Abstract
Following peptide-bond formation, the mRNA:tRNA complex must be translocated within the ribosomal cavity before the next aminoacyl tRNA can be accommodated in the A site. Previous studies suggested that following peptide-bond formation and prior to EF-G recognition, the tRNAs occupy an intermediate (hybrid) state of binding where the acceptor ends of the tRNAs are shifted to their next sites of occupancy (the E and P sites) on the large ribosomal subunit, but where their anticodon ends (and associated mRNA) remain fixed in their prepeptidyl transferase binding states (the P and A sites) on the small subunit. Here we show that pre-translocation-state ribosomes carrying a dipeptidyl-tRNA substrate efficiently react with the minimal A-site substrate puromycin and that following this reaction, the pre-translocation-state bound deacylated tRNA:mRNA complex remains untranslocated. These data establish that pre-translocation-state ribosomes must sample or reside in an intermediate state of tRNA binding independent of the action of EF-G.
Keywords: ribosome, hybrid state, puromycin, translocation
INTRODUCTION
The addition of each amino acid to the growing peptide chain during translation involves three distinct steps: decoding of the incoming aminoacyl tRNA, peptide-bond formation, and translocation of the mRNA:tRNA complex. Translocation of the mRNA:tRNA complex is promoted by the GTPase EF-G in order to vacate the A site so that the next incoming aminoacyl tRNA can be accommodated. Although there is considerable evidence indicating that the elongation factors (and other GTPases involved in the translation cycle) bind to overlapping sites on the ribosome (Cabrer et al. 1972; Miller 1972; Richter et al. 1972; Moazed et al. 1988; Cameron et al. 2002; Frank 2003), there must be structural distinctions between the different functional states of the ribosome that allow for their specificity of action (Richman and Bodley 1972; Wool et al. 1992; Zavialov and Ehrenberg 2003). Traditionally, pre-translocation-state ribosomes have been characterized by the lack of reactivity of bound peptidyl tRNA substrate with incoming aminoacyl tRNA substrates. Conversely, post-translocation-state ribosomes have been characterized by their high level of reactivity with the same aminoacyl tRNA substrates. Defining the structural distinctions between these two states should provide a molecular explanation for these gross differences in the reactivity of the active site of the large ribosomal subunit, as well as provide clues for understanding the specific recognition of the pre-translocation-state ribosome by EF-G.
Chemical modification analysis of pre- and post-translocation-state ribosome complexes provided experimental evidence that the tRNA substrates move in a staggered fashion with respect to the large and small subunits of the ribosome during the translation cycle (Moazed and Noller 1989), as had been predicted earlier (Bretscher 1968). In this model, following peptide-bond formation, the acceptor ends of the tRNAs move with respect to the large subunit to form what is referred to as a hybrid state of tRNA binding, while three-nucleotide movement of the mRNA and (associated anticodon ends of the tRNA) with respect to the small subunit is ultimately catalyzed by EF-G. Thus the hybrid state of tRNA binding can be defined as a state occupied after peptide-bond formation but before translocation, where the tRNAs have “translocated” on the large subunit—occupying the E site and vacating the A site of the large subunit—but have not moved on the small subunit. Other evidence for the staggered movement of tRNAs through the ribosome was obtained from nonradiative fluorescence energy transfer experiments that showed the acceptor end of the tRNA moving substantially following peptide-bond formation but independent of EF-G-catalyzed translocation (Hardesty et al. 1986). Hardesty and colleagues also used fluorescence to establish that two tRNA substrates in addition to the minimal aminoacyl substrate puromycin could be simultaneously bound to the ribosome (consistent with predictions of the hybrid states model; Odom et al. 1990).
However, the existence of such an intermediate or hybrid state of tRNA binding has long been debated, for several fundamental reasons (Wilson et al. 2002). First, the chemical protections of the large subunit by the acceptor ends of the tRNAs seemed to depend on (and thus potentially report on) the three terminal nucleotides of the molecule (the CCA-end; Burkhardt et al. 1998). Indeed, examination of the crystallographic data indicates that the A and P site footprints spatially overlap and only some of the specific protections seem to represent direct tRNA contacts. Second, the chemical footprinting experiments relied on some variation of the ionic conditions, in particular the magnesium concentration, to observe certain changes. These issues have been highlighted by cryoelectron microscopy (cryoEM) studies showing that buffer conditions can have dramatic effects on the extent and sites of tRNA occupancy (Agrawal et al. 1999). In certain polyamine-containing buffers, ribosomes seem to preferentially bind deacylated tRNA substrates (mimicking the deacylated product of the peptide transferase reaction) in a “classical” (P/P) state that is indistinguishable from the state occupied by fMet-tRNAfMet. In contrast, in more simple buffers similar to those used in the original description of the hybrid states model (containing 15 mM Mg(OAc)2 and 160 mM NH4Cl), the same deacylated tRNA substantially occupies a hybrid (P/E) state of binding where the anticodon end of the tRNA is bound in the P site of the 30S subunit while the acceptor end of the tRNA is bound in the E-site (near protein L1) region of the large subunit. In a recent cryoEM study, another glimpse of a P/E hybrid bound tRNA was obtained when ribosomes were loaded with a single peptidyl tRNA that was subsequently deacylated with puromycin and incubated with EF-G:GDPNP (Valle et al. 2003). Although those authors argued that this hybrid state of binding is not naturally occupied following peptide-bond formation in the absence of EF-G, their data do provide support for the proposal that tRNAs move in a staggered fashion with respect to their binding sites on the large and small subunits of the ribosome.
Each of the structural studies described above reported on equilibrium binding states of the tRNAs on the ribosome. One possible interpretation of the structural data is that a hybrid state of binding is not sampled by the tRNAs during the translation cycle and that the tRNAs instead remain in the classical (P/P) state of binding until EF-G promotes the movement of the tRNAs in their entirety to the post-translocation state (Zavialov and Ehrenberg 2003). An alternate interpretation is that a hybrid state of tRNA binding is routinely sampled during the translation cycle, although depending on the in vitro conditions, it may not be a particularly stable (i.e., easily captured) state of binding. Indeed, previous binding studies showed that the transpeptidylation of Phe-tRNAPhe to fMet-Phe-tRNAPhe (to form a pre-translocation state complex) decreased the affinity of the tRNAPhe for the ribosome, thus promoting translocation by ground-state destabilization (Semenkov et al. 2000).
Analysis of the reactivity of the pre- and post-translocation states has not been fully explored and may provide an alternate approach for assessing whether the tRNA substrates sample, even transiently, a hybrid state of binding prior to EF-G-catalyzed translocation. We are aware of several studies that have addressed the question from a functional perspective. Several of these studies began their experiments by forming pre-translocation-state ribosomal complexes on poly-uridylic acid templates (poly-U) using N-Ac-Phe-tRNAPhe as the “initiator” tRNA and supplying Phe-tRNAPhe to form a dipeptidyl-tRNA (N-Ac-Phe-Phe). The earlier study observed that this pre-translocation-state dipeptidyl-tRNA acted as a donor in a peptidyl transferase reaction with puromycin to form the tripeptide product N-Ac-Phe-Phe-Pm (Bergemann and Nierhaus 1983). In interpreting these data, those authors concluded that spontaneous translocation (EF-G-independent) had occurred and that as a result the dipeptidyl-tRNA had become puromycin-reactive. We note, however, that the observed rates of the reaction were much faster than any previously reported rate for spontaneous translocation on untreated ribosomes (Southworth et al. 2002), and no independent method was used to establish that movement of the mRNA:tRNA complex (translocation) had occurred. In a more recent study, Semenkov et al. (1992) performed analogous experiments where the same dipeptidyl-tRNA complex was formed on a poly-U template, and again observed that the dipeptide (N-Ac-Phe-Phe) was reactive with puromycin. Those authors reached a different conclusion and argued that translocation had not occurred, because the reaction was insensitive to translocation-specific antibiotics. They suggested that the pre-translocation-state bound dipeptidyl tRNA was reactive with puromycin (i.e., catalytically competent), but that the rate of catalysis was substantially slower than in a post-translocation-state ribosome. Although these data were compelling, these studies also failed to independently assess translocation. In addition, both studies suffered from using complexes formed on poly-U templates rather than on defined mRNA sequences where distinct tRNA species and initiation factors can confer specific positioning.
We have been interested in similar experiments to determine whether tRNAs sample or reside in an intermediate (perhaps hybrid) state of binding on the ribosome following peptide-bond formation and prior to translocation. What is the stability of such a ribosomal state, and are its structural features those that are recognized by EF-G? Here we describe experiments that establish that an intermediate state of tRNA binding is sampled by a pre-translocation-state ribosome complex. In this intermediate state, the dipeptidyl tRNA reacts efficiently and relatively rapidly with puromycin, thus establishing that on the large subunit, the A site is empty and the P site is occupied by this substrate. Primer extension (toeprinting) analysis confirms that this reactive state is an intermediate by showing that following the peptidyl transferase reaction, the tRNAs remain bound in a pre-translocation state on the small subunit. We are ultimately interested in understanding the functional significance of this hybrid state of tRNA binding during the elongation cycle.
RESULTS
Reactivity of the pre-translocation-state ribosome complexes with puromycin and aminoacyl tRNA substrates
To investigate the behavior of the pre-translocation state, ribosome complexes containing two tRNA substrates and a cognate mRNA were formed in two different ways. In the first case, ribosomes were first programmed with gene 32 mRNA (coding sequence AUG-UUU-AAA, MFK) and then deacylated tRNAMet and N-Ac-Phe-tRNAPhe were sequentially bound (complex I; Fig. 1A ▶). Primer extension (toeprinting) analysis was then used to assess the state of tRNA binding in these ribosomal complexes. In this technique, extension by reverse transcriptase of a radioactively labeled DNA primer annealed to the 3′ end of the mRNA bound in various ribosome complexes is correlated with the position of the mRNA in the ribosome. By binding different tRNAs to the ribosome:mRNA complex (Fig. 1B ▶, lanes 1,2), characteristic toeprints separated by three nucleotides are observed. Interestingly, when two tRNAs are bound to form a pre-translocation-state complex (Fig. 1B ▶, lane 3), a doublet with greater intensity at +1 is observed (Jerinic and Joseph 2000). Thus, using this defined mRNA and sequentially loading the tRNA substrates, translocation of the pre-translocation-state ribosome complex is observed as three-nucleotide movement along the mRNA promoted by EF-G and GTP (Fig. 1B ▶).
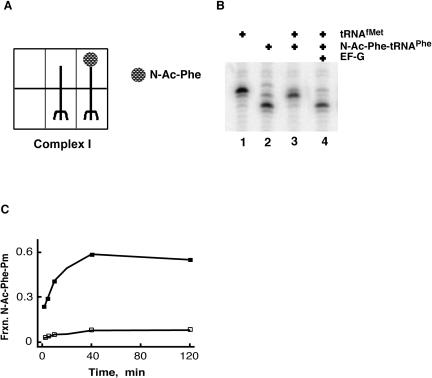
FIGURE 1.
Reactivity of pre-translocation-state ribosome complex I. (A) Complex I was prepared by successive binding of uncharged tRNAMet and N-Acetyl-Phe-tRNAPhe to ribosomes programmed with a gene 32 mRNA variant with AUG UUC UUC (MFF) as the first three codons in its open reading frame. (B) Primer extension analysis of complex I before and after incubation with EF-G and GTP. Ribosome complexes were formed on gene 32 variant mRNA pre-annealed to a [32P]-labeled primer. Markers include tRNAMet (lane 1) which yields a band at position 0, and tRNAPhe (lane 2) which yields a band at position +3. The pre-translocation-state complex (lane 3) has a diagnostic pattern with a doublet heavier at the +1 position. Movement to the +3 position is promoted by EF-G (lane 4). (C) Puromycin reactivity of complex I. Complex I was incubated with 10 mM puromycin at 37°C (open squares), and the fraction N-Ac-Phe converted to N-Ac-Phe-Pm is shown on the y-axis. Puromycin reactivity of the corresponding post-translocation complex (filled squares), formed after incubating complex I with 5 μM EF-G for 5 min at 37°C, is also shown.
We next looked at the reactivity of the pre- and post-translocation-state complexes with the minimal A-site substrate puromycin. The dipeptidyl product (N-acetyl-[14C]-phenylalanine-puromycin) was separated from free N-acetyl-[14C]-phenylalanine and quantitated as described (Southworth et al. 2002). As predicted, in the pre-translocation-state ribosome complex, the N-Ac-Phe-tRNAPhe was not reactive with puromycin, even after prolonged periods of time. However, when EF-G and GTP were added to the pre-translocation complex, essentially all of the N-Ac-Phe-tRNAPhe eventually reacted with puromycin, consistent with the observed movement of the tRNA substrates from a pre- to a post-translocation state (Fig. 1C ▶).
In other experiments, pre-translocation-state ribosome complexes (complex II) were prepared by enzymatically loading each of two aminoacylated tRNA substrates (Semenkov et al. 2000). In this case, ribosomes were programmed with either gene 32 (MFK) mRNA or a variant (AUG-UUC-UUC, MFF), and then fMet-tRNAMet was bound to the AUG-coded P site using initiation factors (IF1, IF2, and IF3) and GTP. Following this more natural initiation step, Phe-tRNAPhe was enzymatically loaded into the ribosome with purified EF-Tu in a ternary complex with GTP. The ribosome catalyzes peptide-bond formation when two competent substrates are loaded into the A and P sites, resulting in a deacylated tRNAMet and a dipeptidyl-tRNA (fMet-Phe-tRNAPhe) on the ribosome. According to convention, this ribosome complex is in a pre-translocation state, awaiting EF-G for elongation to continue (Fig. 2A ▶). Toeprinting experiments confirm that the mRNA:tRNA complexes were positioned as expected prior to translocation, and that three-nucleotide movement along the mRNA is seen following the addition of EF-G and GTP (Fig. 2B ▶).
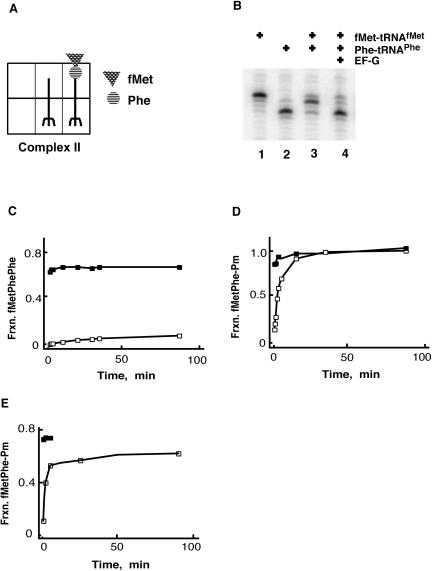
FIGURE 2.
Reactivity of pre-translocation-state ribosome complex II. (A) Complex II was prepared by successive factor-dependent binding of fMet-tRNAMet and Phe-tRNAPhe to the P and A sites of ribosomes programmed with gene 32 variant mRNA (as above) where peptide-bond formation generates deacylated and dipeptidyl tRNA species. (B) Primer extension analysis of complex II before and after incubation with EF-G and GTP. Markers include tRNAMet (lane 1) which yields a band at position 0, and tRNAPhe (lane 2) which yields a band at position +3. The pre-translocation-state complex (lane 3) has a diagnostic pattern with a doublet heavier at the +1 position. Movement to the +3 position is promoted by EF-G (lane 4). (C) Complex II was incubated with equimolar Phe-tRNAPhe in complex with EF-Tu/GTP before (open squares) and after (filled squares) translocation with 5 μM EF-G (as above). The fraction of fMet-Phe converted to fMet-Phe-Phe is shown on the y-axis. (D) Puromycin reactivity of complex II in buffer B with 7 mM MgCl2. Complex II was incubated with 10 mM puromycin, and the fraction of fMet-Phe converted to fMet-Phe-Pm was determined before (open squares) and after (filled squares) translocation with 5 μM EF-G (as above). (E) Puromycin reactivity of complex II in polymix buffer (Jelenc and Kurland 1979). Complex II was incubated with 10 mM puromycin, and the fraction of fMet-Phe converted to fMet-Phe-Pm was determined before (open squares) and after (filled squares) translocation with 5 μM EF-G (as above).
These data are consistent with the idea that well behaved pre-translocation-state ribosome complexes can be formed either by prebinding a deacylated tRNA and then loading a peptidyl-like P-site substrate (N-Ac-Phe-tRNAPhe; complex I) or by enzymatically loading two aminoacylated tRNAs that then react with one another to establish a pre-translocation state (complex II). Both complexes have characteristic toeprints before and after the addition of EF-G and GTP (Figs. 1B ▶, 2B ▶).
We next asked whether the pre-translocation complex with the bound dipeptidyl-tRNA (complex II) was reactive with different aminoacyl tRNA substrates. The third codon in the variant gene 32 mRNA is UUC, allowing us to ask whether the dipeptidyl-tRNA in the pre-translocation state is reactive with Phe-tRNAPhe (bound in a ternary complex with EF-Tu and GTP) in the absence of EF-G. Again, the precursors and products of the reaction were resolved by electrophoresis and quantitated as described (Southworth et al. 2002). As would be predicted, the dipeptidyl-tRNA substrate (fMet-Phe-tRNAPhe) was unreactive with Phe-tRNAPhe unless EF-G was provided to promote movement to a post-translocation state (Fig. 2C ▶). Similar results were observed with the wild-type gene 32 mRNA carrying a lysine codon (AAA) in the third position when Lys-tRNALys was added to react with the pre- and post-translocation-state ribosome (data not shown). However, when the dipeptidyl-tRNA ribosomal complex was incubated with the minimal A-site substrate puromycin, the dipeptidyl-tRNA reacted efficiently to form the tripeptide fMet-Phe-Pm (Fig. 2D ▶). The kinetics of the reaction were uniform, proceeding nearly to completion, and could be fit to a single exponential curve with a rate constant of 0.014 sec−1.
Because previous cryoEM studies indicated that tRNA binding can be affected by buffer choice, we asked whether these dipeptidyl-tRNA pre-translocation-state ribosome complexes were similarly reactive in a variety of conditions. Interestingly, the rate constants and endpoints for the puromycin reaction with pre-translocation-state ribosomes were essentially the same in “simple” buffers containing different amounts of Mg+2, 7 mM or 15 mM, as well as in polyamine buffers, allowing direct comparison with recent studies (Fig. 2E ▶; Agrawal et al. 1999; Zavialov and Ehrenberg 2003).
Pre-translocation-state dipeptidyl-tRNA reacts from an intermediate (A/P) state of binding on the ribosome
Several mechanisms can be proposed to explain the puromycin reactivity of the dipeptidyl-tRNA ribosome pre-translocation complex II. One possibility is that the dipeptidyl-tRNA reacts when stably bound in the pre-translocation state, as has been suggested (Semenkov et al. 1992). Alternatively, the dipeptidyl-tRNA ribosome complex translocates spontaneously (or in a manner dependent on minor EF-G contamination), and it is the post-translocation ribosome complex that actually reacts with puromycin, as has also been suggested (Bergemann and Nierhaus 1983). Similarly, it is possible that puromycin facilitates actual translocation, as was recently observed with the antibiotic sparsomycin (Fredrick and Noller 2003). Finally, the reactivity of the dipeptidyl-tRNA might be explained by dissociation of the dipeptidyl-tRNA and the deacylated tRNAfMet followed by rebinding of the dipeptidyl-tRNA, now in a classical (post-translocation) state of binding. In the latter cases, puromycin would be predicted to react from the classical A site with the dipeptidyl tRNA substrate bound in the classical P site in a post-translocation-state ribosome complex (as has been characterized in detail by Katunin et al. 2002).
We first asked whether the pre-translocation-state ribosome complex translocates slowly over time as a result either of minor levels of EF-G contamination or via factor-independent translocation (Gavrilova et al. 1976; Southworth et al. 2002). If spontaneous translocation does occur, the resulting post-translocation state should be stable. Thus, incubation of the pre-translocation-state ribosome complex (II) for varying amounts of time should yield mixed populations of ribosome states—an original population that reacts with puromycin at a rate constant of 0.014 /sec, and the translocated population that reacts with puromycin at a rate constant of 10/sec (Katunin et al. 2002). These populations can be readily distinguished using kinetic approaches.
The stability of pre-translocation-state ribosome complexes to translocation was tested as follows. Pre-translocation dipeptidyl-tRNA ribosome complexes were formed as above (II) and were incubated at 37°C for varying lengths of time (3, 10, or 45 min) before the addition of puromycin. Following these incubations, homogeneous unaffected rate constants (Fig. 3A ▶) were measured for the puromycin reaction, indicating that the dipeptidyl-tRNA pre-translocation ribosome complexes do not translocate or re-phase (either at 7 mM or 15 mM MgCl2) during the time course of these incubations. These data thus formally exclude potential mechanisms for puromycin reactivity that invoke conversion of the pre-translocation-state complex into a post-translocation-state complex.
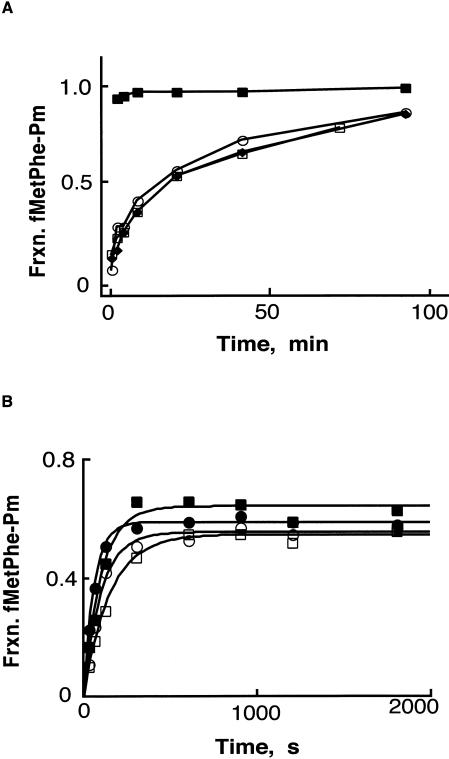
FIGURE 3.
Stability of pre-translocation ribosome complex II. (A) The pre-translocation complex II was incubated at 37°C for 2 (open circles), 10 (filled diamonds) or 45 (open squares) min, before starting the reaction with the addition of puromycin (1 mM). The fraction of fMet-Phe converted to fMet-Phe-Pm was determined where the time of addition of puromycin is taken as time zero in each case. As a control, puromycin reactivity of the post-translocation complex (filled squares) was determined after incubating complex II with 5 μM EF-G (as in Fig. 2 ▶). (B) Chase experiment performed with N-Ac-Phe-tRNAPhe with pre-translocation-state complex II. Reaction was started with the simultaneous addition of 4 equivalents of N-Ac-Phe-tRNAPhe (1 μM final) and 10 mM puromycin, and time points were taken. The fraction of fMet-Phe converted to fMet-Phe-Pm was determined in the absence or presence (filled or open symbols, respectively) of tRNA competitor in buffer B at 7 mM or 15 mM MgCl2 (circles or squares, respectively).
These data argue that the dipeptidyl-tRNA complex reacts with puromycin from a pre-translocation state. However, if there are significant off-rates for the dipeptidyl-tRNA from the pre-translocation ribosome, as has been reported (Semenkov et al. 2000), the rebinding kinetics of the dipeptidyl-tRNA might dominate the measured rate constants for the puromycin reaction. This possibility was tested by performing a chase experiment where excess (fourfold) unlabeled N-Ac-Phe-tRNAPhe was added to the dipeptidyl-tRNA pre-translocation ribosome complex along with puromycin to ask whether the extent or rate of reactivity was affected. If the dipeptidyl-tRNA dissociates during the course of reaction, then the N-Ac-Phe-tRNAPhe will compete for binding to the ribosome with the dipeptidyl-tRNA and thus affect the puromycin reactivity of the complex. The absence of a chase by the N-Ac-Phe-tRNAPhe with the pre-translocation-state dipeptidyl-tRNA complex at both lower and higher magnesium concentrations (Fig. 3B ▶) suggests that the observed rate constants are pseudo-first-order for the reactivity of the pre-translocation-state ribosome complex. When pre-translocation complexes were allowed to sit for extended periods of time at lower Mg+2 concentrations, the N-Ac-Phe-tRNAPhe did effectively chase the reaction, consistent with previously measured off-rates of the dipeptidyl tRNA from this complex (Semenkov et al. 2000).
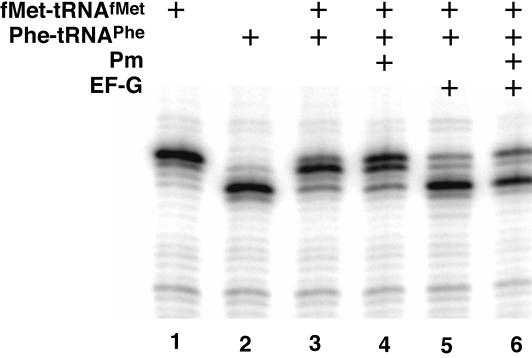
To independently assess the state of binding of tRNAs to the pre-translocation ribosome complex before and after reaction with puromycin, we used toeprinting analysis. These data, shown in Figure 4 ▶, clearly indicate that the dipeptidyl-tRNA is positioned in the A site on the 30S subunit both before and after the reaction with the puromycin substrate. Furthermore, as expected, when EF-G is added to the ribosome complexes that have already reacted with puromycin, the predicted three-nucleotide movement of translocation can still be observed.
FIGURE 4.
Primer extension analysis (toeprinting) of complex II before and after reaction with puromycin. Ribosome complexes were formed on gene 32 variant mRNA pre-annealed to a [32P]-labeled primer. As controls, the first and second codons in the gene 32 variant open reading frame were bound by their cognate tRNAs, fMettRNAMet (lane 1) or Phe-tRNAPhe (lane 2), alone before carrying out the primer extension reaction. Lanes 3 and 4: The toeprint for complex II, before and after reaction with puromycin, respectively. Lanes 5 and 6: The toeprint for complex II, before and after reaction with puromycin, and after incubation with 5 μM EF-G (as above).
Substrate specificity for pre-translocation-state puromycin reactivity mimics that of the post-translocation state
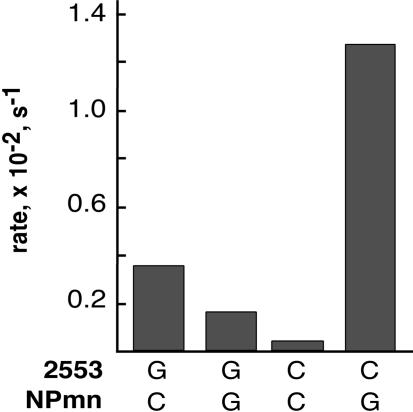
Aminoacyl-tRNAs bound in the A site of the ribosome interact via their CCA-end in a Watson-Crick pairing interaction between C75 of the tRNA and nucleotide G2553 of 23S rRNA (Kim and Green 1999; Nissen et al. 2000). We asked whether the puromycin reaction that was observed with the pre-translocation-state dipeptidyl-tRNA complex depended on the same interactions of the ribosome with the A-site substrate. G2553C mutant ribosomes were produced in Escherichia coli and separated from wild-type ribosomes using a recently developed affinity-tagging procedure (E. Youngman and R. Green, in prep.). Wild-type and variant ribosomes (G2553C) carrying pre-translocation-state bound dipeptidyl-tRNA complex were reacted with CpPm or GpPm substrate, and the rates of the reactions were compared. The data shown in Figure 5 ▶ indicate that the substrate specificity of the wild-type and mutant (G2553C) ribosomes for the cognate Watson-Crick substrate is equivalent to the substrate specificity observed for post-translocation-state ribosomes with the same dipeptidyl-tRNA substrate (Kim and Green 1999). These data confirm that the puromycin reactivity of the pre-translocation-state complexes depends on canonical A-site interactions.
FIGURE 5.
Compensatory base analysis of G2553 mutant ribosomes with C75 tRNA mutant A-site substrates in complex II. The rate of reactivity of the wild-type and mutant ribosome complexes with 8 μM CpPm and GpPm was determined (Kim and Green 1999).
K1/2 and kcat of puromycin reaction of the dipeptidyl-tRNA bound pre-translocation state
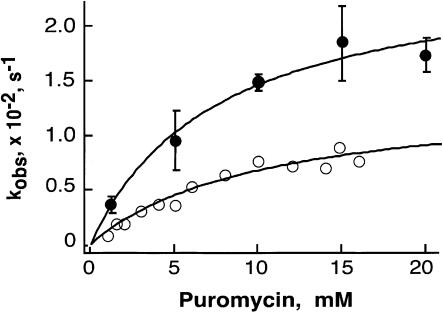
To further characterize the molecular properties of the pre-translocation-state reactivity, we measured the kcat and K1/2 of this reaction at two different magnesium concentrations (7 and 15 mM MgCl2; Fig. 6 ▶; Table 1 ▶). The kinetics of the pre-translocation-state peptide-bond-forming activity were well behaved, and the time courses at different puromycin concentrations were readily fit to single exponential curves. These data were then fit to the Michaelis-Menten equation to derive the kinetic parameters shown in Table 1 ▶. Although the apparent K1/2 of puromycin for the pre-translocation state is comparable to the K1/2 reported for the post-translocation state (2 versus 5.9 or 10.3 mM), the rate constants for these reactions differ by approximately three orders of magnitude (11.6 versus 0.024 or 0.014 sec−1 for the post- and prestates respectively).
FIGURE 6.
Dependence of pre-translocation-state puromycin reactivity on puromycin concentration. The time courses of fMet-Phe-Pm formation by complex II were fitted to single exponential rate equations in order to obtain the kobs for each puromycin concentration in buffer B with either 7 mM or 15 mM MgCl2. Fitting the data to the Michaelis-Menten equation gave a K1/2 of 5.9 or 10.3 mM and a kcat of 0.024 or 0.014 s−1.
TABLE 1.
Kinetic parameters of the puromycin reactivity of pre- and posttranslocation complexes with dipeptidyl fMet-Phe-tRNAPhe
| K1/2 for puromycin (mM) | kcat (s−1) | |
| Pretranslocation (7 mM) | 5.9 ± 1.7 | 0.024 ± 0.003 |
| Pretranslocation (15 mM) | 10.3 ± 2.6 | 0.014 ± 0.002 |
| Posttranslocation | 2.0 ± 0.55 | 11.6 ± 1.0 |
Data for pretranslocation complexes measured in buffer B at either 7 or 15 mM MgCl2.
DISCUSSION
The structural state of the ribosome and the bound mRNA: tRNA complex following peptide-bond formation and preceding EF-G-catalyzed translocation has been actively debated. Chemical modification/protection experiments and fluorescent studies support a model where the tRNAs move through the ribosome in a staggered fashion (Hardesty et al. 1986; Moazed and Noller 1989). As specifically outlined in the hybrid states model, following peptide-bond formation, the acceptor ends of the tRNAs can spontaneously move into the E and P sites of the large subunit (from the P and A sites), whereas the anticodon ends of the tRNAs remain fixed in the small subunit, where they interact with the bound mRNA. This hybrid ribosome complex is then recognized by EF-G to promote the events of translocation, where the anticodon ends of the tRNAs (and the associated mRNA) are moved into their next binding state (the E and P sites on the small subunit). Indeed, structural data have provided compelling evidence that the footprints defined by chemical modification analysis correlate reasonably well with the three observed tRNA binding sites (Yusupov et al. 2001) and that tRNAs can bind in a hybrid state (Agrawal et al. 1999; Valle et al. 2003).
Here we used a biochemical approach to define some of the basic functional properties of the pre-translocation state of the ribosome. First, pre-translocation-state ribosomes occupied with dipeptidyl-tRNA readily react with the minimal aminoacyl substrate puromycin but not with intact aminoacylated tRNAs (Fig. 2 ▶). In contrast, post-translocation-state ribosomes occupied with dipeptidyl-tRNA (i.e., complexes that have been treated with EF-G following peptide-bond formation) readily react with both puromycin and the appropriate intact aminoacyl tRNA (Fig. 2 ▶). The apparent K1/2 for puromycin for the pre-translocation-state ribosome is only fivefold higher than for the post-translocation state, whereas the observed rate constant (kobs) for the peptidyl transferase reaction is substantially lower (decreased ~1000-fold; Fig. 6 ▶; Table 1 ▶). The simplest interpretation of these data is that although binding determinants in the A site of the ribosome seem to be largely unperturbed, moieties critical to the catalysis of peptide-bond formation or substrate positioning are more substantially perturbed in this intermediate state (affecting the rate of catalysis). The stability of the pre-translocation-state ribosome complexes was assessed using tRNA chases and extended incubations of the complexes at 37°C to confirm that the extent and rates of reaction with puromycin were unaffected (Fig. 3 ▶). Primer extension analysis established that following the reaction of the pre-translocation-state bound dipeptidyl-tRNA with puromycin, the tRNA and mRNA substrates remain fixed in a pre-translocation state with respect to the small subunit (Fig. 4 ▶). Finally, experiments using G2553C mutant ribosomes and modified puromycin substrates showed that the substrate binding site that is critical for pre-translocation-state peptidyl transferase reactivity is related to the A site that was characterized previously (Kim and Green 1999), based on its reactivity with substrates bound in the post-translocation state (Fig. 5 ▶). These data support a model where the pre-translocation-state bound peptidyl tRNA must either reside in or sample a hybrid (A/P) state of binding on the ribosome. Further experiments will be critical in defining the stability of this intermediate state of tRNA binding under physiological conditions, to better understand how and when EF-G interacts with the ribosome to promote translocation.
Overall, the data presented here are consistent with experimental observations reported by Semenkov et al (1992). In those studies, a pre-translocation-state ribosome complex was formed on a poly-U messenger RNA with two charged tRNAs (N-Ac-Phe-tRNAPhe and Phe-tRNAPhe) that reacted to form a dipeptidyl-tRNA N-Ac-Phe-Phe-tRNAPhe, and it was reported that this dipeptidyl tRNA substrate reacted efficiently with the minimal A-site substrate puromycin. Semenkov et al. (1992) argued that the reactivity was associated with the pre-translocation state, although they failed to independently assess translocation (e.g., by toeprinting) before and after the reaction with puromycin. Our present data provide convincing evidence that the dipeptidyl-tRNA bound to the ribosome in a pre-translocation state does indeed react with puromycin (meaning that the A site on the large subunit must be accessible to some extent), while the anticodon ends of the tRNAs remain fixed on the small ribosomal subunit in the A site—in an intermediate state of binding. Because our experiments cannot specify whether the deacylated tRNA is bound in the P/E state, we use the term “intermediate” rather than “hybrid” to describe the complex, although chemical footprinting and cryoEM data argue that the intermediate state occupied by both tRNAs just prior to translocation is likely to be hybrid (A/P and P/E; Moazed and Noller 1989; Agrawal et al. 1999).
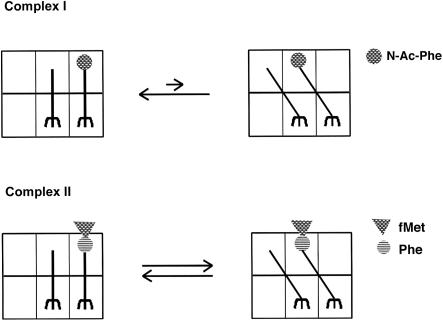
Although the data presented here establish that a hybrid state of tRNA binding is at least sampled by a dipeptidyl-tRNA bound in a pre-translocation state on the ribosome, they do not address the equilibrium binding state of this tRNA species. The strikingly different reactivity profiles of the pre-translocation N-acetylated aminoacyl tRNA and the dipeptidyl tRNA and the strong dependence of the puromycin reactivity on temperature (e.g., even the dipeptidyl tRNA pre-translocation complex is essentially unreactive on ice) may provide clues about the equilibrium binding states of these and other substrates. Perhaps the N-protected aminoacyl tRNA generally occupies the classical (A/A) state (i.e., with only rare excursions into the A/P hybrid state) such that even during extended incubations with puromycin, no peptidyl transfer reaction occurs. According to such a model, the dipeptidyl tRNA more readily samples both the classical (A/A) and hybrid (A/P) binding state following peptide-bond formation (Fig. 7 ▶). And, in the latter case, these excursions of the dipeptidyl tRNA into the hybrid binding state are sufficient to allow puromycin to trap the dipeptidyl moiety and form a peptide bond. Alternatively, the differences in reactivity of the N-Acetyl-Phe-tRNAPhe relative to the dipeptidyl-tRNA might be reconciled by differences in the detailed positioning (for peptide-bond formation) of the “peptidyl” moiety of the tRNA in the A/P state. For example, a more complete peptidyl moiety might allow for more extensive positioning of this substrate in the exit channel so that the aminoacyl ester bond is more optimally exposed for attack. These models might ultimately be distinguished by chemical modification analysis.
FIGURE 7.
Model for sampling of intermediate (hybrid) tRNA binding states by pre-translocation-state bound N-Ac-Phe-tRNAPhe and fMet-Phe-tRNAPhe, where the dipeptidyl tRNA equilibrium favors hybrid-state tRNA binding compared with the equilibrium for N-Acetyl-Phe-tRNAPhe.
Consistent with such a model, it is worth noting that although ribosome complexes containing either the Nacetylated or the dipeptidyl-tRNA substrate are translocated effectively by EF-G, the rates of these reactions are likely to differ substantially. For example, it is known that dipeptidyl tRNA (fMet-Phe-tRNAPhe)-containing pre-translocation ribosome complexes are translocated approximately one order of magnitude more rapidly than complexes containing two deacylated tRNA species (Studer et al. 2003) and more than two orders of magnitude more rapidly than complexes containing Phe-tRNAPhe in the A site (Semenkov et al. 2000). The recent measurements of the binding affinity of pre-translocation-state dipeptidyl tRNA for the ribosome were consistent with these ideas and further suggested that one of the mechanisms utilized by the ribosome to promote translocation is ground-state destabilization (Semenkov et al. 2000).
A unifying interpretation of these and other data is that there are binding determinants for EF-G that are revealed in an unobstructed A site on the large subunit. It follows that translocation is most rapid on ribosome complexes containing peptidyl-tRNAs bound in a hybrid (A/P) state. Thus, the first energetic barrier that must be surmounted during translocation is the movement of the pre-translocation-state tRNAs into a hybrid state of binding, and then in a second step, movement of the mRNA:tRNA complex on the small subunit can take place. Mutations in the ribosome or the tRNA substrates that result in preferential binding of tRNAs in a hybrid state should lead to increased rates of spontaneous translocation and may even affect the rates of EF-G-catalyzed translocation. Preliminary data in our laboratory that are consistent with such a model show that ribosomes covalently modified at their A loop with the minimal A-site substrate 4-thiodT-CPm are refractive to translocation by EF-G (Green et al. 1998; D. Sharma and R. Green, unpubl.). Such a model is also consistent with recent results from Noller and colleagues (Fredrick and Noller 2003), who report that sparsomycin, an antibiotic that binds in the peptidyl transferase center and stabilizes the interaction of the P-site substrate with its binding site, promotes translocation at a substantial rate. A plausible interpretation of these data is that sparsomycin forces binding of the tRNA substrates in a hybrid, somehow “activated” state that pays part of the energetic cost of translocation (Southworth and Green 2003). Occupation of the E site by deacylated tRNAs once translation is past the dipeptide stage is also likely to affect the equilibrium binding states of the various tRNA substrates (Rheinberger and Nierhaus 1986) and thus the rates at which translocation can occur. Interactions between the deacylated P-site tRNA and the E site of the large subunit that appear to increase the efficiency of translocation have been characterized (Lill et al. 1989; Feinberg and Joseph 2001).
Several other published experiments are often cited for their failure to support a hybrid states model for two-step translocation. The data presented here help to resolve some of these specific issues. For example, the failure of Wower et al. (2000) to observe hybrid-state binding in their crosslinking studies can be rationalized by the fact that their pre-translocation state was not composed of a dipeptidyl-tRNA and instead relied on the peptidyl tRNA analog N-Ac-Phe-tRNAPhe, which we find to be unreactive with puromycin, and thus likely not sampling the hybrid state. The fact that Schmeing et al. (2002) did not see minimal tRNA substrates moving in the crystal lattice to a “hybrid state” following peptide-bond formation seems trivially explained either by the constraints of the crystal lattice or, more likely, by the absence of the small subunit and the associated strain of the bound full-length tRNAs. These features seem likely to be critical in stabilizing hybrid-state binding of the tRNA substrates to the ribosome. Indeed, Schmeing et al. simply argued that their structure provided evidence of an intermediate state of tRNA binding that precedes the hybrid state.
The interpretation of recent biochemical and cryoEM data (Valle et al. 2003; Zavialov and Ehrenberg 2003) conflict with some of the views presented here. Those studies looked at the reactivity of pre-translocation-state dipeptidyl-tRNA complexes with puromycin and RF2 (a substrate dependent on binding to a complete A site) and concluded that intermediate, hybrid states of tRNA binding were not spontaneously sampled by pre-translocation-state ribosomes. Instead, those authors proposed that EF-G binding is required to promote movement of both ends of the tRNAs with respect to the ribosome. Although a systematic comparison of their approaches and data suggests that some differences in experimental conditions may be significant, the biggest distinction between these reports and our own is in the interpretation of the data. Although those authors did not provide rate constants for puromycin reactivity with the pre-translocation state, they do indicate that there is “very slow” reactivity of the pre-translocation-state complexes with puromycin in the absence of active EF-G. Indeed, we report pseudo-first-order rate constants for this reaction that are three orders of magnitude slower than those for dipeptidyl-tRNA bound in a post-translocation state. The slower overall rate of the puromycin reaction that they observed can likely be attributed to their use of subsaturating puromycin concentrations of 0.4 mM (relative to 10 mM used in our experiments). Indeed, when we use 0.4 mM puromycin concentrations in polymix buffer (Jelenc and Kurland 1979), our rate constants are at least 10-fold slower than with higher puromcyin concentrations, and the endpoints of the reaction are diminished.
The authors also present cryoEM data showing that the P-site deacylated tRNA can bind in a hybrid state on the ribosome when EF-G is present (Valle et al. 2003; Zavialov and Ehrenberg 2003). From this, they argue that movement of this tRNA to the hybrid state is a consequence of EF-G binding to an “unlocked” ribosome (where the P-site tRNA is deacylated). These conclusions are supported by the observation of increased levels of EF-G binding and EF-G-stimulated GTP hydrolysis on such ribosomes carrying a single deacylated tRNA in the P site (rather than a peptidyl tRNA). However, because translocation does not occur in the absence of at least a minimal A-site substrate (e.g., an ASL; Joseph and Noller 1998), the relevance of EF-G interacting with this particular ribosomal complex is uncertain. Overall, their data (Valle et al. 2003; Zavialov and Ehrenberg 2003) are rather consistent with what one would predict from previous biochemistry suggesting that an authentic pre-translocation-state ribosome carries a deacylated tRNA in the P site (Lill et al. 1989). At issue is whether movement of the ribosome complex into the hybrid state precedes or follows EF-G binding. An alternate view of their data that is easily reconciled with our own data is that hybrid binding states are readily sampled by pre-translocation ribosome complexes and that EF-G binds to and effectively traps this hybrid-state ribosomal complex, eventually promoting movement of the mRNA:tRNA complex on the small subunit.
Thus, we would argue that there remains ambiguity in the ordering of events during translocation—does tRNA movement into hybrid states take place before or after EF-G binds to pre-translocation-state ribosomes? The basic results of our experiments and those of others are similar (Semenkov et al. 1992)—pre-translocation-state ribosome complexes do react with puromycin at a substantial rate (i.e., they do sample a hybrid state), and the question is how significant this sampling of hybrid tRNA binding states might be during the elongation process. In our view, the results are equivocal. However, the fact that the pre-translocation-state ribosome so readily samples a hybrid state of tRNA binding (even in polymix buffers) argues that it is likely to be relevant to the normal cycle of elongation. Further structural and biochemical experiments will be required to establish the actual state of the ribosome that is recognized and acted on by EF-G to promote translocation.
MATERIALS AND METHODS
Assay components
Ribosome 70S particles were purified from E. coli as described (Moazed and Noller 1989). E. coli tRNAPhe and tRNAfMet were purchased from Sigma and aminoacylated with [14C]-phenylalanine and [35S]-methionine, respectively; N-acetylation and formylation were performed as described (Semenkov et al. 2000). The aminoacylation efficiency for the tRNAs ranged from 75% to 95% of the input tRNA as determined by TCA precipitation. The aminoacylated tRNAs were stored in 20 mM potassium acetate, pH 5.5 at −80°C until further use. The gene 32 mRNA templates were prepared by in vitro transcription by phage T7 RNA polymerase (Hartz et al. 1988).
E. coli recombinant His-tagged EF-G was expressed from plasmid pET24b provided by K. Wilson and H. Noller (University of California, Santa Cruz) and purified as described (Wilson and Noller 1998). E. coli recombinant His-tagged EF-Tu and IF2 were provided by C. Merryman (Johns Hopkins University). IF1 and IF3 were overexpressed and purified as described (Pawlik et al. 1981). AMV-RT was purchased from Seikagaku America.
Pre- and post-translocation ribosome complexes
Ribosome pre-translocation complex I was prepared in buffer A (50 mM Tris-HCl, pH 7.6, 15 mM MgCl2, 160 mM NH4Cl, and 5 mM β-mercaptoethanol) at 37°C, if not stated otherwise. The 70S (0.75 μM) ribosomal particles were programmed with a twofold excess of gene 32 (MFK) mRNA (coding sequence Met-Phe-Lys) in the presence of 2.5-fold excess of tRNAfMet for 10 min at 37°C. N-acetyl-[14C]-Phe-tRNAPhe (0.5 μM) was added and incubation continued at 37°C for 15 min as described (Southworth et al. 2002).
Ribosome pre-translocation complex II was prepared in buffer B (50 mM Tris HCl, pH 7.5, 70 mM NH4Cl, 30 mM KCl, 1 mM DTT, and MgCl2 as specified) at 37°C by a modification of the method described by Semenkov et al. (2000). To prepare 70S initiation complexes, 70S ribosomes (1.5 μM) were programmed with a fourfold excess of either MFK or variant MFF gene 32 mRNA, in the presence of 1.5-fold excess of initiation factors 1, 2, and 3, and equimolar N-formyl-[35S]-Met-tRNAMet and 1 mM GTP, in buffer B containing 7 mM Mg2+ for 30 min at 37°C. The ternary complex was prepared by incubating 1.5 μM EF-Tu with 1.5 μM [14C]-Phe-tRNAPhe and 1 mM GTP in buffer B containing 7 mM Mg2+ at 37°C for 15 min. The ternary complex (final concentration 0.5 μM) was added to the initiation complex (final concentration 0.5 μM), and the magnesium concentration (MgCl2) was increased up to 15 mM final, by adding 1/9 volume of 80 mM MgCl2. The initiation complex and the ternary complex were combined, and the incubation continued at 37°C for 2 min. Cellulose TLC electrophoretic analysis (see below) of the reaction indicated that 80%–90% of the f-[35S]-Met-tRNAMet reacted with the [14C]-Phe-tRNAPhe to form the dipeptide product.
Incubation of the pre-translocation-state complexes with EF-G (5 μM) and GTP (1 mM) complex at 37°C for 5 min converted the pre-translocation-state complexes to a post-translocation state, as assessed by primer extension analysis (see below).
Puromycin reactions were carried out by the addition of equal volumes of puromycin solution in the corresponding buffer to the pre- or post-translocation ribosomal complexes I and II. A version of gene 32 mRNA encoding Met-Phe-Phe (MFF) was used to check the reactivity of the ribosome complexes with Phe-tRNAPhe before and after translocation with EF-G. The reactions were stopped by addition of 0.2 N KOH and incubation for 10 min at 37°C. This treatment released the peptides from the tRNAs, allowing them to be resolved electrophoretically. The unreacted f-Met amino acid and the various peptide products were resolved by electrophoresis on cellulose TLC sheets in pyridine:acetic-acid:water (1:40:160) solution. Phosphorimager analysis was done using a Typhoon phosphorimaging system.
Primer extension (toeprint) analysis
Toeprint analysis was carried out essentially as described (Joseph and Noller 1998). Ribosomal complexes were formed as described above, except that the message was provided in equimolar ratios to the ribosomes and was pre-annealed to a cDNA primer (5′-TTT ATCTTCAGAAGAAAAACC-3′) labeled with [32P] at its 5′ end. Translocations with EF-G and puromycin reactions were carried out as described in the previous section. Primer extension on the ribosomal complexes was performed by diluting the reactions into primer extension mix (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 60 mM NH4Cl, 6 mM β-mercaptoethanol, 450 μM dNTPs, and 2 units of AMV-RT) and incubating for 15 min at 37°C. The primer extension products were resolved by electrophoresis on 8% acrylamide, 7 M urea, 1X TBE gels. Markers for P-site occupancy of the first (Met) and second (Phe) codons were obtained by toeprinting the ribosome complexes bound to gene 32 message and only one of the tRNAs, fMet-tRNAMet or Phe-tRNAPhe.
Kinetic experiments
The post-translocation complexes for kinetic analysis were prepared as described above and diluted to 0.25 μM in buffer B. The rates of reaction of the dipeptidyl tRNA, fMet-Phe-tRNAPhe, with puromycin in post-translocation complex II were determined essentially as described (Katunin et al. 2002). Reactions were stopped with 30 mM EDTA, base-treated with 0.2 N KOH, and analyzed on the cellulose electrophoresis system, as described above. The reaction plots were fit to a first-order exponential equation, using KaleidaGraph software. The puromycin concentration-dependence plots were fit to a Michaelis-Menten equation to obtain the K1/2 for puromycin and the kcat for the peptidyl transferase reaction of the pre-translocation-state ribosomes.
Preparation of mutant ribosomes
E. coli ribosomes bearing mutations at position 2553 in the 23S rRNA were prepared by a recently developed affinity-tagging approach (E. Youngman and R. Green, in prep.). The mutations were introduced at 2553 by site-directed mutagenesis of a plasmid-borne version of 23S rRNA carrying an MS2 bacteriophage RNA element under the control of the lambda promoter. The 23S rRNA gene was expressed in an inducible system in E.coli DH10 cells as described. The mutant and wild-type MS2-tagged ribosomes were purified by a single pass over a glutathione column bearing GST-MS2 chimeric protein to greater than 95% homogeneity.
Acknowledgments
We thank S. Dorner, C. Merryman, and other members of our laboratory for discussions, and H.F. Noller and V. Ramakrishnan for helpful comments on the manuscript. This work was supported by grants from the National Institutes of Health and the Howard Hughes Medical Institute. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5148704.
REFERENCES
- Agrawal, R.K., Penczek, P., Grassucci, R.A., Burkhardt, N., Nierhaus, K.H., and Frank, J. 1999. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J. Biol. Chem. 274: 8723–8729. [DOI] [PubMed] [Google Scholar]
- Bergemann, K. and Nierhaus, K.H. 1983. Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J. Biol. Chem. 258: 15105–15113. [PubMed] [Google Scholar]
- Bretscher, M.S. 1968. Translocation in protein synthesis: A hybrid structure model. Nature 218: 675–677. [DOI] [PubMed] [Google Scholar]
- Burkhardt, N., Junemann, R., Spahn, C.M., and Nierhaus, K.H. 1998. Ribosomal tRNA binding sites: Three-site models of translation. Crit. Rev. Biochem. Mol. Biol. 33: 95–149. [DOI] [PubMed] [Google Scholar]
- Cabrer, B., Vazquez, D., and Modolell, J. 1972. Inhibition by elongation factor EF G of aminoacyl-tRNA binding to ribosomes. Proc. Natl. Acad. Sci. 69: 733–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, D.M., Thompson, J., March, P.E., and Dahlberg, A.E. 2002. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 319: 27–35. [DOI] [PubMed] [Google Scholar]
- Feinberg, J.S. and Joseph, S. 2001. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc. Natl. Acad. Sci. 98: 11120–11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J. 2003. Electron microscopy of functional ribosome complexes. Biopolymers 68: 223–233. [DOI] [PubMed] [Google Scholar]
- Fredrick, K. and Noller, H.F. 2003. Catalysis of ribosomal translocation by sparsomycin. Science 300: 1159–1162. [DOI] [PubMed] [Google Scholar]
- Gavrilova, L.P., Kostiashkina, O.E., Koteliansky, V.E., Rutkevitch, N.M., and Spirin, A.S. 1976. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 101: 537–552. [DOI] [PubMed] [Google Scholar]
- Green, R., Switzer, C., and Noller, H.F. 1998. Ribosome-catalyzed peptide-bond formation with an A-site substrate covalently linked to 23S ribosomal RNA. Science 280: 286–289. [DOI] [PubMed] [Google Scholar]
- Hardesty, B., Odom, O.W., and Deng, H.-Y. 1986. The movement of tRNA through ribosomes during peptide elongation: The displacement reaction model. In The structure, function and genetics of ribosomes (eds. B. Hardesty and G. Kramer), pp. 495–508. Springer Verlag, New York.
- Hartz, D., McPheeters, D.S., Traut, R., and Gold, L. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164: 419–425. [DOI] [PubMed] [Google Scholar]
- Jelenc, P.C. and Kurland, C.G. 1979. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. 76: 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerinic, O. and Joseph, S. 2000. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 304: 707–713. [DOI] [PubMed] [Google Scholar]
- Joseph, S. and Noller, H.F. 1998. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 17: 3478–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin, V.I., Muth, G.W., Strobel, S.A., Wintermeyer, W., and Rodnina, M.V. 2002. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10: 1–20. [DOI] [PubMed] [Google Scholar]
- Kim, D.F. and Green, R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4: 859–864. [DOI] [PubMed] [Google Scholar]
- Lill, R., Robertson, J.M., and Wintermeyer, W. 1989. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 8: 3933–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.L. 1972. Elongation factors EF Tu and EF G interact at related sites on ribosomes. Proc. Natl. Acad. Sci. 69: 752–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D. and Noller, H.F. 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Robertson, J.M., and Noller, H.F. 1988. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334: 362–364. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Odom, O.W., Picking, W.D., and Hardesty, B. 1990. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry 29: 10734–10744. [DOI] [PubMed] [Google Scholar]
- Pawlik, R.T., Littlechild, J., Pon, C.L., and Gualerzi, C. 1981. Purification and properties of Escherichia coli translational initiation factors. Biochem Int. 2: 421–428. [Google Scholar]
- Rheinberger, H.J. and Nierhaus, K.H. 1986. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. J. Biol. Chem. 261: 9133–9139. [PubMed] [Google Scholar]
- Richman, N. and Bodley, J.W. 1972. Ribosomes cannot interact simultaneously with elongation factors EF Tu and EF G. Proc. Natl. Acad. Sci. 69: 686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, D. 1972. Inability of E. coli ribosomes to interact simultaneously with the bacterial elongation factors EF Tu and EF G.Biochem. Biophys. Res. Commun. 46: 1850–1856. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Seila, A.C., Hansen, J.L., Freeborn, B., Soukup, J.K., Scaringe, S.A., Strobel, S.A., Moore, P.B., and Steitz, T.A. 2002. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9: 225–230. [DOI] [PubMed] [Google Scholar]
- Semenkov, Y., Shapkina, T., Makhno, V., and Kirillov, S. 1992. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 296: 207–210. [DOI] [PubMed] [Google Scholar]
- Semenkov, Y.P., Rodnina, M.V., and Wintermeyer, W. 2000. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 7: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Southworth, D.R. and Green, R. 2003. Ribosomal translocation: Sparsomycin pushes the button. Curr. Biol. 13: R652–R654. [DOI] [PubMed] [Google Scholar]
- Southworth, D.R., Brunelle, J.L., and Green, R. 2002. EFG-independent translocation of the mRNA:tRNA complex is promoted by modification of the ribosome with thiol-specific reagents. J. Mol. Biol. 324: 611–623. [DOI] [PubMed] [Google Scholar]
- Studer, S.M., Feinberg, J.S., and Joseph, S. 2003. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 327: 369–381. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M., and Frank, J. 2003. Locking and unlocking of ribosomal motions. Cell 114: 123–134. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Blaha, G., Connell, S.R., Ivanov, P.V., Jenke, H., Stelzl, U., Teraoka, Y., and Nierhaus, K.H. 2002. Protein synthesis at atomic resolution: Mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Pept. Sci. 3: 1– 53. [DOI] [PubMed] [Google Scholar]
- Wilson, K.S. and Noller, H.F. 1998. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell 92: 131–139. [DOI] [PubMed] [Google Scholar]
- Wool, I.G., Gluck, A., and Endo, Y. 1992. Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem. Sci. 17: 266–269. [DOI] [PubMed] [Google Scholar]
- Wower, J., Kirillov, S.V., Wower, I.K., Guven, S., Hixson, S.S., and Zimmermann, R.A. 2000. Transit of tRNA through the Escherichia coli ribosome. Cross-linking of the 3′ end of tRNA to specific nucleotides of the 23 S ribosomal RNA at the A, P, and E sites. J. Biol. Chem. 275: 37887–37894. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Zavialov, A.V. and Ehrenberg, M. 2003. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell 114: 113–122. [DOI] [PubMed] [Google Scholar]