Abstract
The class I ligase ribozyme catalyzes a Mg++-dependent RNA-ligation reaction that is chemically analogous to a single step of RNA polymerization. Indeed, this ribozyme constitutes the catalytic domain of an accurate and general RNA polymerase ribozyme. The ligation reaction is also very rapid in both single- and multiple-turnover contexts and thus is informative for the study of RNA catalysis as well as RNA self-replication. Here we report the initial characterization of the three-dimensional architecture of the ligase. When the ligase folds, several segments become protected from hydroxyl-radical cleavage, indicating that the RNA adopts a compact tertiary structure. Ribozyme folding was largely, though not completely, Mg++ dependent, with a K1/2[Mg] < 1 mM, and was observed over a broad temperature range (20°C –50°C). The hydroxyl-radical mapping, together with comparative sequence analyses and analogy to a region within 23S ribosomal RNA, were used to generate a three-dimensional model of the ribozyme. The predictive value of the model was tested and supported by a photo-cross-linking experiment.
Keywords: RNA structure, RNA catalysis, molecular modeling
INTRODUCTION
The class I ligase (Fig. 1A ▶) catalyzes a reaction similar to that of biological RNA polymerases: attack by the 3′-OH of a small substrate RNA on a 5′-triphosphate, forming a new 3′–5′ linkage with concomitant release of pyrophosphate (Ekland et al. 1995). Comparisons between this reaction and RNA polymerization were extended by experiments showing that engineered derivatives of the ligase are able to perform short primer-extension reactions (Ekland and Bartel 1996). More recently, variants of the ribozyme have been shown to catalyze template-directed polymerization of up to 14 nt (Johnston et al. 2001), supporting the idea that early in the origin of life, RNA might have catalyzed its own replication (Bartel 1999; Joyce and Orgel 1999).
FIGURE 1.
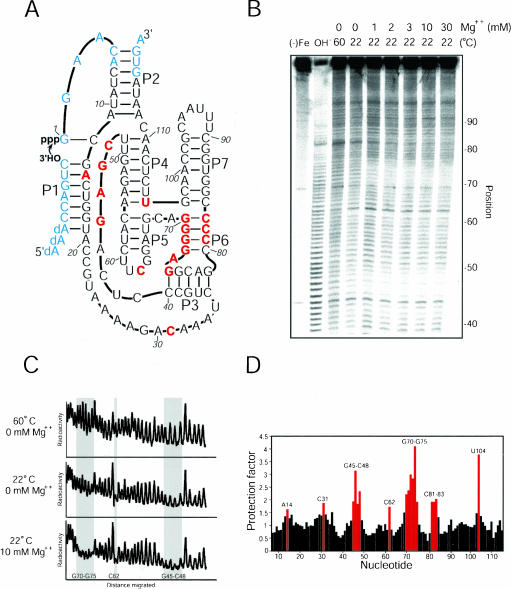
Hydroxyl-radical cleavage of the Class I ligase. (A) The class I ligase. The ligase promotes the attack of the 3′-OH of the substrate on its own 5′-α-phosphate, forming a new 3′–5′ linkage with release of pyrophosphate. Paired regions are designated P1–P7. Residues protected from hydroxyl-radical cleavage are colored red and reflect those highlighted in panel C. Residues colored blue are those for which solvent accessibility was not measured because they were too near to the ends of the RNA. (B) Gel showing hydroxyl-radical cleavage in the presence of different Mg++ concentrations. Background cleavage in the absence of iron is shown [(−)Fe], along with a lane showing cleavage under denaturing conditions (60°C, 0 mM Mg++). The gel was run for 90 min and used to collect data at positions 40–80. Nucleotides were identified by comparison to a ladder generated by partial digest of radiolabeled product using RNase T1 (not shown) and alkaline hydrolysis (OH−). (C) Radioactivity profiles of lanes from the gel in panel B. Regions of substantial protection are shaded. (D) Protection factors for nt 7–115 in the presence of 10 mM Mg++ at 22°C. Protection factors are defined as the ratio of cleavage under denaturing conditions (0 mM Mg++, 60°C) to cleavage under experimental conditions. Protection factors exceeding 1.5 are labeled and colored red.
Recent studies have begun to define the reaction kinetics of the ligase, using ribozymes in both multiple- and single-turnover formats. In a multiple-turnover format, the ribozyme catalyzes ligation with a kcat greater than 2 sec−1 at pH 8.0, a rate exceeding those of other multiple-turnover ribozyme-catalyzed reactions (Bergman et al. 2000). This speed is due in large part to a very fast chemical step (kc). Studies examining kc in a single-turnover format have shown that this step can reach rates exceeding 10 sec−1 at pH 9.0 (N.H. Bergman, C.C. Yen, and D.P. Bartel, in prep.). The studies of self-ligation also showed that the majority of ligase molecules fold accurately and quickly, with a folding rate of about 1.0 sec−1, as measured by attainment of an active structure (Glasner et al. 2002). This suggests that the alternative folding pathways (and accompanying misfolding) seen in some other ribozymes (Pan and Sosnick 1997; Russell and Herschlag 1999) are less prevalent in the case of the ligase.
Like proteinaceous RNA polymerases, the ligase has a near absolute requirement for Mg++ ions (Glasner et al. 2002). Furthermore, these protein enzymes and the ligase show a similar stereospecific response to sulfur substitution at the reactive phosphate, which is consistent with the idea that one of these essential metal ions may be bound in the same position relative to substrate in both catalysts (Eckstein 1985; Glasner et al. 2000). This finding leaves open the possibility that the ligase uses the same mechanism as that proposed for general protein-catalyzed phosphoryl transfer, and prompts questions of whether the ribozyme might also share structural features with analogous protein enzymes.
To better understand the relationship between the structure and function of the ligase, we have begun to characterize its tertiary structure. The solvent accessibility at each position along the sugar–phosphate backbone of the ribozyme was measured using hydroxyl radical probing (Celander and Cech 1990,Celander and Cech 1991). These measurements identified portions of the RNA that are protected from cleavage when the ligase is folded. Together with comparative sequence analysis, these data were used herein to model the ribozyme in three dimensions. A photo-cross-linking experiment showed that the model successfully predicted three-dimensional proximity between ribozyme segments that were far apart in the secondary structure. The model provides a framework for future structural studies and suggests strategies for crystallization of the ligase.
RESULTS AND DISCUSSION
Hydroxyl-radical probing
Hydroxyl radicals, generated by either chemical methods (Tullius et al. 1987; King et al. 1993) or synchrotron radiation (Sclavi et al. 1998), have been used successfully to examine the structure of catalytic RNAs, ribosomal RNAs, and protein–nucleic acid complexes (Tullius and Dombroski 1986; Latham and Cech 1989; Celander and Cech 1991; Joseph and Noller 2000). The radicals, when produced in solution with RNA, attack the ribose moieties in the nucleic acid backbone, causing strand cleavage (Wu et al. 1983; Hertzberg and Dervan 1984). This cleavage is independent of sequence and secondary structure, and is instead dependent on the solvent accessibility of each position in the RNA backbone (Celander and Cech 1990). In beginning investigations of the tertiary structure of the ligase, hydroxyl-radical probing was used to define portions of the ribozyme that are internalized by tertiary structure and thus protected from cleavage.
At 22°C and 10 mM Mg++, a substantial number of nucleotides (17 out of 109 tested) became protected from hydroxyl-radical cleavage (protection factor >1.5; Fig. 1 ▶), indicating that the ligase assumes a compact structure in the presence of Mg++. The most striking protections were seen for nucleotides G45–C48, which make up the most conserved part of a joining region connecting helix P3 with helix P4, and for nucleotides G70–G75, which comprise the 5′ arm of helix P6 (Fig. 1 ▶). Interestingly, the opposite arm of this helix (C81–C83) also had high protection factors, implying that this short helix is nestled within the ribozyme core. In addition to these segments, several isolated nucleotides were protected. Nt A14 and C31 were consistently protected, though at a much weaker level than segments G45–C48 or G70–G75. Nt U104 was strongly protected, perhaps because of the structure inherent in the junction of helices P4, P5, P6, and P7. Finally, nt C62 was strongly protected even in the absence of Mg++, perhaps because of local structure in the UUCG tetraloop (Cheong et al. 1990).
Temperature and Mg++ dependence of ribozyme tertiary structure
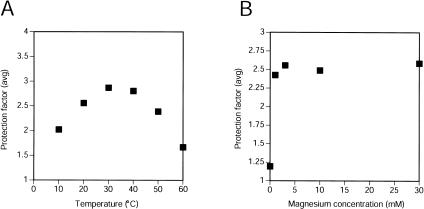
The fold of the ribozyme was assayed with respect to Mg++ and temperature, using the average protection factors from segments G45–C48 and G70–G75 as a measure of the degree to which molecules assumed the native tertiary structure (Fig. 2 ▶). In the presence of 10 mM Mg++, the ligase showed some native structure at 10°C, and was well structured in the range 20°C–40°C. The group I intron shows significant misfolding at lower temperatures (Russell and Herschlag 1999), and the relatively lower protection seen at 10°C might reflect a similar phenomenon. However, the pattern of protection did not change at low temperature, so if a misfolded form is more populated, it is not sufficiently compact to detectably alter the protection pattern. At 50°C, protection factors were again slightly lower, and at 60°C most of the protection from hydroxyl-radical cleavage had disappeared (Fig. 2A ▶). At 60°C and in the absence of Mg++, the ribozyme had no detectable tertiary structure.
FIGURE 2.
Temperature and Mg++ dependence of solvent accessibility in the class I ligase. (A) Protection factors were averaged for G45–C48 and G70–G75, and the average is plotted for experiments in which hydroxyl-radical cleavage was performed at varying temperatures in the presence of 10 mM Mg++. (B) Averaged protection factors are shown for the positions noted in panel A in experiments probing the ligase at different Mg++ concentrations (at 22°C).
When assayed at 20°C, the ligase was essentially completely folded at Mg++ concentrations as low as 1 mM (Fig. 2B ▶), indicating that the [Mg++]1/2(folding) is below 1 mM. (Assaying for folding with less than 1 mM Mg++ could not be performed with this procedure because of trace amounts of free EDTA in probing reactions.) In comparison, the [Mg++]1/2(catalysis) is much higher (40–50 mM), suggesting that a native structure can be achieved without Mg++ ions bound in every catalytically useful binding site (Glasner et al. 2002).
Interestingly, removing Mg++ ions from the ribozyme solution by adding EDTA did not completely denature the ribozyme’s tertiary fold. Rather, the areas protected from cleavage when the ribozyme was folded in the presence of Mg++ were also partially protected by simply dropping the temperature from 60°C to 22°C in the absence of Mg++ (Fig. 1C ▶). The presence of Mg++ changed the magnitude of each protection, but not the overall pattern of protection, suggesting that similar structure was present under both conditions. These data suggest that upon removal of Mg++, the ribozyme stays folded into near native conformations.
Three-dimensional model
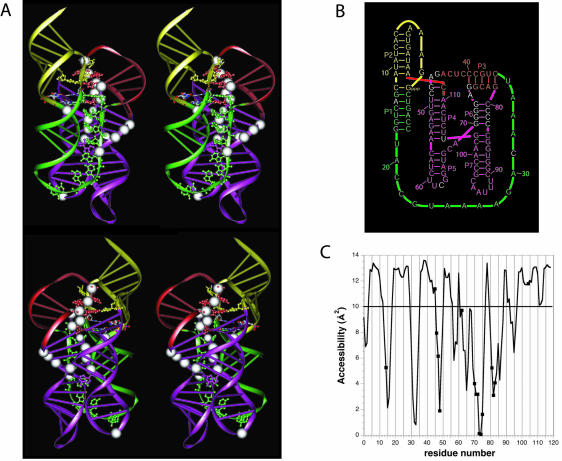
It had been shown that the hairpin loops capping helices P5 and P7 were not important for catalysis and that 5 nt could be removed from the J1/3 junction without substantially impairing catalysis (Ekland and Bartel 1995). Thus, we initiated the modeling with a minimal ligase of 112 nt. Later, the whole ligase was assembled with residues 20–24 added in helical continuity with helix P1. Several models were built that accommodated the stereochemical constraints imposed by the two pseudoknots (helices P2 and P3) present in the secondary structure. Two of these models were consistent with the information contained in the Fe-EDTA protection data. However, one was favored because residues known to be crucial for activity were positioned near the catalytic site and because its topology was more conducive for folding (Fig. 3 ▶). In the alternative model, folding would have been problematic because the path of J3/4 was such that a knot would have been created with the formation of P2.
FIGURE 3.
A three-dimensional model for the class I ligase. (A) Two stereo views of the model. White spheres indicate those residues protected from hydroxyl-radical cleavage. Residues of particular interest are highlighted with all-atom representation. These include the residues of the proposed tandom G•A pairs and residues that both were unpaired in previous representations of the ligase secondary structure (Fig. 1A ▶) and were invariant among 25 active variants of the ligase (Ekland and Bartel 1995), all colored the same as the proximal paired regions. Also shown in all-atom representation are the four residues comprising the 2 bp flanking the ligation junction, colored according to the identity of the atoms. (B) A revised secondary structure diagram of the ligase that better reflects the arrangement of helices proposed by the tertiary model. Watson–Crick pairing is the same as in the original secondary structure (Fig. 1A ▶), except for one additional base pair, G47:C111, near the catalytic site. The color scheme reflects that of the ribbon representations in panel A. (C) Solvent accessibilities of the C4′ atoms in the modeled structure, as calculated using NACCES and a 1.4 Å sphere radius with an averaging over a window of three residues. A cut-off of 10 Å2 between accessibility and nonaccessibility is indicated by a horizontal line.
The key element in the modeling of the ligase is the four-way junction constituted by helices P4, P5, P6, and P7. Numerous possibilities for the arrangement of these helices were explored. A search of the crystal structure of the 50S ribosomal subunit revealed a four-way junction in domain VI of the 23S rRNA of Haloarcula marismortui with similarities to the ligase four-way junction (Ban et al. 2000). The ligase junction is characterized by the presence of 2 nt between helices P5 and P6, the first one being variable and the second one invariably an A in the 25 isolates selected from a diverse pool of class I ligase variants (Ekland and Bartel 1995). The four-way junction in the 23S rRNA also contains single-stranded residues between the two stacks of helices: on one side two As and on the other side a single A (next to a non-Watson–Crick pair). In our favored model of the ligase (Fig. 3 ▶), the four-way junction was based on the framework of the 23S rRNA four-way junction, in that the helices interrupted by the two As correspond to P5 and P6 in the ligase four-way junction. That choice led to P4/P5 and P6/P7 stacks, the four-way junction being crossed so that loops L5 and L7 are brought in the same region of space.
The favored model is consistent with the Fe-EDTA mapping; these protections fit well with a calculated solvent accessibility profile based on the model (Fig. 3C ▶). The model can also rationalize much of the conservation in sequence and pairing potential previously observed among the 25 isolates previously selected from the pool of ligase variants (Ekland and Bartel 1995). For example, the importance of pseudoknot P3 and junction J3/4 in the formation of the catalytic site is apparent.
It was rather natural to form an additional Watson–Crick pair between nt G47 and C111, both invariant among the 25 isolates. To test this predicted pairing possibility, mutants were constructed that disrupted and then restored the potential for Watson–Crick pairing. The ligation rates of the G47U mutant was 140-fold slower than the parent, and the rate of the C111A mutant was 41-fold slower, but the rate of the double mutant (G47U, C111A) was only 17-fold slower, supporting the idea that these residues are paired. Extending P4 with the G47:C111 pairing creates a bulged nucleotide, C48, which was invariant among the 25 isolates. In the model, this residue points towards the reacting G1.
The relative positioning of J3/4 with respect to the 5′ end of the molecule, which carries the reactive 5′-pppG1, suggested the presence of tandem sheared G•A base pairs, G2:A46 and A3:G45, which would constitute the third pseudoknot in the ligase ribozyme. Residues G45 and A46 in J3/4 are conserved among the 25 isolates previously selected from the pool of variants (Ekland and Bartel 1995). Evidence for the importance of G2 and A3 comes from studies of an engineered variant of the ribozyme able to perform short primer-extension reactions (Glasner et al. 2000). For example, this ribozyme utilizes the pppGGA trinucleotide substrate 1300 times more efficiently than the pppG single-nucleotide substrate. G2 and A3 had been removed in the design of this ribozyme and thus the trinucleotide substrate restored in trans analogs of these 2 nt. The proposed G•A tandem explains the increased activity with the trinucleotide substrate, suggesting how G2 and G3 could position and anchor the reactive pppG.
In J1/3, several adenine residues are invariant among the 25 previously selected isolates (Ekland and Bartel 1995). These invariant As were modeled to make contacts in the shallow groove with P1 (A-minor type) and cradle between helices P1 and P3 after touching the 5′ end of helix P5, with residue C31 being the turning residue. The chosen path for J1/3 was such that the invariant adenine A34 faces the reactive pppG from the other side of C48.
Photo-cross-linking
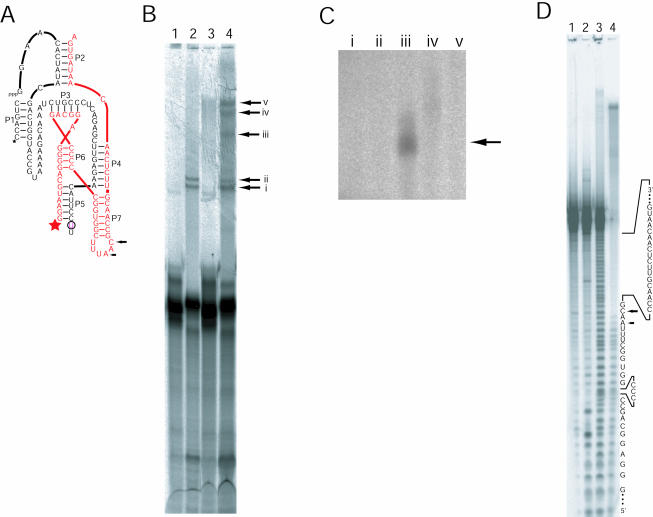
The model predicts that stems P5 and P7 project outward from the core of the ribozyme in a nearly parallel fashion, positioning L5 next to L7. To test this prediction, we investigated whether a single 4-thiouridine (4SU) placed at the position of one loop could produce a cross-link to the adjacent loop. The most straightforward way to incorporate 4SU involves breaking the ribozyme into two strands of RNA at either of the loops. Cross-linking experiments were conducted using a ribozyme containing a break in L5 because a break in this loop leads to only a threefold drop in activity, whereas a break in L7 leads to a sixfold drop. 4SU was incorporated at the 3′ terminus of the upstream fragment (Fig. 4A ▶), then the two-piece ribozyme was reconstituted, folded, and irraditated. Several catalytically active cross-linked molecules were identified based on their ability to ligate a radiolabeled substrate to themselves following irradiation (Fig. 4B ▶). One of these cross-links was to the other strand of the two-piece ribozyme (Fig. 4C ▶). This cross-link mapped to a short segment of L7 (Fig. 4D ▶). Thus, molecules constrained such that the end of P5 is near to L7 retain ligation activity, as predicted by the model.
FIGURE 4.
Crosslinking analyses of the class I ligase. (A) The secondary structure of the two-piece ribozyme used for the cross-linking study. The break in L5 facilitated insertion of a single 4SU residue (circled). The black sequence is fragment A, and the red sequence is fragment B. Stars indicate the sites of radiolabeling on the two different strands comprising the ribozyme (black 33P, red 32P). Arrows mark cross-linking sites, as mapped in panel D. (B) Separation of cross-linked ribozymes. (Lanes 1,3) The two-piece ribozyme lacks 4SU. (Lanes 2,4) The two-piece ribozyme contains 4SU. (Lanes 1,2) The RNA was not irradiated. (Lanes 3,4) Ribozymes were irradiated for 1 h before radiolabeled substrate was added. Lanes 3 and 4 contain about twice as much sample as lanes 1 and 2. The arrows with Roman numerals point to products that were excised and eluted from gel slices for subsequent relabeling. (C) Relabeling of cross-linked RNAs. The lanes marked with Roman numerals correspond to the bands in panel A. Only the RNA in lane iii was appreciably relabeled. (D) Mapping of the cross-links within an active ribozyme. (Lane 1) Unmodified, 5′-labeled fragment B RNA (red strand in A). (Lanes 2,3) Digests of labeled fragment B RNA by RNase T1, and partial alkaline hydrolysis, respectively. (Lane 4) The partial alkaline hydrolysis of the relabeled crosslinked RNA shown in panels B and C. Arrows mark the residues with the most pronounced transitions in ladder intensity, indicating the sites of the cross-links within the sequence of fragment B.
Conclusion
With the data presented here, we have been able to gain the first view of the tertiary structure of the class I ligase. As modeled, the structure of the ligase is quite compact, with extensive coaxial stacking and the P1 substrate helix lying on the four-way junction made of helices P4, P5, P6, and P7. In a general sense, the model is similar to the X-ray structures of several recently characterized RNAs, in that all are based on parallel arrangements of coaxially stacked helices (Cate et al. 1996; Ferre-D’Amare et al. 1998). In the case of the ligase, these stacks seem to be held close by the pseudoknotted secondary structure and conserved joining regions that wrap around much of the ribozyme.
Almost all of the nucleotides that were protected from hydroxyl-radical cleavage are internalized in the modeled tertiary structure (Fig. 3 ▶). In addition, cross-linking data places L5 and L7 close together, supporting the model’s predicted arrangement for the four-helix junction formed by helices P4-P5-P6-P7. The model suggested the presence of an additional Watson–Crick pair, G47:C111, which was subsequently supported experimentally. The model incorporates also a tandem of sheared G•A pairs that can rationalize previous data on the polymerase ribozyme showing the dramatic effect of GA nucleotides in the extended substrate pppGGA compared to the single substrate pppG (Glasner et al. 2000).
Taken together, these results suggest that our view of the architecture of the ligase ribozyme is accurate and should be informative for future studies of the ligase. Indeed, the model was considered when designing the experiments that successfully generated a polymerase ribozyme with the ligase core as its catalytic domain (Johnston et al. 2001). For example, the model predicted that very long primer–template duplexes (analogous to long extensions of P1) would not clash with the ribozyme core. Indeed, the polymerase ribozyme that emerged from these experiments was able to accommodate very long primer–template duplexes, making it a promising starting point for the eventual demonstration of RNA-catalyzed RNA replication.
MATERIALS AND METHODS
Ribozyme and substrate RNAs
The class I ligase ribozyme (GenBank no. U26413) was transcribed in vitro from a plasmid template linearized with EarI as described previously (Bergman et al. 2000). Transcripts were purified on 6% polyacrylamide/8 M urea gels and stored in water at −20°C. The RNA components of ligases containing breaks in either L5 or L7 were transcribed from synthetic DNA templates and purified in 10% polyacrylamide/8 M urea gels.
The ribozyme fragment with a single 4-thiouridine residue (fragment A) was prepared as follows: a synthetic DNA template for transcribing the 5′-terminal 52 nt (fragment A1) of fragment A was synthesized by standard phosphoramidite chemistry, except that 2-O-methyl phosphoramidites were used for the last two positions of the template to reduce the heterogeneity at the 3′-end of the RNA typical of RNA transcribed in vitro (Kao et al. 1999). Radiolabeled fragment A1 RNA was prepared by adding [33P]α-UTP to the transcription reaction. The 3′-terminal 11 nt of fragment A (fragment A2) was purchased as an RNA oligonucleotide (5′-GAACAUUCC-[4SU]-U, Dharmacon Research). It included a terminal U nucleotide because 4SU-Controlled Pore Glass support beads were not available. Fragments A1 and A2 were ligated to form fragment A as follows: fragment A1 (12 μM), fragment A2 (32 μM), and a 20-nt-long DNA oligonucleotide that spans the junction of fragments A1 and A2 (24 μM) were heated to 80°C in 1 mM EDTA and 10 mM Tris-HCl (pH 7.5) for 2 min and allowed to cool slowly to room temperature (Moore and Sharp 1992). T4 DNA ligase buffer, 60 μM ATP, and 3.7 units/μL of T4 DNA ligase (USB) were added, and the ligation reaction was incubated overnight in the dark at room temperature. Fully ligated products were separated in 10% polyacrylamide/8 M urea gels. Ligation efficiency was ~60%, about twice that seen when fragment A was transcribed from a DNA template that lacked the 2′-methoxy substitutions.
The substrate for the ribozyme reaction was a synthetic RNA–DNA hybrid (5′-aaaCCAGUC, DNA bases lowercase; Bergman et al. 2000). It was radiolabeled using T4 polynucleotide kinase and either [32P]γ-ATP or [33P]γ-ATP (NEN). When substrate was used for cross-linking experiments, an additional 15-min “chase” phosphorylation reaction containing excess unlabeled ATP was performed to ensure that nearly all substrate molecules had a 5′-phosphate. RNA concentrations were measured spectrophotometrically at 260 nm, assuming an extinction coefficient that was the sum of those for the individual nucleotides.
Ribozyme assays
Ribozyme reactions in which the parent ligase and derivatives were compared were performed in 50 mM MES (pH 6.0), 60 mM MgCl2, 200 mM KCl, and 600 μM EDTA at 22°C. In all cases, the ribozyme was heated (80°C, 2 min, in H2O) and then cooled (22°C, 2 min) just prior to initiation of the reaction. Ligation reactions were initiated by addition of buffer, salts, and trace 32P-labeled substrate. Aliquots were taken at appropriate time points and added to 2 volumes stop solution containing 120 mM EDTA and 8 M urea. Product and substrate were separated using 20% polyacrylamide gels and quantified by phosphorimaging. Ligation rates were calculated as described previously (Bergman et al. 2000).
Hydroxyl-radical probing
About 30% of the ligase molecules were not active upon initial folding (Schmitt and Lehman 1999; Bergman et al. 2000). To avoid probing the fold of misfolded ribozymes, ligase molecules were incubated with radiolabeled substrate, so that those that had assumed the active fold acquired the radiolabel and became visible in our analysis. Ribozyme (1 μM final concentration) was incubated with trace radiolabeled substrate (<100 nM final concentration) in buffer containing 50 mM BES (pH 7.0), 10 mM MgCl2, 200 mM KCl, and 0.1 mM EDTA. The ribozyme was heated (80°C, 2 min, in H2O) and then cooled (22°C, 2 min) just prior to initiation of the reaction. Ligation reactions were incubated for 10 min at 22°C, at which point the reaction was diluted 10-fold into 50 mM BES (pH 7.0) buffer with MgCl2 sufficient to bring the concentration of Mg++ to that indicated. When a final concentration of 0 mM Mg++ was desired, the RNA was diluted 10-fold into a solution containing 50 mM BES (pH 7.0) and 5 mM EDTA.
Labeled ligation product was subjected to hydroxyl-radical cleavage by adding to the RNA solution 0.1 volumes of a solution containing 20 mM (NH4)2Fe(II)(SO4)2, 20 mM ascorbic acid, and 22 mM EDTA. Solutions of (NH4)2Fe(II)(SO4)2 and ascorbic acid were prepared fresh before each experiment. Cleavage reactions were typically for 15 min at 22°C. For probing at other temperatures, the RNA was also allowed to equilibrate for 15 min at the desired temperature prior to addition of the Fe/Ascorbate/EDTA solution. In the range of 10°C–60°C, changing temperature did not noticeably affect the overall extent of cleavage. After addition of 2 volumes of a solution containing 8 M urea and 25 mM EDTA, cleaved RNAs were separated in 10% polyacrylamide gels and quantified by phosphorimaging (BAS2000, Fuji). Because a single gel provided accurate data on only ~50 nt, reactions were typically loaded several times and electrophoresed for times ranging from 45 min to 4 h to access most portions of the ribozyme. Individual positions were identified by comparison to partial alkaline hydrolysis and partial RNase T1 digestion ladders. Cleavage at each position was normalized to allow for differences in gel loading and cleavage efficiency (although cleavage was always done at levels in which a very small percentage of the ligase molecules were cleaved), and protection factors were calculated for nt 7–115. A protection factor was defined as the amount of cleavage at position N under denaturing conditions (60°C, 0 mM Mg++) divided by the amount of cleavage at position N under folded or experimental conditions (Pan 1995). The level of protection usually varied less than 30% from day to day, and the same positions were protected in each experiment.
Modeling of the class I ligase
Molecular modeling was performed as described previously (Westhof 1993; Massire and Westhof 1998), using the programs MANIP with FRAGMENT. The model was refined with the restrained least-squares program NUCLIN-NUCLSQ. Figures were produced using the program DRAWNA (Massire et al. 1994). The coordinates of the modeled structure of the ribozyme have been deposited in the Protein Data Bank and are available under accession number 1QXI (RCSB020188).
Isolation and mapping of active, 4SU–cross-linked ribozymes
The 4SU cross-linking agent was chosen because it has been used successfully in a variety of settings (Sontheimer and Steitz 1993; Dontsova et al. 1994; Christian and Harris 1999) and because it generates photo-dependent cross-links without a long linker arm (for review, see Favre et al. 1998). A 4SU was appended to the 3′ end of the upstream half of the ribozyme (fragment A), and the phosphates were removed from the downstream half of the ribozyme (fragment B; Fig. 4A ▶). Full ribozymes were assembled by mixing equimolar fragment A and fragment B RNAs, then heating to 80°C in water and cooling to 22°C. Reaction buffer (10 mM MgCl2, 200 mM KCl, 600 μM EDTA, and 50 mM MES at pH 7.0) was added (bringing the ribozyme concentration to 1.2 μM), and solutions containing split ribozymes were then placed in a microtiter plate that was cooled to 4°C and irradiated with a UV transilluminator (UVP) set at 302 nM. A polystyrene petri dish was used to filter out wavelengths lower than 300 nM. After irradiation for 1 h, 33P-labeled substrate was added (200 nM final concentration) and ribozymes were allowed to react for 1 h in the dark at room temperature. Note that this substrate was prepared with a final phosphorylation step using an excess of unlabeled ATP to block the free 5′-OH of the substrate. After allowing the ligation reaction to proceed for 1 h at 22°C, 2 volumes of a stop solution containing 8 M urea and 25 mM EDTA were added, and the RNAs were resolved in a 10% or 12% polyacrylamide/8 M urea gel. Control experiments were performed in parallel omitting irradiation and/or the 4SU substitution.
When the 4SU-containing ribozyme mixture was analyzed following irradiation, five bands were detected above the major band corresponding to the uncross-linked, reacted ribozyme (Fig. 4B ▶, lane 4). Quantitation of the five bands indicated that each band contained approximately the same amount of radioactivity, and each was less than 1% of counts corresponding to the uncross-linked, ligated product. Two of the five bands were observed in the control reaction in which UV irradiation was omitted (Fig. 4B ▶, lane 2), and the counts from these bands did not increase with irradiation. No bands were seen in control lanes showing ribozymes constructed without a 4SU residue, so it appeared that the three slowest migrating bands contained UV- and 4SU-dependent cross-links. All five cross-linked RNAs were excised from the gel, eluted, and precipitated in ethanol and then used as substrates in a kinase reaction, this time using [32P]-γATP instead of [33P]-γATP. This second radiolabeling identified products that cross-linked to the downstream RNA strand (fragment B), because only this type of cross-link would contain a free 5′-hydroxyl for labeling.
Only one of the five isolated RNAs was efficiently radiolabeled (Fig. 4C ▶, lane iii), and this product was purified from a 6% polyacrylamide/8 M urea gel. Inactive, cross-linked molecules should migrate differently in gels of different acrylamide percentages, so changing the gel percentage between the first and second purifications allowed the active cross-linked ribozymes to be better separated from inactive molecules. The relabeled product was then subjected to partial alkaline hydrolysis, and this reaction was run in a 10% polyacrylamide/8 M urea gel (Fig. 4D ▶). The point at which cross-linking occurred was mapped by comparison to partial alkaline hydrolysis and partial RNase T1 ladders generated from uncross-linked fragment B labeled with [32P]γ-ATP (Fig. 4D ▶).
Acknowledgments
We thank Chuck Merryman, Peter Unrau, Craig Peebles, Wendy Johnston, James Berger, and Benoît Masquida for valuable discussions. We thank also Wendy Johnston for performing the experiment supporting the G47:C111 pairing and Benoît Masquida for the plot of the accessibilities.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5177504.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 1999. Re-creating an RNA replicase. In The RNA world, 2nd ed. (eds. R.F. Gesteland et al.), pp. 143–162. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bergman, N.H., Johnston, W.K., and Bartel, D.P. 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Celander, D.W. and Cech, T.R. 1990. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: Similar reactivity toward single- and double-stranded forms. Biochemistry 29: 1355–1361. [DOI] [PubMed] [Google Scholar]
- ———. 1991. Visualizing the higher order folding of a catalytic RNA molecule. Science 251: 401–407. [DOI] [PubMed] [Google Scholar]
- Cheong, C., Varani, G., and Tinoco Jr., I. 1990. Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature 346: 680–682. [DOI] [PubMed] [Google Scholar]
- Christian, E.L. and Harris, M.E. 1999. The track of the pre-tRNA 5′ leader in the ribonuclease P ribozyme–substrate complex. Biochemistry 38: 12629–12638. [DOI] [PubMed] [Google Scholar]
- Dontsova, O., Tishkov, V., Dokudovskaya, S., Bogdanov, A., Doring, T., Rinke-Appel, J., Thamm, S., Greuer, B., and Brimacombe, R. 1994. Stem-loop IV of 5S rRNA lies close to the peptidyltransferase center. Proc. Natl. Acad. Sci. 91: 4125–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein, F. 1985. Nucleoside phosphorothioates. Annu. Rev. Biochem. 54: 367–402. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1995. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 23: 3231–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1996. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382: 373–376. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370. [DOI] [PubMed] [Google Scholar]
- Favre, A., Saintome, C., Fourrey, J.L., Clivio, P., and Laugaa, P. 1998. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid–protein interactions. J. Photochem. Photobiol. B 42: 109–124. [DOI] [PubMed] [Google Scholar]
- Ferre-D’Amare, A.R., Zhou, K., and Doudna, J.A. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395: 567–574. [DOI] [PubMed] [Google Scholar]
- Glasner, M.E., Yen, C.C., Ekland, E.H., and Bartel, D.P. 2000. Recognition of nucleoside triphosphates during RNA-catalyzed primer extension. Biochemistry 39: 15556–15562. [DOI] [PubMed] [Google Scholar]
- Glasner, M.E., Bergman, N.H., and Bartel, D.P. 2002. Metal ion requirements for structure and catalysis of an RNA ligase ribozyme. Biochemistry 41: 8103–8112. [DOI] [PubMed] [Google Scholar]
- Hertzberg, R.P. and Dervan, P.B. 1984. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): Reaction conditions and product analyses. Biochemistry 23: 3934–3945. [DOI] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., and Bartel, D.P. 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319– 1325. [DOI] [PubMed] [Google Scholar]
- Joseph, S. and Noller, H.F. 2000. Directed hydroxyl radical probing using iron(II) tethered to RNA. Methods Enzymol. 318: 175–190. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. and Orgel, L.E. 1999. Prospects for understanding the origin of the RNA world. In The RNA world, 2nd ed. (eds. R.F. Gesteland et al.), pp. 49–77. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kao, C., Zheng, M., and Rudisser, S. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, P.A., Jamison, E., Strahs, D., Anderson, V.E., and Brenowitz, M. 1993. ‘Footprinting’ proteins on DNA with peroxonitrous acid. Nucleic Acids Res. 21: 2473–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham, J.A. and Cech, T.R. 1989. Defining the inside and outside of a catalytic RNA molecule. Science 245: 276–282. [DOI] [PubMed] [Google Scholar]
- Massire, C. and Westhof, E. 1998. MANIP: An interactive tool for modelling RNA. J. Mol. Graph. Model 16: 197–205, 255–257. [DOI] [PubMed] [Google Scholar]
- Massire, C., Gaspin, C., and Westhof, E. 1994. DRAWNA: A program for drawing schematic views of nucleic acids. J. Mol. Graph 12: 201–206. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. and Sharp, P.A. 1992. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science 256: 992–997. [DOI] [PubMed] [Google Scholar]
- Pan, T. 1995. Higher Order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 34: 902–909. [DOI] [PubMed] [Google Scholar]
- Pan, T. and Sosnick, T.R. 1997. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 4: 931–938. [DOI] [PubMed] [Google Scholar]
- Russell, R. and Herschlag, D. 1999. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 291: 1155–1167. [DOI] [PubMed] [Google Scholar]
- Schmitt, T. and Lehman, N. 1999. Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chemis. Biol. 6: 857–869. [DOI] [PubMed] [Google Scholar]
- Sclavi, B., Woodson, S., Sullivan, M., Chance, M., and Brenowitz, M. 1998. Following the folding of RNA with time-resolved synchrotron X-ray footprinting. Methods Enzymol. 295: 379–402. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J. and Steitz, J.A. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. [DOI] [PubMed] [Google Scholar]
- Tullius, T.D. and Dombroski, B.A. 1986. Hydroxyl radical “footprinting”: High-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc. Natl. Acad. Sci. 83: 5469–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius, T.D., Dombroski, B.A., Churchill, M.E., and Kam, L. 1987. Hydroxyl radical footprinting: A high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 155: 537–558. [DOI] [PubMed] [Google Scholar]
- Westhof, E. 1993. Modeling the three-dimensional structure of ribonucleic acids. J. Mol. Struct. 286: 203–210. [Google Scholar]
- Wu, J.C., Kozarich, J.W., and Stubbe, J. 1983. The mechanism of free base formation from DNA by bleomycin. A proposal based on site specific tritium release from Poly(dA.dU). J. Biol. Chem. 258: 4694–4697. [PubMed] [Google Scholar]