Abstract
Telomerase is a ribonucleoprotein complex responsible for maintaining telomere length of eukaryotic chromosomes. Human telomerase has two main components, the human telomerase reverse transcriptase and the human telomerase RNA (hTR). Two domains of hTR essential for telomerase activity are the template domain, comprised of an 11-nt templating and alignment sequence, and the CR4/CR5 domain. Highly conserved residues in the CR4/CR5 domain form the stem–loop P6.1, which is important for assembly and activity of mammalian telomerase. Here, we have determined that stem–loop P6.1 can participate in a long-range RNA–RNA interaction with the template region of hTR. We characterized this interaction through mobility shift assays, mutation analysis, and UV cross-linking experiments. Mutation analysis revealed that the P6.1 loop nucleotides participate in the interaction with the template. The site of interaction at the template domain was determined via UV cross-linking experiments. These data show that an RNA–RNA interaction exists between two highly conserved regions of hTR that are critical for the higher order folding of telomerase RNA. This interaction argues for the proximity of the template and the CR4/CR5 domain, and provides the basis for a revised model of hTR, partitioning the RNA into a catalytic domain and a localization domain.
Keywords: telomerase RNA, RNA–RNA interaction, RNA structure, mobility shift assay, UV cross-linking
INTRODUCTION
Telomeres, the protective termini of eukaryotic chromosomes, are comprised of G/T-rich sequences (TTAGGG in vertebrates) and their associated proteins (Blackburn 1991; reviewed in Blackburn 2000 and Cech 2000). Telomerase is a reverse transcriptase responsible for the synthesis and addition of telomeric repeats to chromosomal ends (Greider and Blackburn 1985). This ribonucleoprotein complex acts to balance the effects of natural chromosomal shortening that occurs through incomplete DNA replication. Though telomerase activity is not detectable in normal human cells, telomerase is active in roughly 85%–90% of oncogenic cells (Kim et al. 1994). In 1999, Weinberg and coworkers showed that the oncogenic transformation of human cells required human telomerase reverse transcriptase (hTERT; Hahn et al. 1999). These observations have led researchers to believe that telomerase is a promising target for the development of anticancer therapies (Corey 2002; Rezler 2002).
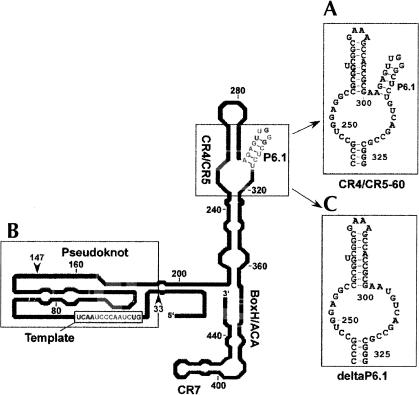
The two main components of telomerase are the protein (TERT) and the RNA (TR). Telomerase reverse transcriptase (TERT) contains reverse transcriptase motifs essential for enzymatic activity as well as a telomerase specific T motif (Lingner et al. 1997; Nakamura et al. 1997). The stable association of TERT with a telomerase RNA containing the template for telomeric repeat synthesis is a unique feature of the telomerase reverse transcriptase (Greider and Blackburn 1989). Vertebrate telomerase RNA varies significantly in sequence and length (Chen et al. 2000). However, phylogenetic and covariation analysis of 35 vertebrate telomerase RNAs revealed that vertebrate telomerase RNAs share a similar secondary structure composed of highly conserved domains. The four conserved domains found in vertebrate telomerase RNAs, including hTR are: (1) the pseudoknot domain, (2) the box H/ACA domain, (3) the CR4/CR5 domain, and (4) the CR7 domain (Chen et al. 2000; Fig. 1 ▶).
FIGURE 1.
Human telomerase RNA constructs utilized to demonstrate RNA–RNA interactions via mobility shift assays. The secondary structure of human telomerase RNA is adapted with permission (from Elsevier © 2000, Chen et al. 2000) with the following RNA regions indicated: (A) CR4/5–60, (B) RNA33–147, and (C) deltaP6.1. Bases in gray are 100% conserved in vertebrate telomerase RNA.
Human telomerase RNA (hTR) is a 451-nt RNA containing an 11-nt templating region (5′-CUAACCCUAAC-3′) located near its 5′ terminus. The templating region is comprised of the template sequence for the synthesis of d(GGTTAG) as well as an alignment domain (italicized; Feng et al. 1995; Gavory et al. 2002). It has been proposed that the alignment domain hybridizes to the 3′ terminus of the DNA substrate, and positions the telomere for processive synthesis of telomeric repeats (Greider and Blackburn 1989; Shippen-Lentz and Blackburn 1990; Autexier and Greider 1994).
The structurally conserved domains have specific functions in active telomerase. The box H/ACA and CR7 domains are dispensable for reconstitution of telomerase activity in vitro (Bachand and Autexier 2001). However, in vivo, the box H/ACA motif is required for the 3′ terminal formation of the mature telomerase RNA (Mitchell et al. 1999). Additionally, the box H/ACA and CR7 domains participate in nucleolar localization of telomerase RNA (Lukowiak et al. 2001). It has been shown that the CR4/CR5 and pseudoknot domains are essential for catalytic activity (Tesmer et al. 1999; Beattie et al. 2000; Mitchell and Collins 2000; Martín-Rivera and Blasco 2001). In vitro studies indicate that these two domains can reconstitute telomerase activity in the presence of the protein component (Tesmer et al. 1999). Further studies have indicated that nt 33–147 and 163–330 contain hTERT-binding sites critical for telomerase activity (Bachand and Autexier 2001). Based on this evidence, we reasoned that RNA–RNA interactions, possibly critical for telomerase function, might be present in hTR involving these two conserved domains.
The data presented here show that an RNA–RNA interaction can exist between two hTR domains distant in primary sequence. We utilized separate domains of hTR in our structural characterization of the RNA. Gel mobility shift assays, mutation analysis, and UV cross-linking identified the interacting residues as the P6.1 stem–loop in the CR4/CR5 domain and the template domain. Previous studies (Mitchell and Collins 2000; Chen et al. 2002) have indicated that P6.1 is a functionally important stem–loop in the CR4/CR5 domain in mammalian telomerase RNA. Recent NMR data from Varani and coworkers indicates that formation of P6.1 positions the exposed loop bases for possible RNA–RNA or RNA–protein interactions (Leeper et al. 2003). Our findings show that P6.1 participates in a long-range RNA–RNA interaction with residues in the template region of hTR. Because the regions participating in this interaction are highly conserved, this interaction may play a critical role in the catalytic activity of the telomerase complex.
RESULTS
hTR RNA–RNA interaction revealed by mobility shift assay
Because the pseudoknot and the CR4/CR5 domains have been implicated in the catalytic activity of telomerase, we examined the possible existence of a long-range RNA–RNA interaction between these two highly conserved domains. RNA constructs containing the two domains were synthesized in vitro for use in RNA–RNA gel mobility shift assays. CR4/5–60 contains the CR4/CR5 domain (nt 243–326, hTR numbering) with the L6 loop (nt 266–291) mutated to a GAAA tetraloop (Fig. 1A ▶). RNA33–147 is comprised of the template region and two strands of the pseudoknot domain (Fig. 1B ▶).
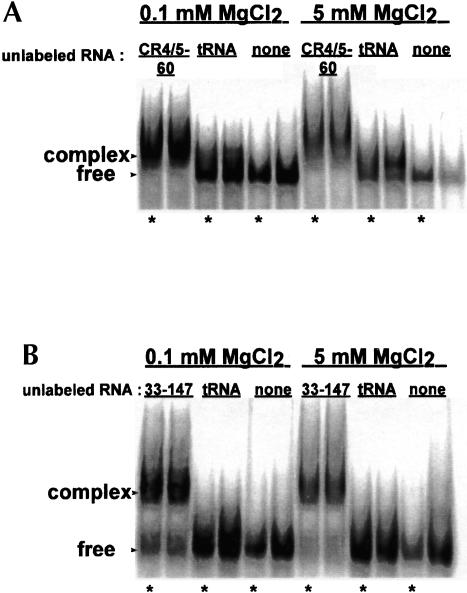
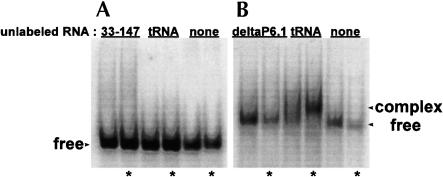
In gel mobility shift experiments, we find that RNA33–147 forms complex with CR4/5–60 (Fig. 2 ▶). When RNA33–147 is end labeled and in the presence of excess CR4/5–60, a slower migrating band, indicating occurrence of an RNA–RNA interaction, is observed (Fig. 2A ▶). To confirm that RNA33–147 and CR4/5–60 interact, we performed the complementary experiment in which CR4/5–60 is radioactively labeled and in the presence of excess RNA33–147 (Fig. 2B ▶). Our results indicate that these two distal domains within hTR can form a gel-stable RNA–RNA interaction.
FIGURE 2.
Native gel mobility shift assays reveal complex formation between CR4/5–60 and RNA33–147. (A) 32P 5′-end labeled RNA33–147 in the presence of excess unlabeled RNA. (B) 32P 5′-end labeled CR4/5–60 in the presence of excess unlabeled RNA. For both A and B, the indicated MgCl2 concentrations correspond to MgCl2 binding buffer concentrations (see Materials and Methods). Because minimal differences are observed between the two binding buffers, all following gels show samples assayed in 5 mM MgCl2. Bands with reduced mobility in each gel correspond to an RNA–RNA complex whereas the faster migrating bands correspond to free RNA. Samples in lanes marked by an asterisk (in this and all following mobility shift gels) were incubated for 30 min at 37°C prior to loading on 6% native gels.
We performed the interaction assays under both low (0.1 mM) and high (5 mM) magnesium ion concentrations to examine the magnesium dependence of the interaction. We find that band shifts do not depend significantly on the magnesium concentration in the binding buffer. However, the ability to detect interactions does depend on the presence of 5 mM MgCl2 in the gel and the gel running buffer. When the preformed complexes are run on a native gel in 1× TBE (90 mM Tris-borate, 2 mM EDTA), a shift is no longer observed due to the magnesium-ion-dependent nature of the interaction (data not shown). The mobility shift gels contained in this study show discrete bands, indicating that the RNA constructs and the complexes formed between them are well behaved under native conditions.
Identifying the site of interaction in the CR4/CR5 domain
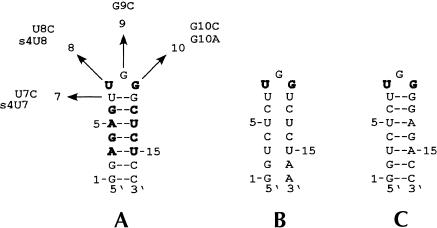
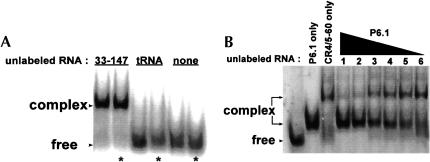
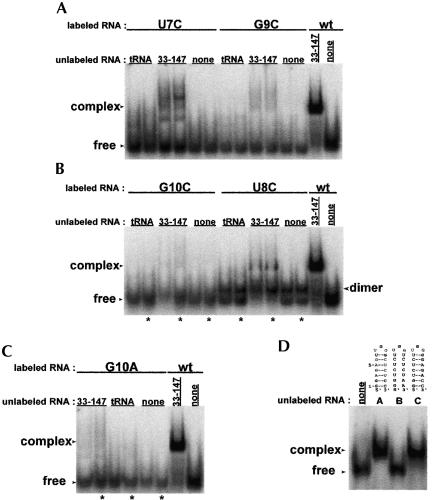
We were next interested in defining the nucleotides within the CR4/CR5 domain that participate in the long-range interaction. Nucleotides 302–314 form the stem–loop P6.1 within the CR4/CR5 domain (Fig. 1 ▶). According to covariation analysis, these residues are highly conserved. Additionally, in mouse telomerase RNA, formation of this stem–loop was found to be important for catalytic activity (Chen et al. 2002). We examined the possibility that this stem–loop participates in the RNA–RNA interaction observed between the larger domain constructs. The P6.1 hairpin construct (Fig. 3A ▶) was synthesized and used in a mobility shift assay with RNA33–147 (Fig. 4A ▶). The P6.1 band is shifted in the presence of RNA33–147. We used a competition mobility shift assay between P6.1 and CR4/5–60 to determine if the two RNAs bind the template in the same manner (Fig. 4B ▶). P6.1 does effectively compete with CR4/5–60 for the template, indicating that the smaller hairpin construct and the larger domain construct interact similarly with the template. A CR4/5–60 variant with P6.1 deleted (deltaP6.1; Fig. 1C ▶) does not form complex with RNA33–147 (Fig. 5 ▶), indicating that this stem–loop is essential for the RNA–RNA interaction and confirming that P6.1 is the site of interaction. In mobility shift assays, we determined that the Kd of RNA33–147 and P6.1 association is roughly 4 μM (data not shown).
FIGURE 3.
Secondary structure and mutation constructs of the P6.1 hairpin. Numbering corresponds to the nucleotide position in the hairpin beginning at the 5′ end. Bases in bold are 100% conserved in vertebrate telomerase RNAs (Chen et al. 2000). Hairpin A is wild-type P6.1 with the indicated mutations U7C, U8C, G9C, G10C, and G10A. UV cross-linking employed 4-thio-U modified RNA hairpins (designated as s4U7 and s4U8). Base pairing in the stem of hairpin B is disrupted whereas the loop bases remained unchanged. Hairpin C restores base pairing in the stem with an altered stem sequence.
FIGURE 4.
P6.1, a small hairpin located in the CR4/CR5 domain, participates in an RNA–RNA interaction with RNA33–147. (A) The P6.1 hairpin is 32P 5prime;-end labeled and shifted with excess unlabeled RNA. (B) P6.1 and CR4/5–60 interact similarly with the template. The starting complex contains 32P 5′-end labeled RNA44–57 in the presence of excess CR4/5–60 (10 μM). Lanes 1–6 contain the starting complex and decreasing amounts of P6.1 in the following concentrations: (1) 10 μM, (2) 5 μM, (3) 2 μM, (4) 1 μM, (5) 500 nM, and (6) 100 nM.
FIGURE 5.
Deletion of the P6.1 stem–loop abrogates association of CR4/5–60 with RNA33–147. (A) 32P 5′-end labeled deltaP6.1 in the presence of excess unlabeled RNA. (B) 32P 5′-end labeled RNA33–147 in the presence of excess unlabeled RNA. The data show that no band shift is observed when the P6.1 hairpin is deleted from the CR4/CR5 domain.
Variants of the P6.1 hairpin were synthesized with mutations at the 7, 8, 9, and 10 positions indicated in Figure 3A ▶. According to covariation analysis (Chen et al. 2000), residues 8 and 10 are 100% conserved in vertebrate telomerase RNAs whereas residues 7 and 9 are variable across the different species. We therefore constructed the following individual mutants to determine which bases in the loop are essential for the RNA–RNA interaction: (1) U8C, (2) G10A, (3) G10C, (4) U7C, and (5) G9C. U8C, G10A, and G10C do not produce appreciable shifts (Fig. 6A ▶–C). This result was expected because the U8 and G10 positions are 100% conserved and interfering with highly conserved residues is likely to disrupt the higher order structural interactions. U7C and G9C are able to interact with RNA33–147, though the amount of stable complex is reduced in comparison to that produced with wild-type hairpin. The more conservative U7C mutation leads to partial interaction whereas the less conservative mutation G9C results in lower binding affinity. Although U7 and G9 are not 100% conserved, mutation of U7 likely introduces a structural change in P6.1, leading to weaker RNA–RNA interaction. Mutation of G9 likely disrupts the interaction because it lies directly in the center of the loop, occupying an important point of contact. These mutation results indicate that the loop residues (5′-UGG-3′) are necessary for the RNA–RNA interaction.
FIGURE 6.
Mutation of the P6.1 hairpin loop sequence interferes with RNA complex formation between P6.1 and RNA33–147. (A) U7C and G9C introduce mutations at bases that are variable across vertebrate telomerase RNAs (Chen et al. 2000). RNA complex formation is observed, but at levels much lower than with wild-type P6.1 (wt), indicating that the RNA–RNA interaction is much weaker in the presence of the mutated hairpin. (B) G10C and U8C introduce mutations at 100% conserved bases and also show greatly reduced complex formation. The hairpin with mutation U8C dimerizes under nondenaturing conditions, indicated by bands with mobility between the free RNA and the complexed RNAs. (C) Mutation at P6.1 position G10 disrupts formation of the RNA–RNA complex. (D) A base-paired stem is required for interaction. RNA44–57 is 5′-end labeled. Mutant hairpin B does not form complex whereas hairpin C, with a restored stem, is able to interact with the template.
P6.1 stem mutants (Fig. 3B,C ▶) were also utilized in mobility shift assays. Formation of a stem of the correct length is important for telomerase activity (Chen et al. 2002). The following stems were assayed: a P6.1 mutant that does not form a stem (Fig. 3B ▶) and a P6.1 with a mutant stem (Figs. 3C ▶, 6D ▶). The P6.1 mutant that does not form a stem is unable to produce a shift in the presence of the template. The P6.1 with a mutant stem is able to interact with the template, indicating that although the formation of the stem is required, the sequence of the stem can be variable. These data show that the P6.1 loop sequence as well as the formation of the stem are essential for the long-range interaction.
Defining the site of interaction in RNA33–147
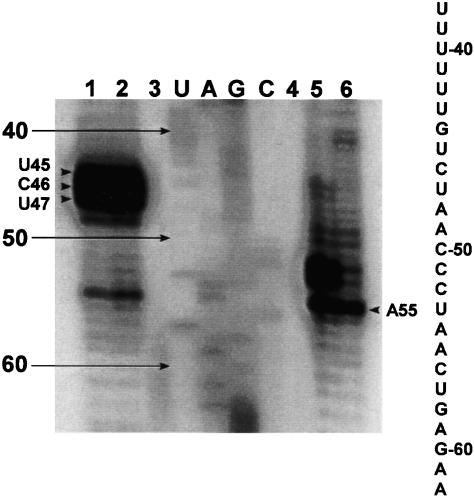
We next investigated what positions within RNA33–147 are participating in the RNA–RNA interaction. To do this, we employed 4-thiouracil (4-thio-U) cross-linking, which would elucidate which bases within RNA33–147 are in close enough proximity to the loop region of the P6.1 hairpin to form cross-links. Two modified RNAs were synthesized; one contained a 4-thio-U modification at the U7 position (s4U7) and the other contained a 4-thio-U modification at the U8 position (s4U8; Fig. 3A ▶). Gel shift analysis shows that the 4-thio-U-modified P6.1 gives a comparable band shift with RNA33–147 (data not shown). Sequencing of the cross-linked products showed that the site of interaction within RNA33–147 occurs near the template sequence of hTR (Fig. 7 ▶). In the s4U7 experiments, the cross-linking occurs at positions G44, U45, and C46. In the s4U8 cross-linking experiment, the cross-linking occurs at A54. The locations of the cross-links are summarized in Figure 10 ▶, below.
FIGURE 7.
UV cross-linking reveals that the site of interaction within RNA33–147 is at the template domain. In the sequencing gel of the cross-linking reaction between 4-thio-U hairpins and RNA33–147, each lane contains a reverse transcriptase reaction as described in Materials and Methods. Lanes 1 and 2 are the s4U7 cross-linked products run in duplicate. Lane 3 is RNA33–147 reverse transcribed with only dNTPs. Lanes U, A, G, and C correspond to the sequencing ladder produced by dideoxy sequencing methods. Lane 4 is the reverse transcriptase reaction (only dNTPs) of UV-irradiated RNA33–147. Lanes 5 and 6 are the s4U8 cross-linked products run in duplicate. Arrows indicate stops due to cross-linking. The hTR sequence of the template region is indicated alongside the gel.
FIGURE 10.
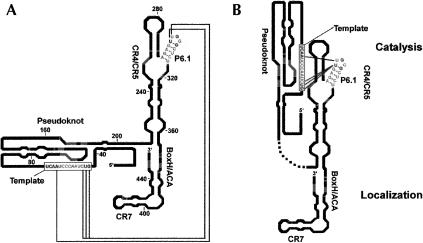
Models of human telomerase RNA. (A) Secondary structure of human telomerase RNA (adapted with permission from Elsevier © 2000, Chen et al. 2000) with the proposed long-range interactions revealed in this study indicated by the connecting lines. (B) A new model for the secondary structure of hTR with interacting residues shown by gray connecting lines. The catalytic domains (pseudoknot, CR4/CR5, and template) and localization domains (box H/ACA and CR7) are indicated.
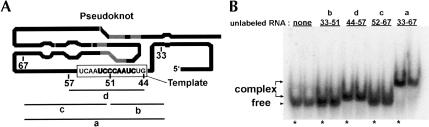
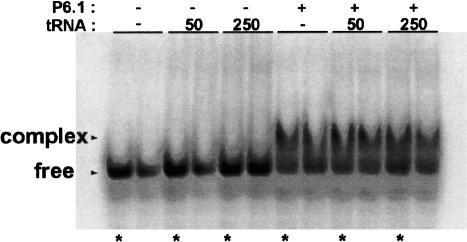
To further verify the site of interaction, mobility shift experiments were employed with small RNA fragments from the RNA33–67 region (Fig. 8 ▶). Initially, RNA33–147 was reduced to RNA33–67 to determine if the RNA–RNA interaction could be detected in the absence of any part of the pseudoknot. RNA33–67, encompassing only bases 5′ of the pseudoknot region, shifts the P6.1 hairpin (Fig. 8 ▶). To further isolate the site of interaction, three short RNA fragments were synthesized: (1) RNA33–51, (2) RNA44–57, and (3) RNA52–67 (Fig. 8A ▶). Two of the RNAs, RNA33–51 and RNA52–67, contain only a portion of the template region, whereas the third, RNA44–57, contains the entire template region. Only RNA44–57 shifts P6.1, indicating that the entire template is necessary for interaction with the hairpin (Fig. 8B ▶). A further truncation of RNA44–57, involving only the 11-nt templating domain, 5′-CUAACCCUAAC-3′, shifts the P6.1 hairpin (Fig. 9 ▶). In a competition assay, we show that tRNA (at concentrations of either 50 μg/mL or 250 μg/mL) cannot compete with P6.1 for binding of the template (Fig. 9 ▶). This result argues for the specificity of the interaction revealed in this study.
FIGURE 8.
RNA–RNA interaction requires the entire hTR template domain. (A) Human telomerase RNA truncations analyzed for long-range RNA–RNA interaction with the P6.1 hairpin. Numbering corresponds to the hTR nucleotide sequence. The truncated RNA regions are: (a) RNA33–67, (b) RNA33–51, (c) RNA52–67, and (d) RNA44–57. Bases in bold are 100% conserved nucleotides (Chen et al. 2000). (B) Gel shift assay with 32P 5′-end labeled P6.1 hairpin and truncated RNA pieces from the 5′ region of hTR. RNA33–51 and RNA52–67 contain sections of the template and show no complex formation. Only RNA33–67 and RNA44–57, which contain the entire template, produce complexes with the hairpin.
FIGURE 9.
The 11-nt templating and alignment domain (5′-CU AACCCUAAC-3′) is minimally required for long-range interaction with the P6.1 hairpin. The 11-nt templating region is 32P 5′-end labeled. Competition with tRNA at concentrations of 50 μg/mL and 250 μg/mL did not alter the mobility shift, indicating that the interaction is specific. The numbers 50 and 250 indicate tRNA concentrations.
DISCUSSION
The studies presented here argue for a new model of telomerase RNA architecture that involves higher order folding of hTR (Fig. 10 ▶). We propose that the CR4/CR5 and pseudoknot domains are close in space, though distal in primary sequence. Our data argue for the presence of long-range RNA–RNA interactions possibly essential for overall architecture and function.
Studies by Mitchell and Collins (2000) show that deletion of residues 303–315 (P6.1) interferes with proper hTR and hTERT interaction and abrogates telomerase activity. Each of the 35 vertebrate RNAs has the capability of forming this short stem, indicating its importance in telomerase structure and function. Because deletion of this stem–loop interferes with the RNA–RNA interaction observed in our study, this higher order RNA–RNA interaction may have functional significance. Enzymatic mapping analysis shows that the loop bases of P6.1 (U307, G308, and G309) are accessible in vitro but inaccessible in vivo (Antal et al. 2002). Because these bases become protected in the presence of hTERT, it has been suggested that they associate with the protein. The recent NMR structure of P6.1 from Varani and coworkers shows that these bases are part of a well-defined structure that allows the exposed loop to participate in tertiary interactions with other portions of hTR or RNA–protein interactions with hTERT (Leeper et al. 2003). Here, we show that P6.1 can participate in RNA–RNA interactions with the template of hTR. Based on our mutation analysis, the residues involved in the long-range RNA–RNA interactions are in the loop of P6.1.
Interaction between P6.1 and the template is mediated by the loop sequence and the formation of a stem. No complex forms when the stem is unable to base pair. However, RNA–RNA interactions are maintained as long as the P6.1 stem forms, regardless of the stem sequence. Our P6.1 stem mutant analysis is consistent with studies on mouse telomerase RNA that showed that disruption of the P6.1 base pairing in the stem abolishes telomerase activity (Chen et al. 2002).
Phylogenetic studies performed by Chen et al. (2000) show that positions 8 and 10 are 100% conserved. Therefore, these mutant hairpins are not expected to shift RNA33–147. Positions 7 and 9 are not 100% conserved; however, the shifts produced in the presence of the single-base mutated hairpins are minimal compared to the shifts observed in the presence of the wild-type hairpin. Because the structure of the hairpin is critical for the RNA–RNA interaction, as evidenced by the stem mutant data, altering the P6.1 structure can lead to instability or abolishment of the interaction. The U7C mutation would alter the structure by replacing a U-G wobble pair with a G-C pair. U7 may be essential for the proper formation of the hairpin. G9 is positioned directly in the middle of the loop, suggesting that this exposed base would be a key participant in the interaction, and disruption of this critical base would lead to reduced binding affinity. Our results indicate that conservation of the loop residues (5′-UGG-3′) is critical for the formation of the RNA–RNA interaction. This data is consistent with work accomplished by Greider and coworkers (Chen et al. 2002). Mutation of the loop sequence in the P6.1 stem–loop in mammalian telomerase RNA reduces telomerase activity, suggesting that mutation of this loop could disturb important RNA–RNA or RNA–protein interactions that contribute to the functional conformation at or near the catalytic center of telomerase.
Telomerase RNA structure has been characterized in ciliates (Romero and Blackburn 1991; Bhattacharyya and Blackburn 1994) as well as in humans (Chen et al. 2000; Antal et al. 2002). Studies on the Tetrahymena thermophila telomerase RNA suggest that the template region of telomerase RNA is mostly single stranded but possibly constrained to some conformation by folding of the rest of the RNA (Bhattacharyya and Blackburn 1994). Enzymatic mapping studies from the Kiss group (Antal et al. 2002) show that U47 and C50–C52 are not modified, whereas C46 and U53 are weakly modified. These results indicate that these bases participate in base-pairing interactions, protein–RNA interactions, or tertiary RNA structure. Additionally, bases adjacent to the template region are necessary for catalytic activity (Gavory et al. 2002; Miller and Collins 2002). When bases 54 and 56 are mutated, enzyme processivity is greatly reduced, indicating the importance of the conservation of these nucleotides (Chen and Greider 2003). We propose that these bases participate in critical long-range RNA–RNA interactions with residues in the CR4/CR5 domain.
Because both the P6.1 hairpin and the template region contain highly conserved bases, we believe that the interaction determined in our study has biological significance. In the current model of telomerase activity, telomeric repeats are added via a primer recognition step at the template, elongation, and translocation in preparation for the addition of the following repeat (Shippen-Lentz and Blackburn 1990). A regulatory mechanism involving the pseudoknot domain has recently been proposed (Comolli et al. 2002; Theimer et al. 2003). Both groups propose a molecular switch mechanism involving an equilibrium between the pseudoknotted and hairpin forms of the pseudoknot domain. For telomerase to be active, the pseudoknot domain must form.
We propose that other intramolecular RNA–RNA interactions, highlighted in Figure 10 ▶, are involved in the higher order folding and consequently proper function of human telomerase RNA. Our results argue for the close proximity of the CR4/CR5 domain and the template. Because both are important for catalytic activity, the 5′ portion of the RNA is likely closer to the CR4/CR5 domain than to the portions of the RNA important for nuclear localization of the RNA (Lukowiak et al. 2001). As shown in Figure 10B ▶, the CR4/CR5, pseudoknot domain, and template form the catalytic domain whereas the localization domain is composed of the box H/ACA and CR7 domains.
To investigate the link between the interaction observed in our study and its functional significance, we utilized mouse telomerase RNA constructs in gel mobility shift assays. Based on functional data from Chen and Greider (2003), we expected mouse P6.1 to shift the human template because mouse CR4/CR5 and human pseudoknot, in the presence of hTERT or mTERT, can reconstitute activity. However, an interaction is not observed by gel mobility shift assay (data not shown). Mouse P6.1 is also unable to shift the mouse template. Functionally, mouse and human telomerase differ in that human telomerase is processive, whereas mouse telomerase exhibits only a low level of processivity (Morin 1989; Prowse et al. 1993). If the P6.1/template interaction is involved in processivity, then mouse P6.1 would not be expected to interact similarly with the template. The telomerase protein may be able to compensate for the reduced binding affinity caused by the differences in sequence and structure between human P6.1 and mouse P6.1.
The entire template and alignment domain are necessary for the observed interaction. No shift is observed in the interaction assay between the P6.1 hairpin and RNAs containing only 5′ or 3′ portions of the template domain. This result, along with our cross-linking data, suggests that the hairpin may be interacting at multiple sites in the template domain. If the interaction is dynamic, the hairpin may be involved in the mechanism for translocation of the template during reverse transcription. In Tetrahymena telomerase, it has recently been shown that RNA sequences distant from the template are involved in mediating nucleotide and repeat addition processivity (Lai et al. 2003). The conserved hairpin in the CR4/CR5 domain may play a similar role in human telomerase enzyme processivity. P6.1 may ready the template domain by either stabilizing the template or positioning it for proper reverse transcription by the protein. Further studies involving the protein will aid in the characterization of the functional significance of the RNA–RNA interaction revealed in our study.
MATERIALS AND METHODS
Synthesis of RNA
RNA was transcribed in vitro in transcription buffer (80 mM HEPES-KOH at pH 7.5, 2 mM spermidine, 40 mM DTT, 25 mM MgCl2), 4 mM of each nucleotide triphosphate (CTP, ATP, UTP, GTP), RNAsecure (to prevent RNA degradation), 500 nM bottom strand (DNA template), 600 nM top strand (T7 primer), and T7 RNA polymerase for 4–6 h at 37°C. The following are the template sequences used for transcription of the indicated RNAs (in bold):
CR4/5–60 (5′-GGGCGGCTGACAGAGCCA ACTTTGGTGGCTTTCGCCGACCGCG GCCTCCAGGCCCTATAGTGAGTCGT ATTACGAATT-3′);
deltaP6.1 (5′-GGGCGGCTGACATTCGCGGTGGCTTTCGCCG ACCGCGGCCTCCAGGCCCTATAGTGAGTCGTATTACGAA TT-3′);
RNA33–67 (5′-CGCCCTTCTCAGTTAGGGTTAGACAAAAAAT GGCCTATAGTGAGTCGTATACGAATT-3′);
RNA33–51 (5′-GGTTAGACAAAAAATGGCCTATAGTGAGTCG TATTACGAATT-3′);
RNA44–57 (5′-AGTTAGGGTTAGACTATAGTGAGTCGTATTA CGAATT-3′);
RNA52–67 (5′-CGCCCTTCTCAGTTAGTATAGTGAGTCGTAT TACGAATT-3′);
RNAP6.1 (5′-GGAGAGCCCAACTCTCCTATAGTGAGTCGTAT TACGAATT-3′).
The P6.1 hairpin mutants were transcribed like the wild-type hairpin with the single base or stem mutations as indicated in the Results section.
RNA33–147 was synthesized by transcription off a template that was PCR generated from a pUC19 plasmid containing the sequence encoding for RNA33–147. The PCR template (100 nM) was used for in vitro transcription with T7 RNA polymerase.
The following RNA oligonucleotides were ordered from the Caltech Oligonucleotide Synthesis Facility:
4-thio-U7 (5′-GGAGAG(s4U)UGGGCUCUCC-3′);
4-thio-U8 (5′-GGAGAGU(s4U)GGGCUCUCC-3′);
hTRtemplate (11-nt) (5′-CUAACCCUAAC-3′).
All RNA synthesized by the synthesis facility required 2′-OTBDMS deprotection by use of triethylamine-3HF (Aldrich). After overnight deprotection in HF, the RNA samples were ethanol precipitated, dried, and resuspended in double-distilled water.
All RNA products were gel purified by denaturing polyacrylamide gel electrophoresis (PAGE) in 1× TBE (90 mM Tris-borate, 2 mM EDTA). An electroelution apparatus was used to elute the RNA into 0.5× TBE. The RNA products were ethanol precipitated, dried to pellets, and resuspended in double-distilled water. The concentrations were determined by UV spectroscopy and the biopolymer calculator developed by the Schepartz lab (Palmer 1998). Purity was assayed by analytical PAGE, mass spectrometry, or both.
32P 5′-end labeling
The terminal triphosphates of the transcribed RNA products were removed using calf intestinal phosphatase (Roche Diagnostics). The RNAs and DNA sequencing primers [33–147seq (5′-AAGG CGGCAGGCCGAGGC-3′) and tempseq (5′-AGTCAGCGAGAA AAACAGCG-3′)] were kinased with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (NEN Life Science). The 5′-end labeled products were purified by NAP-25 columns (Amersham Pharmacia Biotech). Concentrations were assayed by scintillation counts based on an ATP standard, and adjusted to 20 nM.
Gel shift assay
The final concentrations of unlabeled RNA used in the gel shift assays were all 10 μM except in competition assays (concentrations indicated in the figure legends). The final concentration of the 5′-end labeled RNA was 5 nM. RNA binding buffers were adapted from Ferrandon et al. (1997). Each RNA was annealed by incubation in 1× binding buffer (high salt: 300 mM KCl, 5 mM MgCl2, 50 mM sodium cacodylate at pH 7.5; or low salt: 40 mM KCl, 0.1 mM MgCl2, 50 mM sodium cacodylate at pH 7.5) at 90°C for ~1–2 min and then placed on ice for 5 min. The RNAs were then mixed and either placed on ice for 30 min or incubated at 37°C for 30 min. Five microliters of gel loading buffer (5× binding buffer, 50% glycerol) were added to each mixture before running on a 6% native polyacrylamide gel in 1× TBM (90 mM Tris-borate, 5 mM MgCl2) maintained at 3–5°C. The gels were run between 2 and 3 h at 2–3 W, dried, and exposed to a PhosphorImager screen overnight. The gels were analyzed on a STORM860 and by ImageQuant.
4-thio-U cross-linking
The following 4-thiouridine cross-linking protocol was adapted from established protocols (Dubreuil et al. 1991). The 4-thio-U-modified RNAs were 5′-end labeled with [γ-32P]ATP. The modified hairpin (10 μM) and RNA33–147 (10 μM) were separately incubated in 1× high salt binding buffer for 2 min at 90°C and set on ice for 5 min. The RNAs were mixed together, set on ice, and irradiated with a handheld 365-nm UV light for 30 min. The RNAs were then ethanol precipitated, dried, and resuspended in double-distilled water. The cross-linked sample was separated from uncross-linked material by running the reaction on a 12% denaturing PAGE in 1× TBE. The cross-linked product was excised from the gel, eluted into 0.5× TBE, ethanol precipitated, dried, and resuspended in double-distilled water. Concentrations were determined by means of UV spectrometry (Palmer 1998).
Sequencing reactions
The cross-linked products and RNA33–147 were sequenced by Moloney Murine Leukemia Virus (MMLV) reverse transcription (Invitrogen) and the 5′-end labeled primers 33–147seq and tempseq. The sequencing reaction and UV cross-linking mapping protocols were adapted from established procedures (Lowe and Eddy 1999). For the RNA33–147 sequencing reaction, 2 μg RNA and 1.5 pmoles labeled sequencing primer were incubated for 2 min at 90°C in 1× annealing buffer (250 mM Tris-HCl at pH 8.0, 375 mM KCl, 50 mM dithiothreitol) in a 10-μL annealing reaction. The annealed product was set on ice for 2 min.
For the primer extension reaction, four tubes (A, T, C, and G) were prepared. In each of the four tubes (A, T, C, or G) was placed: 2 μL of the annealed product (from above), 330 μM of each dNTP (A, T, C, and G), 1 mM of the appropriate ddNTP (A, T, C, or G), and MMLV RT buffer [250 mM Tris-HCl at pH 8.0, 375 mM KCl, 15 mM MgCl2, 50 mM dithiothreitol, 1× Superase-In (Ambion, proprietary); 100 U MMLV reverse transcriptase] to a total volume of 5 μL. The extension reactions were incubated for 1 h at 42°C, and then placed at 65°C to heat inactivate the reverse transcriptase. Ten microliters of sequencing dye (1× TBE, 80% formamide, bromophenol blue, xylene cyanol) were added to each tube and 5 μL of each reaction was loaded onto a 10% denaturing PAGE. The gel was run at 50 W for 2–2.5 h in 1× TBE. The sequencing gel was fixed with destain solution (40% methanol, 10% acetic acid, 50% water) for 30 min prior to drying. The dried gels were exposed to screens overnight and imaged by the PhosphorImager.
The cross-linked products were sequenced as above with the following changes. Only one tube was necessary for the primer extension reaction. This tube contained no ddNTPs. The stops occurred as a result of the cross-links. The sequencing reaction and the mapping reaction of the cross-linked products were run adjacently on a sequencing gel as described above. The cross-links were identified by stops in the mapping reaction ladder and the base of interaction read from the sequencing ladder.
Acknowledgments
We thank Adam Frankel, Terry Takahashi, and Tianbing Xia for helpful discussion. This work was supported by a National Institutes of Health grant NIH R01 GM60416 (R.W.R) and an Alfred P. Sloan Research Fellowship BR-4008 (R.W.R).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5118104.
REFERENCES
- Antal, M., Boros, E., Solymosy, F., and Kiss, T. 2002. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 30: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier, C. and Greider, C.W. 1994. Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes & Dev. 8: 563–575. [DOI] [PubMed] [Google Scholar]
- Bachand, F. and Autexier, C. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol. Cell Biol. 21: 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie, T.L., Zhou, W., Robinson, M.O., and Harrington, L. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11: 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, A. and Blackburn, E.H. 1994. Architecture of telomerase RNA. EMBO J. 13: 5721–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, E.H. 1991. Structure and function of telomeres. Nature 350: 569–573. [DOI] [PubMed] [Google Scholar]
- ———. 2000. The end of the (DNA) line. Nat. Struct. Biol. 7: 847–850. [DOI] [PubMed] [Google Scholar]
- Cech, T.R. 2000. Life at the end of chromosomes: Telomeres and telomerase. Angew. Chem. Int. Ed. 39: 34–43. [DOI] [PubMed] [Google Scholar]
- Chen, J.L. and Greider, C.W. 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 22: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.L., Blasco, M.A., and Greider, C.W. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514. [DOI] [PubMed] [Google Scholar]
- Chen, J.L., Opperman, K.K., and Greider, C.W. 2002. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 30: 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli, L.R., Smirnov, I., Xu, L., Blackburn, E.H., and James, T.L. 2002. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. 99: 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey, D.R. 2002. Telomerase inhibition, oligonucleotides, and clinical trials. Oncogene 21: 631–637. [DOI] [PubMed] [Google Scholar]
- Dubreuil, Y.L., Expert-Benzancon, A., and Favre, A. 1991. Conformation and structural fluctuations of a 218 nucleotides long rRNA fragment: 4-thiouridine as an intrinsic photolabeling probe. Nucleic Acids Res. 19: 3653–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., Funk, W.D., Wang, S., Weinrich, S.L., Avilion, A.A., Chiu, C., Adams, R.R., Chang, E., Allsopp, R.C., Yu, J., et al. 1995. The RNA component of human telomerase. Science 269: 1236–1241. [DOI] [PubMed] [Google Scholar]
- Ferrandon, D., Koch, I., Westhof, E., and Nüsslein-Volhard, C. 1997. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16: 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavory, G., Farrow, M., and Balasubramanian, S. 2002. Minimum length requirement of the alignment domain of human telomerase RNA to sustain catalytic activity in vivo. Nucleic Acids Res. 30: 4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C.W. and Blackburn, E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- ———. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomerase repeat synthesis. Nature 337: 331–337. [DOI] [PubMed] [Google Scholar]
- Hahn, W.C., Counter, C.M., Lundberg, A.S., Beijersbergen, R.L., Brooks, M.W., and Weinberg, R.A. 1999. Creation of human tumour cells with defined genetic elements. Nature 400: 464–468. [DOI] [PubMed] [Google Scholar]
- Kim, N. W., Piatyszek, M.A., Prowse, K.R., Harley, C.B., West, M.D., Ho, P.L., Coviello, G.M., Wright, W.E., Weinrich, S.L., and Shay, J.W. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Lai, C.K., Miller, M.C., and Collins, K. 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 11: 1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper, T., Leulliot, N., and Varani, G. 2003. The solution structure of an essential stem-loop of human telomerase RNA. Nucleic Acids Res. 31: 2614–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Hughes, T.R., Shevchenko, A., Mann, M., Lundblad, V., and Cech, T.R. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Lukowiak, A.A., Narayanan, A., Li, Z., Terns, R.M., and Terns, M.P. 2001. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA 7: 1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Martín-Rivera, L. and Blasco, M. 2001. Identification of functional domains and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J. Biol. Chem. 276: 5856–5865. [DOI] [PubMed] [Google Scholar]
- Miller, M.C. and Collins, K. 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. 99: 6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R. and Collins, K. 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6: 361–371. [DOI] [PubMed] [Google Scholar]
- Mitchell, J.R., Cheng, J., and Collins, K. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell Biol. 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, G.B. 1989. The human telomerase terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59: 521–529. [DOI] [PubMed] [Google Scholar]
- Nakamura, T.M., Morin, G.B., Chapman, K.B., Weinrich, S.L., Andrews, W.H., Lingner, C.B., Harley, C.B., and Cech, T.R. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959. [DOI] [PubMed] [Google Scholar]
- Palmer, C.R. 1998. Biopolymer Calculator. http://paris.chem.yale.edu/extinct.html.
- Prowse, K.R., Avilion, A.A., and Greider, C.W. 1993. Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl. Acad. Sci. 90: 1493–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezler, E.M., Bearss, D.J., and Hurley, L.H. 2002. Telomeres and telomerase as drug targets. Curr. Opin. Pharm. 2: 415–423. [DOI] [PubMed] [Google Scholar]
- Romero, D.P. and Blackburn, E.H. 1991. A conserved secondary structure for telomerase RNA. Cell 67: 343–353. [DOI] [PubMed] [Google Scholar]
- Shippen-Lentz, D. and Blackburn, E.H. 1990. Functional evidence for an RNA template in telomerase. Science 247: 546–552. [DOI] [PubMed] [Google Scholar]
- Tesmer, V.M., Ford, L.P., Holt, S.E., Frank, B.C., Yi, X., Aisner, D.L., Ouellete, M., Shay, J.W., and Wright, W.E. 1999. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell Biol. 19: 6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., Trantirek, L., and Feigon, J. 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 100: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]