Abstract
We have previously shown that alpha/beta interferon (IFN-α/β) and gamma interferon (IFN-γ) inhibit hepatitis B virus (HBV) replication by eliminating pregenomic RNA containing viral capsids from the hepatocyte. We have also shown that HBV-specific cytotoxic T lymphocytes that induce IFN-γ and tumor necrosis factor alpha (TNF-α) in the liver can inhibit HBV gene expression by destabilizing preformed viral mRNA. In order to further study the antiviral activity of IFN-α/β, IFN-γ, and TNF-α at the molecular level, we sought to reproduce these observations in an in vitro system. Accordingly, hepatocytes were derived from the livers of HBV-transgenic mice that also expressed the constitutively active cytoplasmic domain of the human hepatocyte growth factor receptor (c-Met). Here, we show that the resultant well-differentiated, continuous hepatocyte cell lines (HBV-Met) replicate HBV and that viral replication in these cells is efficiently controlled by IFN-α/β or IFN-γ, which eliminate pregenomic RNA-containing capsids from the cells as they do in the liver. Furthermore, we demonstrate that IFN-γ, but not IFN-α/β, is capable of inhibiting HBV gene expression in this system, especially when it acts synergistically with TNF-α. These cells should facilitate the analysis of the intracellular signaling pathways and effector mechanisms responsible for these antiviral effects.
Hepatitis B virus (HBV) is a hepatotropic DNA virus that causes acute and chronic hepatitis and hepatocellular carcinoma (5). We have previously shown that the intrahepatic induction of alpha/beta interferon (IFN-α/β), gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) by various stimuli can inhibit HBV gene expression and/or HBV replication in the livers of HBV-transgenic mice (3, 9, 10, 23). More recently, we demonstrated that similar noncytopathic antiviral events probably occur in the livers of chimpanzees acutely infected with HBV (15).
Although it is now well documented that HBV replication can be inhibited by these cytokines (11, 19), in vivo systems are by nature difficult to manipulate and control. Therefore, studies designed to determine which cytokines primarily inhibit HBV replication and to decipher the antiviral intracellular mechanism(s) they induce would be greatly facilitated by an in vitro cell culture system that accurately reflects the cytokine-dependent control of HBV observed in vivo. Unfortunately, primary cultures of HBV-transgenic hepatocytes are not suitable for these studies because they rapidly become less well differentiated in vitro. Recently, however, well-differentiated immortalized mouse hepatocyte cell lines were established from transgenic mice expressing the constitutively active cytoplasmic domain of the human hepatocyte growth factor receptor (c-Met) in their livers (2). Cells prepared from these mice are not transformed, and they can be induced to a stable, highly differentiated hepatocyte phenotype by dimethyl sulfoxide (DMSO) (29).
In this report, we describe the establishment and characterization of an in vitro cell culture system (HBV-Met) based on immortalized, highly differentiated hepatocytes prepared from mice transgenic for both c-Met and HBV. We show that HBV-Met cells, induced to a highly differentiated hepatocyte phenotype by DMSO, support HBV gene expression, HBV replication, and virus secretion. Importantly, HBV replication in this in vitro system is inhibited by the intracellular antiviral mechanism(s) induced by either IFN-α/β or IFN-γ, but not by TNF-α. Moreover, longer treatment with IFN-γ leads to a reduction in the intracellular level of HBV transcripts, especially when it acts synergistically with TNF-α.
MATERIALS AND METHODS
Transgenic mice.
HBV-transgenic mouse lineage 1.3.46 (official designation, Tg[HBV 1.3 genome] Chi32) has been previously described (14). These mice replicate HBV in the hepatocyte from an integrated greater-than-genome-length HBV transcriptional template. The level of HBV replication in the livers of these mice is comparable to that seen in the livers of infected patients with chronic hepatitis, and there is no evidence of cytopathology (14). Cyto-Met transgenic mice which express the constitutively active cytoplasmic domain of the human hepatocyte growth factor receptor (c-Met) in their livers have also been previously described (1). HBV-Met double-transgenic mice were produced by mating homozygous 1.3.46 mice with homozygous cyto-Met transgenic mice. F1 progeny were tested for HBV gene expression and replication by detection of hepatitis B e antigen (HBeAg) in the serum (EBK 125I RIA kit; DiaSorin Inc., Stillwater, Minn.) according to the manufacturer's instructions.
Primary hepatocyte culture.
Livers from 3-week-old F1 mice were perfused and collagenase digested (collagenase D; Roche Molecular Biochemicals, Indianapolis, Ind.) as described previously (22). The cells were further purified over a 60% Percoll gradient (Pharmacia Biotech, Uppsala, Sweden). Hepatocyte viability was assessed by trypan blue dye exclusion to be >80%. The cells were plated at high density on collagen I-coated Biocoat dishes (Becton Dickinson, Franklin Lakes, N.J.) in RPMI 1640 (Gibco Invitrogen Corp., Carlsbad, Calif.) supplemented with 10% fetal calf serum (Gibco Invitrogen Corp.), 55 ng of epidermal growth factor (Becton Dickinson)/ml, 16 ng of IGF-II (Calbiochem, San Diego, Calif.)/ml, 10 μg of insulin (Sigma, St. Louis, Mo.)/ml, and penicillin-streptomycin-glutamine (100×; liquid) (Gibco Invitrogen Corp.) (complete medium), as previously described (2a, 29). After overnight incubation, the majority of the cells had attached. The semiconfluent cultures were washed and maintained without transfer for several weeks.
HBV-Met cell lines and clones.
All subsequent cell maintenance was performed using collagen-coated plasticware and complete RPMI medium as described above for the primary hepatocyte cultures. Individual epithelial cell islands were expanded slowly in dishes of increasing size and frozen after 10 generations. Eight immortalized HBV-transgenic hepatocyte lines were established. One of these HBV-Met lines is characterized in the present work. To obtain HBV-Met cell clones, serial dilutions of an early passage of HBV-Met cells were plated in collagen I-coated 96-well plates. Twelve cell clones were established based on their hepatocyte-like morphology as described in Spagnoli et al. (29). These 12 clones were subsequently tested for HBV gene expression by Northern blot analysis and for HBV replication by Southern blot analysis as described below. Clone HBV-Met.4 was chosen for the studies presented in this work. Aliquots of 5 × 106 cells of early passages of the HBV-Met.4 cells were frozen in 90% serum and 10% DMSO. Otherwise, HBV-Met.4 cells were propagated in complete medium until they reached confluence and then passaged at a 1:15 dilution. All experiments were performed with cells that underwent <40 passages. For experiments, typically 0.5 × 106 to 1 × 106 HBV-Met.4 cells were seeded into 60-mm-diameter dishes.
Cytokine treatment.
Prior to each experiment, immortalized hepatocytes were grown to confluence and subsequently kept in complete medium supplemented with 2% DMSO for 10 days, at which time cytokine treatments were started. Recombinant murine IFN-β (mIFN-β; provided by M. Moriyama, Toray Industries, Tokyo, Japan), recombinant mIFN-γ, or recombinant mTNF-α (provided by S. Kramer, Genentech, South San Francisco, Calif.) was added at the indicated concentrations. During the whole time of DMSO addition and cytokine treatment, the medium was replaced every other day with fresh medium, supplemented with DMSO and cytokines when necessary.
HBV DNA analysis.
Cells were lysed in the culture dish by adding 500 μl of DNA lysis buffer (50 mM Tris-HCl [pH 8.0], 20 mM EDTA, and 1% sodium dodecyl sulfate). Samples were then digested overnight at 37°C with proteinase K (1 mg/ml), and total DNA was extracted as described previously (14). Twenty micrograms of total DNA was analyzed by Southern blotting with a 32P-labeled full-length HBV DNA probe after HinDIII digestion (14). All quantifications were done with a Cyclone storage phosphor system (Packard Instrument Company, Meriden, Conn.).
HBV DNA in the cell culture supernatant.
Two days after medium change, 400 μl of HBV-Met cell culture supernatant was collected and centrifuged at 14,000 × g for 10 s to remove cell debris. The supernatant was then digested with 1 mg of proteinase K/ml in a total volume of 500 μl containing 50 mM Tris base (pH 8.0) and 1% sodium dodecyl sulfate at 37°C overnight. Nucleic acids were extracted by phenol-chloroform extraction and precipitated after the addition of 10 μg of Escherichia coli tRNA with isopropanol. Nucleic acids were dissolved in 30 μl of Tris-EDTA; 15 μl was loaded onto a 1.3% agarose gel (1× Tris-acetate-EDTA) and electrophoresed for 1 h at 5 V/cm. The gel was then blotted in 1.5 M NaCl-0.5 M NaOH by vacuum blotting it (VacuGene; Amersham Pharmacia Biotech AB, Uppsala, Sweden) for 1 h onto a nylon membrane (Magnagraph; Osmonics Laboratory Products, Minnetonka, Minn.). Subsequent Southern blotting was performed as described previously (14). Alternatively, HBV in the cell culture supernatant was quantified by subjecting 0.5 μl of extracted nucleic acids to HBV (genotype ayw)-specific TaqMan PCR in a 50-μl reaction volume using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, Calif.), 200 nM upper primer HBV469U. The probe was labeled with fluorescein (6-FAM) at the 5′ end and with the black hole quencher (BHQ-1; Bioresearch Technologies, Novato, Calif.) at the 3′ end (5′-CCCGTTTGTCCTCTAATTCC-3′), 200 nM lower primer HBV569L (5′-GTCCGAAGGTTTGGTACAGC-3′), and 100 nM TaqMan probe HBV495P [5′-6-FAMd(CTCAACAACCAGCACGGGACCA)BHQ-1-3′]. Tenfold serial dilutions (108 to 100 copies) of plasmid DNA containing a monomeric HBV insert were used as standards in parallel HBV-specific PCRs.
HBV RNA analysis.
Total cellular RNA was isolated by the guanidine thiocyanate method using standard protocols (6). Encapsidated RNA was extracted from cells grown in a 60-mm-diameter culture dish. Briefly, cells were scraped from the dish into 1 ml of PBS and pelleted by brief centrifugation in a tabletop microtube centrifuge. The pelleted cells were lysed in 0.3 ml of lysis buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris base [pH 8.0], 0.5% NP-40). Nuclei were pelleted by centrifugation for 5 min at 12,000 × g and 4°C in a microtube centrifuge. Encapsidated RNA in the supernatant was extracted as previously described (33). The RNA was dissolved in 50 μl of H2O, and 15 μl was used for Northern blot analysis as described previously (14).
Immunohistochemical analysis.
HBV-Met cells were grown in collagen I-coated Biocoat eight-chamber culture slides. The cells were fixed as previously described (13). The intracellular distribution of HBV core antigen (HBcAg) was detected by an avidin-biotin detection system as described elsewhere (13). This method sequentially uses a primary rabbit anti-HBcAg antiserum (Dako, Carpinteria, Calif.), a secondary biotin-conjugated goat antiserum specific for rabbit immunoglobulin G [IgG(Fab′)2; Sigma Chemical Co.], a streptavidin-horseradish peroxidase conjugate (Extravidin; Sigma), and 3-amino-9-ethyl carbazole (Shandon-Lipshaw, Pittsburgh, Pa.) as a coloring substrate.
RESULTS
Efficient HBV replication in an HBV-Met transgenic hepatocyte cell line.
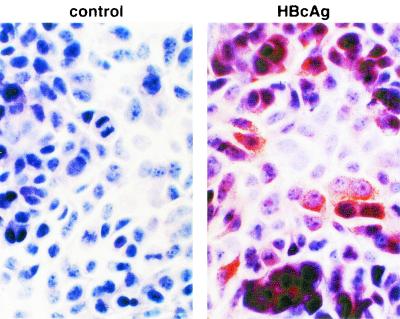
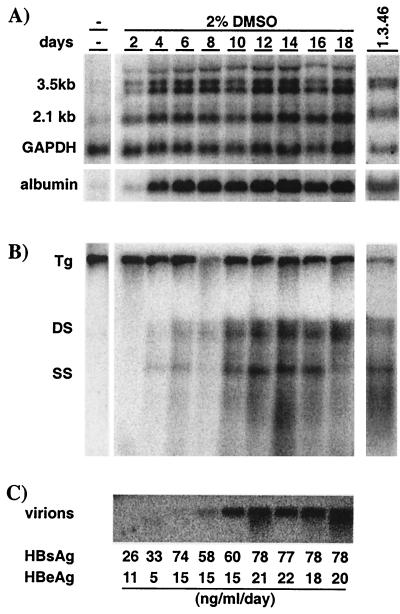
An immortalized hepatocyte cell line, HBV-Met, was derived from primary mouse hepatocyte cultures isolated from 1.3.46/c-Met double-transgenic mice as described above. As shown in Fig. 1, these cells grow as a cobblestone epithelial monolayer. They require collagen for attachment and, when provided with serum, epidermal growth factor I, IGF II, and insulin, they have a doubling time of approximately 24 h (data not shown). In the absence of DMSO, dividing cells expressed low levels of HBV RNA transcripts as measured by Northern blot analysis (Fig. 2A, far left); however, no HBV replicative intermediates were detectable by Southern blot analysis of total cellular DNA (Fig. 2B, far left). Because it has been shown that treatment of c-Met immortalized hepatocyte cells with 2% DMSO induces hepatocellular differentiation (29), we incubated confluent monolayers of HBV-Met cells with DMSO and monitored the effect on HBV gene expression and replication for the next 18 days. Consistent with differentiation, cellular expression of the liver-specific gene for albumin was observed to increase during DMSO treatment (Fig. 2A, bottom), and this was accompanied by an increase in HBV transcripts (Fig. 2A, days 2 to 18) and cytoplasmic HBV DNA replicative intermediates (Fig. 2B, days 2 to 18). HBV replication reached a steady-state level after approximately 10 days of DMSO treatment, which was comparable to the level of HBV replication in the livers of the parental 1.3.46 HBV-transgenic mice (Fig. 2B, far right). Consistent with active replication, significant amounts of cytoplasmic HBcAg were detectable by immunohistochemistry at this time (Fig. 1, right panel). As viral replication increased, there was also a corresponding secretion of HBV virions, indicated by the presence of mature HBV DNA in the cell culture supernatant (Fig. 2C). Likewise, secretion of HBsAg and HBeAg into the cell media was detected (Fig. 2C).
FIG. 1.
Immortalized HBV-transgenic hepatocyte cell lines express viral antigen. HBV-Met cells were grown to confluence and then cultured in 2% DMSO. After 10 days of DMSO treatment, HBV-Met cells were analyzed for intracellular HBcAg by immunohistochemistry in the absence (left panel) or presence (right panel) of HBcAg-specific primary antibody as described in Materials and Methods.
FIG. 2.
Efficient HBV gene expression and replication in immortalized HBV-transgenic hepatocytes. HBV-Met cells were grown to confluence and then cultured in 2% DMSO for an additional 18 days. Total cellular RNA and DNA were harvested prior to and every 2 days during DMSO treatment. For comparison, total liver RNA and DNA from 1.3.46 HBV-transgenic mice were extracted and analyzed as previously described (14) (far right). (A) Northern blot analysis for HBV, GAPDH, and albumin gene expression. (B) Southern blot analysis of intracellular HBV DNA replicative intermediates. (C) HBV virion DNA was extracted from cell culture supernatant and analyzed by Southern blotting as described in Materials and Methods. Levels of HBeAg and HBV surface antigen (HBsAg) secreted into the culture supernatant are indicated at the bottom in nanograms per milliliter per day. Abbreviations: Tg, HBV transgene; DS, HBV dsDNA; SS, HBV ssDNA.
Inflammatory cytokines eliminate cytoplasmic HBV DNA replicative intermediates from HBV-Met cells.
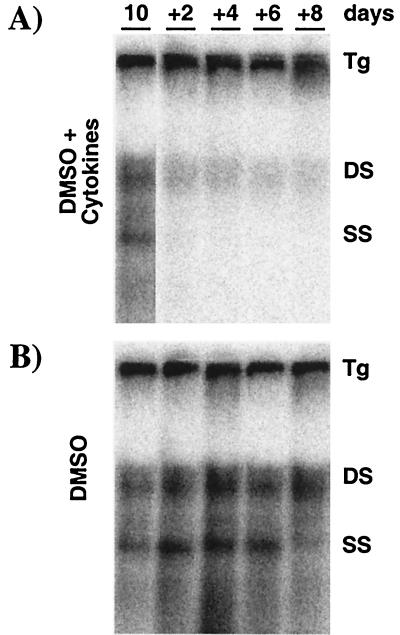
To determine whether HBV replication in the HBV-Met cells is inhibited by inflammatory cytokines, we treated the cultures with a pool of cytokines and measured the level of HBV replicative intermediates over time by Southern blot analysis. For these and all subsequent experiments, confluent HBV-Met cells were incubated for 10 days with 2% DMSO, allowing HBV replication to reach steady-state levels (Fig. 3A, lane 1). The cultures were then maintained in 2% DMSO alone or in DMSO plus a pool of mIFN-β, mIFN-γ, and mTNF-α at 1,000 U/ml each. DNA was extracted from the cells, and the level of intracellular HBV DNA intermediates was monitored beginning at 2 days and continuing up to 8 days after cytokine addition.
FIG. 3.
Inflammatory cytokines inhibit HBV replication in hepatocyte cell cultures. HBV-Met cells were grown to confluence and then cultured in 2% DMSO. Starting at day 10 of DMSO treatment, the culture medium was replaced every other day (+2 to +8) by complete medium containing 2% DMSO. At the same time points, total cellular DNA was harvested and subjected to HBV-specific Southern blot analysis. (A) Starting at day 10, the medium was supplemented with a pool of mIFN-β, mIFN-γ, and mTNF-α (1,000 U/ml each) at every medium change. (B) Control cultures were treated exactly as described for panel A except for omitting cytokines from the culture medium. For abbreviations, see the legend to Fig. 2.
The morphology, cell count, and viability of the cells were not affected by the cytokine treatment over the time course of the experiment (data not shown). However, Fig. 3A shows that cytokine treatment of the HBV-Met cells profoundly reduces cytoplasmic HBV DNA replicative intermediates as early as 2 days after cytokine addition compared to untreated cells harvested at the same time points (Fig. 3B). As observed after cytokine induction in HBV-transgenic mice (33), single-stranded DNA (ssDNA) is completely abolished, while trace amounts of double-stranded DNA (dsDNA) remained throughout the experiment (Fig. 3A). Therefore, in vitro cytokine treatment of HBV-Met cells induces an antiviral activity that appears to mimic the noncytopathic clearance of HBV replication observed in vivo.
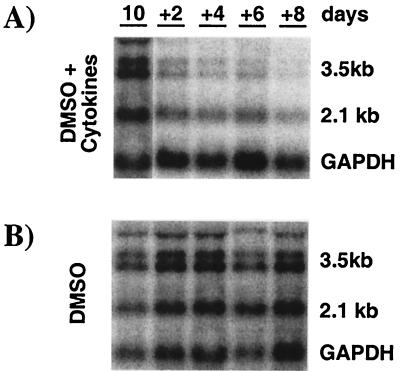
The cellular level of HBV transcripts is controlled by inflammatory cytokines.
We have previously shown that the intrahepatic induction of cytokines in HBV-transgenic mice after stimuli such as injection of virus-specific cytotoxic T-lymphocytes (12), infection with murine cytomegalovirus (3), or persistent infection with lymphocytic choriomeningitis virus (9) suppresses HBV gene expression. Accordingly, the level of HBV RNA was monitored in total RNA extracted from parallel cell cultures in the experiment shown in Fig. 3. In DMSO-treated control cell cultures, the cellular content of HBV RNA did not change significantly during the course of the experiment (Fig. 4B). In contrast, 2 days after cytokine addition, the levels of the HBV 3.5- and 2.1-kb transcripts were reduced threefold when normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (Fig. 4A; lane +2). With continuous cytokine treatment, HBV RNA levels further decreased over time (Fig. 4A; lanes +2 to +8). By day 8 of cytokine treatment, the 3.5- and 2.1-kb HBV transcript levels were reduced 30-fold and 5-fold, respectively (Fig. 4A, lane +8), compared to pretreatment levels. In summary, the magnitude and kinetics of the cytokine-mediated inhibition of HBV replication and gene expression observed in the HBV-Met cell culture system are strikingly similar to our previous findings in HBV-transgenic mice after injection of virus-specific cytotoxic T lymphocytes, where viral DNAs are cleared first, followed by a reduction in viral RNAs (12).
FIG. 4.
Inflammatory cytokines inhibit HBV gene expression in hepatocyte cell cultures. HBV-Met cells were treated exactly as described in the legend to Fig. 3. Total RNA was extracted from the cells at the indicated time points and analyzed for HBV and GAPDH gene expression by Northern blotting. (A) Starting at day 10, the medium was supplemented with a pool of mIFN-β, mIFN-γ, and mTNF-α (1,000 U/ml each) at every medium change. (B) Control cultures were treated exactly as described for panel A except for omitting cytokines from the culture medium. For abbreviations, see the legend to Fig. 2.
Inflammatory cytokines eliminate pgRNA-containing capsids from the hepatocyte cytoplasm.
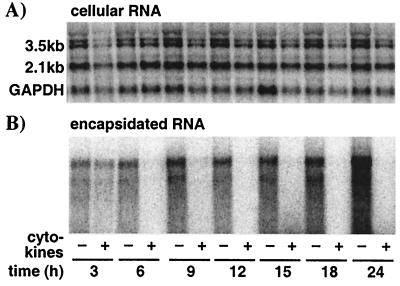
Previously, we have shown that IFN-α/β induction in the livers of HBV-transgenic mice eliminates pregenomic RNA (pgRNA)-containing capsids from the hepatocyte cytoplasm within 6 to 12 h without affecting the level of HBV-specific mRNA (33). In this set of experiments, we wanted to determine whether the cytokines affect the same step(s) in HBV replication in the HBV-Met cell culture model. Therefore, we treated HBV-Met cells with cytokines as described in the legend to Fig. 3 and monitored the intracellular level of HBV transcripts, as well as encapsidated HBV RNA isolated from cytoplasmic HBV capsids, every 3 h during the first 24 h of the treatment.
Figure 5A shows the intracellular level of HBV RNA in the HBV-Met cells in the presence and absence of cytokine treatment at the indicated time points after cytokine addition. Quantification of the Northern blot signals revealed no change in total HBV RNA in the cells over the course of the experiment. In contrast, encapsidated HBV RNA is cleared from the HBV-Met cell cytoplasm as early as 6 h after cytokine addition, despite the fact that encapsidated HBV RNA is still increasing in the control samples harvested at the same time points (Fig. 5B). The disappearance of encapsidated HBV RNA from the cells by 6 h indicates that the cytokines induce an intracellular mechanism that eliminates pgRNA-containing capsids very rapidly. Furthermore, these results are consistent with a hypothesis proposed based on in vivo data (33), in which inflammatory cytokines actively eliminate pgRNA-containing capsids from the cell while the derivative HBV DNA replicative intermediates are passively cleared from the cells as a result of capsid maturation and viral export.
FIG. 5.
Inflammatory cytokines eliminate pgRNA-containing capsids from the hepatocyte cytoplasm. HBV-Met cells were grown to confluence and then cultured in 2% DMSO for an additional 10 days. At that time, a pool of mIFN-β, mIFN-γ, and mTNF-α (1,000 U/ml each) was added to the culture medium. (A) Total cellular RNA was isolated from untreated (−) and cytokine-treated (+) HBV-Met cells at the indicated time points and analyzed for intracellular HBV and GAPDH transcripts by Northern blotting. (B) Encapsidated RNA was extracted from cytoplasmic HBV capsids and subjected to HBV-specific Northern blot analysis as described in Materials and Methods.
Establishment and characterization of HBV-Met clones.
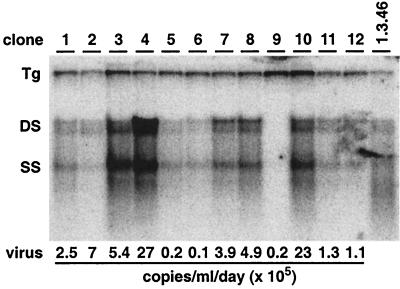
To ensure that the HBV-Met cell culture system reflects only events that occur in hepatocytes and not nonparenchymal cells that might have contaminated the HBV-Met cell line, we established 12 clones that cytologically resembled hepatocytes (29). The clones were treated with DMSO for 10 days and then analyzed for HBV replication by Southern blot analysis of total cellular DNA (Fig. 6). The levels of HBV replication varied considerably in different clones (Fig. 6), consistent with the differences in HBcAg expression in different cells in the HBV-Met cell line (Fig. 1, right panel). Quantitative real-time PCR of viral DNA in the cell culture supernatant collected at the same time point showed that the amount of virus secreted from the cells reflects the level of intracellular HBV replication (Fig. 6). For two clones (HBV-Met.4 and HBV-Met.7), it was also verified that HBV replication was equally sensitive to cytokine treatment as in the cell lines (data not shown). All subsequent studies described were performed with the HBV-Met cell clone no. 4 (HBV-Met.4) (Fig. 6, clone 4).
FIG. 6.
HBV replication in HBV-Met cell clones. HBV-Met clones were established by limiting dilution of the HBV-Met cell line. Individual clones were grown to confluence and then maintained in 2% DMSO for an additional 10 days. At that time, total DNA was extracted from the cells and analyzed for HBV DNA replicative intermediates by Southern blotting. Viral DNA extracted from the cell culture supernatant was quantified by real-time PCR and is indicated as viral copies per milliliter per day. For abbreviations, see the legend to Fig. 2.
Both mIFN-β and mIFN-γ induce a hepatocellular mechanism(s) that inhibits HBV replication.
Clone HBV-Met.4 was used to assess the individual effects of mIFN-β, mIFN-γ, and mTNF-α on HBV replication. For these experiments, the cells were grown and differentiated as described in the legend to Fig. 2, and parallel dishes were treated with either mIFN-β (100 U/ml), mIFN-γ (1,000 U/ml), mTNF-α (1,000 U/ml), or a combination of mIFN-γ and mTNF-α (1,000 U/ml each). Total cellular DNA was harvested at the indicated time points after cytokine addition and analyzed for intracellular HBV DNA replicative intermediates by Southern blot analysis.
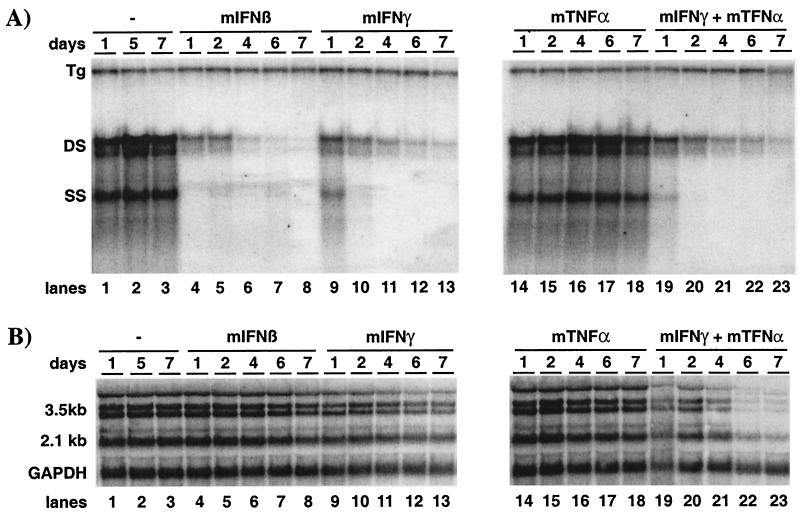
Figure 7A, left, lane 4 shows that HBV replication in the HBV-Met.4 cells was reduced 18-fold at the level of ssDNA and reduced 5-fold at the level of dsDNA after only 1 day of 100-U/ml mIFN-β treatment alone. HBV DNA continued to decrease thereafter, by more than 90-fold at day 7 after mIFN-β addition (Fig. 7A, lanes 5 to 8) compared to the untreated control samples (Fig. 7A, lanes 1 to 3). Similarly, mIFN-γ treatment of HBV-Met.4 cells resulted in a 25-fold inhibition of HBV DNA replication on day 7 (Fig. 7A, lane 13), whereas HBV ssDNA and dsDNA were reduced only 4-fold on day 1 (Fig. 7A, lane 9). This suggests that mIFN-γ is less potent than mIFN-β in this system and that its antiviral effect is delayed. In fact, titration experiments showed that as little as 10 U of mIFN-β/ml could inhibit HBV replication whereas at least 100 U of mIFN-γ/ml was needed to induce an antiviral response (data not shown). Figure 8A, right, lanes 14 to 18, shows that HBV DNA replication was not controlled by mTNF-α under these experimental conditions. When mTNF-α was combined with mIFN-γ, HBV replication was inhibited (Fig. 7A, lanes 19 to 23), but the kinetics and extent of inhibition mirrored the antiviral activity seen with mIFN-γ alone. Similarly, no other combination of these cytokines enhanced the effect of the most potent one in the mixture at the level of HBV replication (data not shown).
FIG. 7.
Both type I and type II cytokines induce a hepatocellular mechanism(s) inhibiting HBV replication. The HBV-Met.4 clone was grown to confluence and then maintained in 2% DMSO. After 10 days of culture in DMSO, parallel dishes were treated with either 100 U of mIFN-β/ml, 1,000 U of mIFN-γ/ml, 1,000 U of mTNF-α/ml, or a combination of mIFN-γ and mTNF-α at 1,000 U/ml each. (A) Southern blot analysis for HBV DNA replicative intermediates at the indicated time points during cytokine treatment or from untreated control cultures. (B) Total cellular RNA harvested at the same time points and analyzed for HBV and GAPDH transcripts by Northern blot analysis. For abbreviations, see the legend to Fig. 2.
FIG. 8.
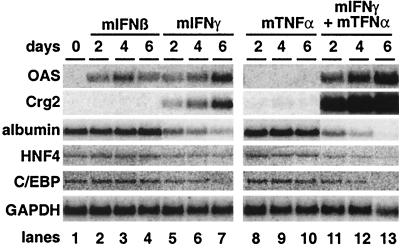
Changes in cellular gene expression induced by inflammatory cytokines. Total cellular RNA from the experiment shown in Fig. 7 was subjected to Northern blot analysis specific for the indicated genes. Transcript levels at days 0, 2, 4, and 6 of cytokine treatment are shown.
Synergistic antiviral activity of mIFN-γ and mTNF-α on HBV gene expression.
To determine which of the cytokines was responsible for the antiviral activity that inhibited HBV gene expression as shown in Fig. 3, total cellular RNA isolated from parallel dishes in the experiment shown in Fig. 7A was subjected to Northern blot analysis. To control for loading differences, the membranes were also probed for GAPDH.
Although mIFN-β rapidly and profoundly reduced the level of intracellular HBV DNA replicative intermediates (Fig. 7A, lanes 4 to 8), extended treatment with mIFN-β did not affect the steady-state level of HBV transcripts (Fig. 7B, lanes 4 to 8). Prolonged treatment with mTNF-α also failed to reduce HBV mRNA levels (Fig. 7B, lanes 14 to 18), while mIFN-γ caused a slight (twofold) reduction of HBV transcripts in the HBV-Met.4 cells by day 7 (Fig. 7B, lanes 9 to 13). Importantly, the combination of mIFN-γ and mTNF-α reduced the HBV mRNA content approximately 17-fold on day 6 (Fig. 7B, lanes 19 to 22), suggesting that these two cytokines act synergistically to inhibit HBV gene expression. In contrast, the combination of mTNF-α with mIFN-β did not inhibit HBV gene expression (data not shown).
The synergy between mIFN-γ and mTNF-α was also evident at the level of liver-specific cellular gene expression. As shown in Fig. 8, while GAPDH mRNA levels remained relatively constant under all conditions (Fig. 8, bottom panel), the steady-state transcript level of several hepatocellular genes varied depending on the particular cytokine administered. As expected, transcript levels for 2′,5′-oligoadenylate synthetase (OAS) were induced by mIFN-β (Fig. 8, lanes 1 to 4) and by mIFN-γ (Fig. 8, lanes 5 to 7) but not by mTNF-α (Fig. 8, lanes 8 to 10). However, the combination of mTNF-α and mIFN-γ induced OAS expression to levels 5-fold higher than mIFN-γ alone (Fig. 8, lanes 11 to 13). Similarly, expression of the chemokine Crg-2 was induced to 100-fold-higher levels by the combination of mTNF-α and mIFN-γ (Fig. 8, lanes 11 to 13) compared to mIFN-γ alone (Fig. 8, lanes 5 to 7), whereas mTNF-α alone (Fig. 8, lanes 8 to 10), like mIFN-β alone, had no effect (Fig. 8, lanes 2 to 4).
In the opposite manner, some hepatocellular genes were subject to synergistic down-regulation by mTNF-α and mIFN-γ. Expression of the negative acute-phase gene for albumin was reduced sevenfold after 6 days of mIFN-γ treatment (Fig. 8, lane 7), while mIFN-β alone (Fig. 8, lanes 2 to 4) or mTNF-α alone (Fig. 8, lanes 8 to 10) had no effect. The combination of mIFN-γ and mTNF-α, however, enhanced albumin down-regulation 50-fold (Fig. 8, lanes 11 to 13). The same pattern of change in gene expression was seen for the liver transcription factor hepatocyte nuclear factor 4 (HNF4) and C/EBPα, whose transcripts were minimally reduced by mIFN-γ alone, not reduced by mIFN-β or mTNF-α, and virtually eliminated by the combination of mIFN-γ and mTNF-α.
DISCUSSION
In this report, we describe an immortalized HBV-transgenic mouse hepatocyte cell culture system that replicates HBV and is sensitive to the antiviral activities of cytokines, reflecting the behavior of the virus and the cytokines in vivo (3, 4, 9, 11, 12, 15, 33). Immortalized hepatocyte cultures were established from mice transgenic for HBV (14) and the constitutively active cytoplasmic fragment of the human hepatocyte growth factor receptor (c-Met [1]). Cell lines immortalized by c-Met have been shown to display a liver-specific gene expression profile which can be further enhanced by cultivating the cells in the presence of the well-known differentiation agent DMSO (2, 29, 35). Consistent with this finding, DMSO induced the expression of hepatocyte-specific genes, such as those for albumin and transthyretin (data not shown), in our HBV-Met cells. Robust HBV gene expression was also dependent on DMSO-induced hepatocyte differentiation, perhaps due to the activation of hepatocyte-specific transcription factors, which have been shown to regulate HBV promoters (18, 30). Notably, however, HBV DNA replication does not seem to be dependent on hepatocyte differentiation, since it is easily supported in nondifferentiated HBV-Met cells upon transfection of constructs that express the viral transcripts from constitutively active promoters (data not shown). Consistent with this finding, it has recently been shown that expression of certain liver-specific transcription factors that transcriptionally activate HBV allows for HBV replication in nonhepatic cells (31). Therefore, the strong restriction of HBV to the liver in vivo is, at least at one level, the consequence of the dependence of HBV promoter and enhancer sequences on hepatocyte-specific transcription factors.
One advantage of the in vitro HBV replication model system presented here is the availability of a corresponding in vivo system. Hence, we have shown that the profound cytokine-induced inhibition of HBV replication we observed in vitro quantitatively and qualitatively reflects the events that occur after cytokine induction in vivo in the HBV-transgenic mouse model (11, 12, 33). Additionally, our observations correspond well with studies performed with duck HBV-infected duck hepatocytes (28). Taken together, the experiments presented strongly support the use of the HBV-Met cell line to study the mechanisms responsible for the cytokine-dependent noncytopathic inhibition of HBV gene expression and replication previously documented in vivo using HBV-transgenic mice (12, 33).
Previously, we showed that HBV DNA replication in vivo in HBV-transgenic mice is regulated by inflammatory cytokines (3, 4, 9, 12, 16, 33) and that under certain conditions, HBV gene expression is also subject to cytokine control (3, 9, 12, 16). While it has been possible to some degree to attribute these antiviral activities to specific cytokines in vivo (3, 4, 9, 12, 23), the HBV-Met cell culture system permits direct analysis of the individual antiviral activities of these cytokines on both HBV DNA replication and gene expression. The results reveal several interesting new facts.
First, we have demonstrated that inflammatory cytokines act directly on hepatocytes to induce an intracellular mechanism(s) that inhibits HBV replication.
Second, we found that mIFN-β, as well as the universal IFN-α/β (chimeric human IFN-αA/D) alone (data not shown), can profoundly inhibit HBV DNA replication, while the level of HBV transcripts is not affected during up to 6 days of cytokine treatment. Although similar findings have been obtained by others (17), and IFN-α/β has been reported to inhibit HBV gene expression in other systems in vitro (7, 26, 27, 32), all previous studies have been performed with poorly differentiated, transformed hepatoma cell lines that may not reflect the intracellular events triggered by cytokines in well-differentiated hepatocytes.
Third, we observed that, of the three cytokines tested, only mIFN-γ could independently reduce the steady-state level of HBV transcripts in the HBV-Met cells. In Fig. 8 we show that expression of albumin, HNF4, and C/EBPα is also reduced in mIFN-γ-treated hepatocytes. These results suggest that cytokine-induced changes in hepatocyte-specific transcription factors known to control HBV gene expression (31) might be responsible for the observed inhibition of HBV gene expression, and further experiments are being done to determine the precise mechanism mediating this inhibition.
Fourth, the mIFN-γ effect on the steady-state level of HBV transcripts was synergistically enhanced when HBV-Met cells were simultaneously treated with mIFN-γ and mTNF-α. Synergistic antiviral activity of mIFN-γ and mTNF-α has long been recognized (34). Indeed, synergistic activity of these cytokines has been shown to affect early steps in herpes simplex virus replication at the level of early gene transcription and translation (8), while they inhibit murine cytomegalovirus late gene transcription and DNA replication (20). Our results add HBV to the list of viruses whose gene expression is synergistically suppressed by IFN-γ and TNF-α. The synergistic activity of these cytokines on HBV-Met cells was also evident from the enhanced activation of the IFN-responsive genes, such as OAS and chemokine Crg-2/IP-10, which has been previously shown to be synergistically induced by IFN-γ and TNF-α (24; reviewed in reference 25). At the same time, there was also synergy in the repression of albumin, HNF4, and C/EBPα gene expression. The fact that in the HBV-Met cells efficient inhibition of HBV gene expression requires synergistic action of cytokines whereas HBV DNA replication is efficiently controlled by mIFN-β alone or mIFN-γ alone may explain why in vivo HBV DNA replication is more sensitive to cytokine control then HBV RNA.
Fifth, while it has been shown that IFN-γ and TNF-α can synergistically block adenovirus capsid formation (21), we do not find synergistic inhibition of HBV DNA replication, even though this occurs at the level of formation or stability of pgRNA-containing capsids (Fig. 5) (33). In fact, mIFN-β is as potent as mIFN-γ in inhibiting HBV DNA replication, and we do not observe any enhancement of this effect when mIFN-β and mTNF-α or mIFN-γ are combined (data not shown).
Taken together, the HBV-Met cell lines should prove to be a very useful model system to study the effects of individual cytokines and other agents on HBV gene expression and replication. Accordingly, we have recently performed gene profiling experiments on these cells to identify the hepatocellular genes associated with the antiviral effects of these cytokines, and as a direct result of those studies, we are now using the HBV-Met cells to explore the molecular basis for the cytokine-induced antiviral effect (S. Wieland, M. Robek, and F. V. Chisari, unpublished data).
Acknowledgments
V.P. and S.F.W. contributed equally to this work.
This work was supported by grant CA40489 from the National Institutes of Health (F.V.C.). V.P. was supported by a fellowship from the SKAGGS Institute. S.L.U. was supported by NIH fellowship AI49670. M.T. was supported by the Associazione Italiana Ricerca sul Cancro (AIRC), the Ministero della Sanità, and cofin-MIUR Italy.
We thank Toray Industries for providing the recombinant mIFN-β and Genentech for providing the recombinant mIFN-γ and mTNF-α. We thank Luca Guidotti for valuable discussion and advice; Carl Colburn, Amber Morris, and Angelina Eustaquio for excellent technical assistance; and Andrea Achenbach for assistance with manuscript preparation.
Footnotes
This is manuscript no. 14574-MEM from the Scripps Research Institute.
REFERENCES
- 1.Amicone, L., M. A. Galimi, F. M. Spagnoli, C. Tommasini, V. De Luca, and M. Tripodi. 1995. Temporal and tissue-specific expression of the MET ORF driven by the complete transcriptional unit of human A1AT gene in transgenic mice. Gene 162:323-328. [DOI] [PubMed] [Google Scholar]
- 2.Amicone, L., F. M. Spagnoli, G. Spath, S. Giordano, C. Tommasini, S. Bernardini, V. De Luca, C. Della Rocca, M. C. Weiss, P. M. Comoglio, and M. Tripodi. 1997. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J. 16:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bellovino, D., Y. Lanyau, I. Garaguso, L. Amicone, C. Cavallari, M. Tripodi, and S. Gaetani. 1999. MMH cells: an in vitro model for the study of retinol-binding protein secretion regulated by retinol. J. Cell. Physiol. 181:24-32. [DOI] [PubMed] [Google Scholar]
- 3.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1998. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J. Virol. 72:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1997. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J. Virol. 71:3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 13:29-60. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Davis, M. G., and R. W. Jansen. 1994. Inhibition of hepatitis B virus in tissue culture by alpha interferon. Antimicrob. Agents Chemother. 38:2921-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feduchi, E., M. A. Alonso, and L. Carrasco. 1989. Human gamma interferon and tumor necrosis factor exert a synergistic blockade on the replication of herpes simplex virus. J. Virol. 63:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidotti, L. G., P. Borrow, M. V. Hobbs, B. Matzke, I. Gresser, M. B. Oldstone, and F. V. Chisari. 1996. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc. Natl. Acad. Sci. USA 93:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidotti, L. G., and F. V. Chisari. 1999. Cytokine-induced viral purging-role in viral pathogenesis. Curr. Opin. Microbiol. 2:388-391. [DOI] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 12.Guidotti, L. G., T. Ishikawa, M. V. Hobbs, B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 13.Guidotti, L. G., V. Martinez, Y. T. Loh, C. E. Rogler, and F. V. Chisari. 1994. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J. Virol. 68:5469-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 16.Guilhot, S., L. G. Guidotti, and F. V. Chisari. 1993. Interleukin-2 downregulates hepatitis B virus gene expression in transgenic mice by a posttranscriptional mechanism. J. Virol. 67:7444-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, Y., and K. Koike. 1989. Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J. Virol. 63:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosovsky, M. J., I. Qadri, and A. Siddiqui. 1998. The regulation of hepatitis B virus gene expression: an overview of the cis- and trans-acting components, p. 21-50. In R. Koshy and W. H. Caselmann (ed.), Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. Imperial College Press, London, United Kingdom.
- 19.Lin, O. S., and E. B. Keeffe. 2001. Current treatment strategies for chronic hepatitis B and C. Annu. Rev. Med. 52:29-49. [DOI] [PubMed] [Google Scholar]
- 20.Lucin, P., S. Jonjic, M. Messerle, B. Polic, H. Hengel, and U. H. Koszinowski. 1994. Late phase inhibition of murine cytomegalovirus replication by synergistic action of interferon-gamma and tumour necrosis factor. J. Gen Virol. 75:101-110. [DOI] [PubMed] [Google Scholar]
- 21.Mayer, A., H. Gelderblom, G. Kumel, and C. Jungwirth. 1992. Interferon-gamma-induced assembly block in the replication cycle of adenovirus 2: augmentation by tumour necrosis factor-alpha. Virology 187:372-376. [DOI] [PubMed] [Google Scholar]
- 22.Mazier, D., R. L. Beaudoin, S. Mellouk, P. Druilhe, B. Texier, J. Trosper, F. Miltgen, I. Landau, C. Paul, O. Brandicourt, et al. 1985. Complete development of hepatic stages of Plasmodium falciparum in vitro. Science 227:440-442. [DOI] [PubMed] [Google Scholar]
- 23.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohmori, Y., and T. A. Hamilton. 1995. The interferon-stimulated response element and a kappa B site mediate synergistic induction of murine IP-10 gene transcription by IFN-γ and TNF-α. J. Immunol. 154:5235-5244. [PubMed] [Google Scholar]
- 25.Paludan, S. R. 2000. Synergistic action of pro-inflammatory agents: cellular and molecular aspects. J. Leukoc. Biol. 67:18-25. [DOI] [PubMed] [Google Scholar]
- 26.Rang, A., S. Gunther, and H. Will. 1999. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J. Hepatol. 31:791-799. [DOI] [PubMed] [Google Scholar]
- 27.Romero, R., and J. E. Lavine. 1996. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology 23:17-23. [DOI] [PubMed] [Google Scholar]
- 28.Schultz, U., J. Summers, P. Staeheli, and F. V. Chisari. 1999. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J. Virol. 73:5459-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spagnoli, F. M., L. Amicone, M. Tripodi, and M. C. Weiss. 1998. Identification of a bipotential precursor cell in hepatic cell lines derived from transgenic mice expressing cyto-Met in the liver. J. Cell Biol. 143:1101-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang, H., K. E. Banks, A. L. Anderson, and A. McLachlan. 2001. Hepatitis B virus transcription and replication. Drug News Perspect. 14:325-334. [PubMed] [Google Scholar]
- 31.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tur-Kaspa, R., L. Teicher, O. Laub, A. Itin, D. Dagan, B. R. Bloom, and D. A. Shafritz. 1990. Alpha interferon suppresses hepatitis B virus enhancer activity and reduces viral gene transcription. J. Virol. 64:1821-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, G. H., and D. V. Goeddel. 1986. Tumour necrosis factors alpha and beta inhibit virus replication and synergize with interferons. Nature 323:819-822. [DOI] [PubMed] [Google Scholar]
- 35.Zaret, K. S. 1996. Molecular genetics of early liver development. Annu. Rev. Physiol. 58:231-251. [DOI] [PubMed] [Google Scholar]