjork, G.R. 1997. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 271: 209–221. [DOI] [PubMed] [Google Scholar]
- Moazed, D. and Noller, H.F. 1986. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994. [DOI] [PubMed] [Google Scholar]
- ———. 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons Jr., W.M., Tarry, M.J., Carter, A.P, and Ramakrishnan, V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., and Rodnina, M.V. 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17: 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 7: 104–107. [DOI] [PubMed] [Google Scholar]
- Perret, V., Garcia, A., Puglisi, J., Grosjean, H., Ebel, J.P., Florentz, C., and Giege, R. 1990. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie 72: 735–743. [DOI] [PubMed] [Google Scholar]
- Petrullo, L.A. and Elseviers, D. 1986. Effect of a 2-methylthio-N6-isopentenyladenosine deficiency on peptidyl-tRNA release in Escherichia coli. J. Bacteriol. 165: 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V. and Wintermeyer, W. 1995. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. 92: 1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Fricke, R., and Wintermeyer, W. 1994a. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33: 12267– 12275. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Semenkov, Y.P., and Wintermeyer, W. 1994b. Purification of fMet-tRNAfMet by fast protein liquid chromatography. Anal. Biochem. 219: 380–381. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Matassova, N.B., Katunin, V.I., Semenkov, Y.P., and Wintermeyer, W. 1999. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. 96: 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. 1984. Principles of nucleic acid structure. Springer-Verlag, New York, Berlin, Heidelberg, Tokyo.
- Sampson, J.R. and Uhlenbeck, O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov, Y.P., Rodnina, M.V., and Wintermeyer, W. 2000. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 7: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Shi, H. and Moore, P.B. 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited. RNA 6: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, J.W., Gdaniec, Z., Guenther, R., Marszalek, M., Sochacka, E., Malkiewicz, A., and Agris, P.F. 2000. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39: 13396–13404. [DOI] [PubMed] [Google Scholar]
- Sundaram, M., Durant, P.C., and Davis, D.R. 2000. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39: 12575–12584. [DOI] [PubMed] [Google Scholar]
- Vicens, Q. and Westhof, E. 2001. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure 9: 647–658. [DOI] [PubMed] [Google Scholar]
- Wilson, R.K. and Roe, B.A. 1989. Presence of the hypermodified nucleotide N6-(delta 2-isopentenyl)-2-methylthioadenosine prevents codon misreading by Escherichia coli phenylalanyl-transfer RNA. Proc. Natl. Acad. Sci. 86: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian, C., Marszalek, M., Sochacka, E., Malkiewicz, A., Guenther, R., Miskiewicz, A., and Agris, P.F. 2000. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry 39: 13390–13395. [DOI] [PubMed] [Google Scholar]
- Yarian, C., Townsend, H., Czestkowski, W., Sochacka, E., Malkiewicz, A.J., Guenther, R., Miskiewicz, A., and Agris, P.F. 2002. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277: 16391–16395. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S. and Nishimura, S. 1995. Modified nucleosides and codon recognition. In tRNA: Structure, biosynthesis, and function (eds. D. Söll and U.L. RajBhandary), pp. 207–233. ASM Press, Washington, DC.
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]