Abstract
RNA containing 5-fluorouridine has been assumed to inhibit strongly or irreversibly the pseudouridine synthases that act on the RNA. RNA transcripts containing 5-fluorouridine in place of uridine have, therefore, been added to reconstituted systems in order to investigate the importance of particular pseudouridine residues in a given RNA by inactivating the pseudouridine synthase responsible for their generation. In sharp contradiction to the assumption of universal inhibition of pseudouridine synthases by RNA containing 5-fluorouridine, the Escherichia coli pseudouridine synthase TruB, which has physiologically critical eukaryotic homologs, is not inhibited by such RNA. Instead, the RNA containing 5-fluorouridine was handled as a substrate by TruB. The E. coli pseudouridine synthase RluA, on the other hand, forms a covalent complex and is inhibited stoichiometrically by RNA containing 5-fluorouridine. We offer a hypothesis for this disparate behavior and urge caution in interpreting results from reconstitution experiments in which RNA containing 5-fluorouridine is assumed to inhibit a pseudouridine synthase, as normal function may result from a failure to inactivate the targeted enzyme rather than from the absence of nonessential pseudouridine residues.
Keywords: RNA modification, 5-fluorouridine, 5-fluorouracil, dyskeratosis congenita, TruB, RluA
INTRODUCTION
The conversion of uridine to pseudouridine, Ψ, occurs in all three domains of life and is the most common post-transcriptional modification of RNA (Grosjean and Benne 1998). Many RNA species have highly conserved Ψ residues, including a cluster around the peptidyl transfer site of the ribosome and the nearly universally conserved Ψ in the TΨC loop of tRNA. The rearrangement of U to Ψ is catalyzed by the Ψ synthases, enzymes that fall into five families that share no statistically significant global sequence similarity (Gustafsson et al. 1996; Koonin 1996; Kaya and Ofengand 2003; Ma et al. 2003). The reported crystal structures of Ψ synthases (Foster et al. 2000; Hoang and Ferre-D’Amare 2001; Sivaraman et al. 2002) share a core β-sheet fold that strongly indicates a common ancestry for at least four of the families (Hoang and Ferre-D’Amare 1991; Mueller 2002; Sivaraman et al. 2002). Although the importance of Ψ at many positions in diverse RNAs remains unknown, several are physiologically critical. Lack of Ψ at positions 1911, 1915, and 1917 of 23S rRNA in Escherichia coli causes severe growth retardation (Raychaudhuri et al. 1998). Mutations in the genes encoding the orthologous Ψ synthases Cbf5p in Saccharomyces cerevisiae (Zebarjadian et al. 1999) and Mfl in Drosophila melanogaster (Phillips et al. 1998; Giordano et al. 1999) lead, respectively, to markedly reduced growth rates and miniflies that suffer other abnormalities. Disruption of the gene encoding the human homolog, dyskerin, causes the X-linked form of the disease dyskeratosis congenita (Heiss et al. 1998).1 In nuclear extracts, the inhibition of the Ψ synthases that handle U2 snRNA prevents snRNP formation and hence blocks the splicing of pre-mRNA (Lenz et al. 1994; Yu et al. 1998).
This last set of studies and mechanistic work regarding the Ψ synthases relied on the inhibition of the enzymes by RNA containing 5-fluorouridine in place of U, [f5U]RNA. Such RNA was assumed to inhibit all Ψ synthases strongly, by either very tight binding (Samuelsson 1991) or the irreversible formation of a covalent enzyme–RNA adduct (Gu et al. 1999). Partially purified yeast Ψ synthases were very potently inhibited by [f5U]tRNA (complete inhibition at <100 nM), but inhibition was reversed by nuclease treatment and therefore presumed to arise from noncovalent complexes (Samuelsson 1991), although formation of a reversible covalent adduct cannot be excluded by the data. Irreversible inhibition of the E. coli Ψ synthase TruA by [f5U]tRNA likely arises from the Michael addition of a catalytically essential aspartic acid residue at C6 of the pyrimidine ring to form an ester intermediate, and the hydrated fluoronucleoside product that was isolated was reasonably inferred to result from ester hydrolysis of the covalent adduct (Gu et al. 1999). A structure of the E. coli Ψ synthase TruB cocrystallized with [f5U]RNA was recently solved (Hoang and Ferre-D’Amare 1991). It was hoped that the ester intermediate would be observed, but the f5U was surprisingly rearranged to a C-glycoside in the same way that U would be to form Ψ. The pyrimidine ring was also hydrated, and the hydration was postulated to result from ester hydrolysis of the covalent intermediate over the course of the crystallographic study (Hoang and Ferre-D’Amare 1991).
To test the supposition that a covalent adduct forms and subsequently undergoes hydrolysis, we directly probed the effect of [f5U]RNA on TruB. Unexpectedly, we found no evidence for a covalent adduct nor strong inhibition when TruB was incubated with [f5U]RNA. In sharp contrast, we tested a second Ψ synthase and found that it was irreversibly inhibited upon incubation with such RNA by the formation of a covalent adduct. The surprising findings with TruB do not compromise the studies cited above (Lenz et al. 1994; Yu et al. 1998; Gu et al. 1999) that relied on [f5U]RNA, but they do strongly impact the design and interpretation of future experiments using inhibitory RNA.
RESULTS AND DISCUSSION
Effect of [f5U]RNA on TruB
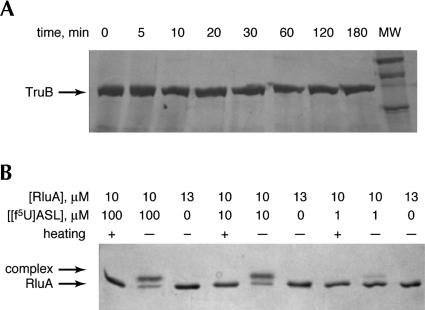
A stem–loop corresponding to the T-arm stem-loop (TSL) of yeast tRNAPhe with f5U in place of the isomerized U, [f5U]TSL, was used for these studies. In the same stem–loop context, U is isomerized by TruB as efficiently as U55 in full-length tRNA as judged by kcat and Km values (Gu et al. 1998), so it is unlikely that the use of the shorter substrate rather than full-length tRNA will significantly impact the results of these studies. In the reported crystal structure, TruB was cocrystallized with the same stem–loop (Hoang and Ferre-D’Amare 1991). We pursued two lines of investigation. First, we probed for the formation of a covalent complex between TruB and [f5U]TSL as was observed for the E. coli Ψ synthase TruA and tRNA containing f5U (Huang et al. 1998; Gu et al. 1999). That complex formed over the course of several hours (k = 0.44 h−1) when the substituted tRNA (0.5 μM) was incubated with enzyme (12.5 μM), so that all RNA was bound (Kd = 93 nM), and the complex was stable to SDS-PAGE conditions unless the sample was heated before electrophoresis (Gu et al. 1999). To test for the formation of a similar covalent intermediate, TruB (12.5 μM) was incubated with [f5U]TSL (125 μM); these conditions should assure that all enzyme is bound, as Km = 800 nM for TSL without f5U (Gu et al. 1998). Aliquots were withdrawn at various times and analyzed by SDS-PAGE (Fig. 1A ▶). No band other than that of TruB alone appeared either at early (5–30 min) or late (1–3 h) time points. Either no complex forms, or the complex is noncovalent or a covalent adduct that readily collapses under the SDS-PAGE conditions.
FIGURE 1.
Complex formation between TruB or RluA and RNA stem–loops containing f5U by gel shift in SDS-PAGE. (A) Incubation of TruB (37.3 kD; marked with arrow) with [f5U]TSL does not result in a shifted band over a broad time range; TruB was present at 12.5 μM in all incubations; (MW) molecular weight standards. (B) Incubation of RluA (26.2 kD; marked with arrow) with [f5U]ASL produces a complex that survives denaturation by formamide and SDS (shifted band), with the amount of complex reflecting the ratio of enzyme to RNA. If the samples are heated before electrophoresis, the complex decomposes, and some decomposition is apparent even without heating.
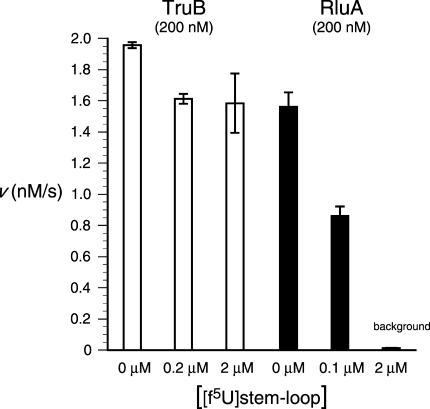
Because inactivation could be achieved by the formation of a covalent adduct that remains intact when the enzyme is in its native conformation, the ability of [f5U]TSL to inhibit TruB irreversibly was examined directly. TruB was incubated with 0, 1.0, or 10 equivalents of [f5U]TSL for 3 h at 37°C and then diluted 10-fold into an assay solution containing tRNA substrate at a concentration of 2 μM. Because Km = 150 nM for this tRNA (Ramamurthy et al. 1999b), substrate is saturating under these assay conditions, and the measured rates provide the specific activity of TruB after incubation with [f5U]TSL. Only a very modest decrease in activity was observed when TruB was treated with either 1.0 or 10 equivalents of [f5U]TSL (Fig. 2 ▶), which eliminates the irreversible formation of a covalent adduct that does not survive SDS denaturation. The observed loss of activity (18%) is the same with 10-fold different concentrations of [f5U]TSL, suggesting that reversible inhibition (competitive, noncompetitive, or uncompetitive) by [f5U]TSL or its products is not the cause of the reduced activity. Instead, we considered the possibility that TruB is less stable under the incubation conditions in the presence of RNA, either because of differences in the bound and free forms of the enzyme or a more general effect that a polyanion might have on the aggregation behavior of this protein. To test this possibility, the experiments were repeated with TSL (with no f5U), and the same loss of activity (17%) was observed relative to TruB incubated in the absence of RNA (data not shown). When TruB was preincubated with the nonsubstrate anticodon stem–loop (as described below, but with U rather than f5U), a smaller but significant reduction in activity was observed (8.9%). Although further experiments are required to establish its exact cause, the loss of activity when TruB is incubated with [f5U]TSL clearly arises from a more general effect due to the presence of RNA rather than a specific effect resulting from the presence of f5U in the RNA.
FIGURE 2.
Inhibition of TruB and RluA by RNA stem–loops containing f5U. TruB is only very modestly inhibited by preincubation with [f5U]TSL (open bars); the same degree of inhibition (18%) is seen with two concentrations of [f5U]TSL that differ by 10-fold, which is inconsistent with the product stem–loop serving simply as a competitive, noncompetitive, or uncompetitive inhibitor. RluA is inhibited by preincubation with [f5U]ASL in essentially stoichiometric fashion (solid bars), which is consistent with irreversible inhibition by the formation of a covalent adduct. The concentrations given are those in the preincubation mixtures, which were diluted 10-fold for the activity assays.
Inhibition of RluA by [f5U]RNA
As a control for our methodology and to extend our characterization of the inhibitory effect of [f5U]RNA on Ψ synthases, we performed the same set of reactions using E. coli RluA, which isomerizes U746 in 23S rRNA and U32 in tRNAPhe, both of which are found in the loop of stem– loop structures (Wrzesinski et al. 1995). Preliminary reports (Horne et al. 1997) and work in our lab (T. Greco, unpubl.) show that the isolated stem–loops from both 23S rRNA and tRNAPhe are good substrates for RluA. We chose to use the anticodon stem-loop (ASL) from tRNAPhe with U32 replaced by f5U, [f5U]ASL. In sharp contrast to the results with TruB and [f5U]TSL, incubation of RluA with [f5U]ASL resulted in both irreversible enzyme inhibition (Fig. 2 ▶) and a complex that survives SDS-PAGE conditions but is not heat stable (Fig. 1B ▶), as was observed for the similar TruA complex (Huang et al. 1998; Gu et al. 1999). Both effects show the expected dependence on the stoichiometry of RluA and [f5U]ASL. Complex formation was also demonstrated using urea-PAGE, in which case the [f5U]ASL band was greatly retarded after incubation with RluA, and differential staining showed the presence of both RNA and protein in the shifted band (supplementary material). Two-dimensional gel analysis confirmed that the shifted band in the urea–polyacrylamide gel was the same species observed by SDS-PAGE (supplementary material). The methodology, therefore, should have sufficed to reveal complex formation and inactivation of TruB by [f5U]TSL if either were occurring.
[f5U]TSL is a substrate for TruB
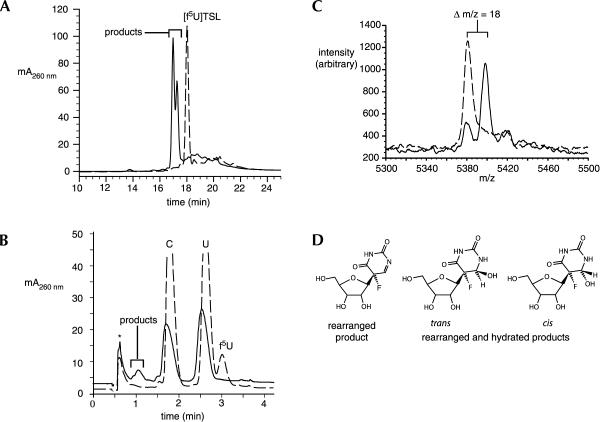
Given the lack of strong inhibition of TruB by [f5U]TSL, we hypothesized that the rearranged f5U observed in the crystal structure of TruB (Hoang and Ferre-D’Amare 1991) was generated much more quickly than had been assumed; if the ester intermediate formed, it hydrolyzed within the 3-h period of the preincubation with TruB. To test this hypothesis, [f5U]TSL was examined for its ability to serve as a substrate for the enzyme. TruB and [f5U]TSL were incubated at 37°C for 3 h, and the [f5U]TSL was then analyzed by HPLC over a C18 column. After incubation with TruB, the retention time of [f5U]TSL was reduced and its peak doubled (Fig. 3A ▶). The earlier elution is consistent with the greater polarity expected for either the rearranged pyrimidine (with one more imino group than f5U) or the rearranged and hydrated product observed in the crystal structure (with one more imino group and an additional hydroxyl group at C6). The peak doubling likely results from either a mixture of the hydrated and unhydrated forms of the rearranged f5U or from a mixture of the cis and trans isomers of the hydrated form (Fig. 3D ▶). To ensure that the difference in chromatographic behavior arose from alteration of the f5U, [f5U]TSL was subjected to total enzymatic degradation and resolution of the resulting nucleosides by HPLC (Mueller et al. 1998) both before and after incubation with TruB (Fig. 3B ▶). Before incubation, peaks corresponding to C, U, f5U, G, and A were observed (in that elution order); after incubation, the C, U, G, and A peaks remained, but the f5U peak was replaced with a doubled peak that eluted before C (as does Ψ). To ascertain whether or not hydration occurred as a result of incubation, mass spectrometry (MALDI-TOF MS) was used. Before incubation with TruB, [f5U]TSL gave rise to the expected peak at 5380.3 m/z, which shifted 18 m/z to 5398.3 m/z after incubation, as expected for the addition of a water molecule (Fig. 3C ▶). A small residual peak at 5380.3 m/z may be due to dehydration under the MALDI sample conditions or reflect the presence of both the hydrated and unhydrated forms of the rearrangment product of f5U. To help distinguish between these possibilities, a HETCOSY two-dimensional NMR experiment (Collier et al. 1996) was used to ascertain the chemical shift of the proton attached to C6 of f5U by virtue of its coupling to the fluorine nucleus. The HETCOSY spectrum has a single, broad feature at Hδ 4.3 after [f5U]TSL is incubated with TruB (C. Spedaliere, unpubl.), which is consistent with a proton attached to an sp3-hybridized carbon rather than an sp2 center, and thus favors a mixture of the cis and trans hydration isomers. Given the presence of the cis isomer of the rearranged and hydrated f5U within the TSL in the crystal structure of TruB, it seems highly likely that the enzyme generates the cis isomer as one of the products and that it equilibrates with the other product in solution, or that the rearranged product is released and becomes hydrated in solution. In either case, TruB bound with the cis isomer of the hydrated product crystallizes preferentially to the other product, which is present at a comparable concentration as judged by HPLC (Fig. 3A ▶).
FIGURE 3.
Conversion of [f5U]TSL into products upon incubation with TruB. (A) A portion of the C18 HPLC traces of intact stem–loop before (broken line) and after (solid line) incubation with TruB showing quantitative conversion of [f5U]TSL into products. (B) A portion of the C18 HPLC traces of the nucleosides resulting from digestion of [f5U]TSL before (broken line) and after (solid line) incubation with TruB; the void peak is labeled with an asterisk. (C) MALDI-TOF mass spectrum of intact [f5U]TSL before (broken line) and after (solid line) incubation with TruB; the shift of 18 m/z is that expected for hydration. (D) The structures of the likely products from f5U; the stereochemistry at C5 is assigned on the basis of the appearance of the depicted cis isomer (within the RNA stem–loop) in the cocrystal structure of TruB (Hoang and Ferre-D’Amare 1991).
Although a definitive chemical characterization remains to distinguish between the possible chemical products and thus provide insight into the reaction mechanism of the Ψ synthases, the data clearly show that TruB converts [f5U]TSL into a mixture of two products on a time scale of hours. TruB acts on the f5U residue, and at least one of the products has been hydrated, which is fully consistent with the rearranged and hydrated product of f5U observed in the cocrystal of TruB and [f5U]TSL (Hoang and Ferre-D’Amare 1991). Because the mixture of products, at most, inhibits TruB to the same extent as regular TSL product (with Ψ rather than a fluorinated derivative), the assumed universality of potent inhibition of Ψ synthases by [f5U]RNA must be discarded. RluA can now be added to the number of Ψ synthases that are inhibited by such RNA. What is the cause of the difference in behavior toward [f5U]RNA exhibited by TruB vesus TruA, RluA, and the yeast Ψ synthases responsible for Ψ13 and Ψ55?
TruB belongs to a different family of Ψ synthases than either RluA or TruA, both of which form inhibitory covalent complexes with [f5U]RNA, so the differences in behavior may fall along family lines. However, E. coli RluD is a Ψ synthase of the RluA family, and it does not appear to form covalent complexes with [f5U]RNA (A. Matte, pers. comm.). The yeast enzyme that isomerizes U55 of tRNA, now named Pus4p (Becker et al. 1997), is a member of the TruB family that does not form a detectable covalent adduct, but is reported to be strongly inhibited by [f5U]tRNA (Samuelsson 1991).2 Given the indistinguishable values of kcat and Km for TSL and full-length tRNA displayed by TruB (Gu et al. 1998), it seems unlikely that our use of [f5U]TSL rather than [f5U]tRNA can account for the disparate inhibition results from TruB and its yeast counterpart. Instead, it appears more likely that subtle differences in active site geometry may account for the differences in behavior.
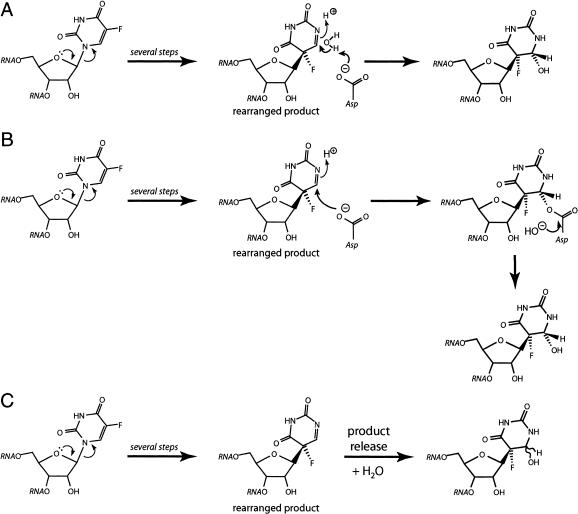
It is possible that seemingly minor variations in the microenvironment of Ψ synthase active sites allow f5U to proceed along the reaction coordinate to different extents. TruB catalyzes the rearrangement to the C-glycoside, for example, but TruA and RluA get stuck at an intermediate. The mechanism followed by the Ψ synthases has not been definitively established, although the work with TruA and [f5U]tRNA favors the mechanism in which the essential aspartic acid residue serves as a nucleophile in a Michael addition at C6 to make the ester intermediate (Gu et al. 1999) that was proposed to give rise to the rearranged and hydrated product observed in the structure of the TruB•[f5U]TSL complex (Hoang and Ferre-D’Amare 1991). In the alternative mechanism, the essential aspartic acid residue makes a covalent adduct with the ribose moiety at C1′ (Huang et al. 1998) when the glycosidic bond is broken to start the isomerization of U to Ψ. As pointed out earlier (Mueller 2002), the paucity of candidate acid/base residues strongly suggests that the essential aspartic acid residue serves as the general base to deprotonate C5 in the last step of Ψ generation. After it is freed from its covalent attachment (to either C5 or C1′), the essential aspartic acid residue is in the deprotonated form and lies in the vicinity of C6. Once the rearrangment to the C-glycoside has occurred by either mechanism, the essential aspartic acid residue would be positioned to serve as a general base to activate water for attack at C6 to generate the rearranged and hydrated product (Fig. 4A ▶). Alternatively, the essential aspartic acid residue could add to C6 to make an ester intermediate with the rearranged product (whether or not that ester is an intermediate of the regular catalytic pathway), giving rise to the formation of a covalent intermediate that may hydrolyze quickly or slowly (Fig. 4B ▶). It is also possible that some Ψ synthases will release the rearranged product, which could undergo hydration in solution (Fig. 4C ▶).
FIGURE 4.
Reaction routes that could explain the disparate behavior of Ψ synthases toward [f5U]RNA and the rearranged and hydrated product observed in the TruB•[f5U]TSL cocrystal structure (Hoang and Ferre-D’Amare 1991). In all three depicted schemes, the reaction can proceed by either proposed mechanism to generate the rearranged product. (A) The essential aspartic acid serves as a general base in the direct hydration of the rearranged product. (B) The essential aspartic acid residue performs a Michael addition at C6, followed by ester hydrolysis, which could be fast or slow; it is also possible that the ester forms as part of the mechanism involving Michel addition at C6, so that the depicted rearranged product never exists. (C) The rearranged product is released by the enzyme into solution and becomes hydrated, likely in a stereorandom manner.
Subtle differences in active-site geometry among the Ψ synthases, then, could give rise to disparate results with [f5U]RNA. Some active sites afford enough room for water to fit between the essential aspartic acid residue and C6, leading to direct hydration. Other active sites with a shorter distance between the essential aspartic acid residue and C6 favor ester formation, and the stability of that linkage to reversal and hydrolysis is governed by the access of water and the ability of the active site to stabilize the transition state for hydrolysis. Although they are remarkably informative and provide great mechanistic insight, the current Ψ synthase structures cannot resolve this issue. The TruB•[f5U]TSL complex (Hoang and Ferre-D’Amare 1991) is an unproductive one with C6 distant from the essential aspartic acid residue due to a rotation of ~180° about the glycosidic bond. The structure of RsuA was solved with uracil and UMP rather than an RNA bound (Sivaraman et al. 2002), and the structure of TruA was solved with no active site ligands (Foster et al. 2000). None of these structures, therefore, can address fine points in the relative positioning of the essential aspartic acid residue and C6 in a productively bound reaction intermediate or product. The full chemical characterization of the products from f5U after incubation of [f5U]RNA with TruB and RluA are already underway and promise to help discern between these various scenarios for the disparate behavior of Ψ synthases toward [f5U]RNA.
Conclusions
Regardless of the eventual outcome of studies to elucidate the mechanism of the Ψ synthases and the products generated from [f5U]RNA, the ability of TruB to handle [f5U]TSL as a substrate on a time scale of hours sounds a strong cautionary note for future investigations using [f5U]RNA to examine the physiological importance of particular Ψ residues. The lack of an effect may not be due to a dispensable Ψ, but instead due to a Ψ synthase that is not strongly inhibited by the inhibitory RNA. Until a particular Ψ synthase has been investigated, we urge caution in interpreting the results of negative experiments with [f5U]RNA. On the other hand, the disruption of a process by such RNA, as has been shown elegantly for U2 snRNA function (Lenz et al. 1994; Yu et al. 1998), constitutes strong evidence for the importance of particular Ψ residues.
MATERIALS AND METHODS
General
E. coli TruB and RluA were overexpressed and purified as described previously (Ramamurthy et al. 1999a); both bear the amino-terminal His6•tag encoded by pET15b (Novagen). Chemicals were purchased from Fisher Scientific or its Acros Chemicals division unless otherwise noted. Prime RNase inhibitor was purchased from Eppendorf–5′; nuclease P1 was purchased from Roche Applied Science; calf intestinal alkaline phosphatase was purchased from Promega; ammonium citrate (dibasic) was purchased from Sigma. RNA oligonucleotides were purchased from Dharmacon, Inc.; the RNA was deprotected according to the manufacturer’s instructions, and concentrations were determined using the provided extinction coefficients. The RNA oligonucleotide corresponding to the T-arm stem-loop (TSL) of Saccharomyces cerevisiae tRNAPhe with f5U in place of the U isomerized by TruB was denoted [f5U]TSL and had the sequence CUGUGUf5UCGAUCCACAG. The RNA oligonucleotide corresponding to the anticodon stem-loop (ASL) of E. coli tRNAPhe with f5U in place of the U isomerized by RluA was denoted [f5U]ASL and had the sequence GGGGAf5UUGAAAAUCCCC. Standard assay buffer is 50 mM HEPES buffer (pH 7.5), containing ammonium chloride (100 mM), DTT (5 mM), EDTA (1 mM), and Prime RNAse inhibitor (30 U). HPLC was performed using a Beckman System Gold instrument equipped with a System 125 solvent module, a Rheodyne 7725i injector, and a Beckman 168 diode array detector; a Higgins Analytical CLIPEUS C18 5-μm column (50 × 4.6 mm) was purchased from Bodman Industries and used for all HPLC analysis.
Complex formation
TruB (12.5 μM) was incubated at 25°C with [f5U]TSL (125 μM) in standard assay buffer (200 μL). At various times, aliquots (10 μL) were mixed with an equal volume of Laemmli sample buffer and analyzed by SDS-PAGE (10%–20% gradient gel) without heating the samples; protein was detected by staining with Coomassie Blue. RluA (10 or 70 μM) was incubated at 25°C with [f5U]ASL (7, 10, or 100 μM) in standard assay buffer (40 μL); the concentrations were matched to afford stoichiometries of 10:1, 1:1, and 1:10 RluA:RNA. After 3 h, two aliquots (10 μL) were withdrawn from each incubation mixture and mixed with formamide (10 μL) and Laemmli sample buffer (10 μL)3; one sample was incubated 5 min at 100°C, whereas the other was not. These samples were analyzed by SDS-PAGE (10% gel), and protein was detected by staining with Coomassie Blue. A similar series of incubations were run with stoichiometries of 5:1, 1:1, and 1:5 RluA:RNA, but the two aliquots were each mixed with an equal volume of neat formamide, and one sample was incubated 5 min at 70°C. The samples were analyzed by PAGE in a 15% gel containing urea (7 M). The gel was stained with Radiant Red (Bio-Rad Laboratories) to detect RNA, and then with Coomassie Blue to detect protein.
Enzyme assays
The activity of TruB and RluA after preincubation with [f5U]TSL and [f5U]ASL, respectively, was measured using the standard tritium release assay for Ψ formation (Ramamurthy et al. 1999a); this assay measures the liberation of tritium from tRNA transcript containing [5-3H]uridine, which occurs when U is isomerized to Ψ. TruB (200 nM) or RluA (200 nM) was incubated with [f5U]TSL (0–2 μM) or [f5U]ASL (0–2 μM), respectively, in standard assay buffer. After 3 h at 37°C, an aliquot (15 μL) was withdrawn and added to a solution (135 μL) of the same buffer containing tritiated tRNA (2 μM); aliquots were withdrawn periodically to measure the rate of Ψ formation as described previously (Ramamurthy et al. 1999a). All assays were performed in duplicate, except for the incubations of TruB with 10 equivalents of [f5U]TSL and RluA with a 0.5 equivalent of [f5U]ASL, which were performed in triplicate.
Alteration of [f5U]TSL by TruB
TruB (200 nM) and [f5U]TSL (20 μM) were incubated at 37°C for 3 h in standard assay buffer (100 μL). Protein was extracted with phenol saturated with TE buffer (pH 4.3). After the aqueous phase was washed with chloroform:isoamyl alcohol (24:1), the RNA was ethanol precipitated, and the pellet was washed twice with 70% ethanol and air dried. Intact [f5U]TSL that had or had not been incubated with TruB was then analyzed by HPLC over a C18 column, eluting with a gradient of acetonitrile according to the following program (the first number is the percentage of aqueous acetonitrile (40% vol/vol) in 25 mM ammonium acetate buffer (pH 6.0); the second number is the elapsed time in minutes): 0, 0; 0, 3; 5, 10; 25, 25; 75, 27; 75, 30; 100, 31; 100, 36; 0, 38; 0, 43. A linear gradient was used except from 0%–5%, which was accomplished with a concave gradient. To localize the changes in [f5U]TSL, the RNA oligonucleotide before and after incubation with TruB was digested to component nucleosides by the sequential action of nuclease P1 and calf intestinal alkaline phosphatase (Gehrke et al. 1982; Mueller et al. 1998). A gradient of acetonitrile (0%–40% vol/vol) in 0.25 M ammonium acetate buffer (pH 6.0), was used to resolve the free nucleosides (Buck et al. 1983; Mueller et al. 1998).
To characterize the altered [f5U]TSL, matrix-assisted laser desorption/time of flight mass spectrometry (MALDI-TOF MS) was used. TruB (200 nM) and [f5U]TSL (20 μM) were incubated at 37°C for 3 h in standard assay buffer (100 μL). Protein was extracted with phenol and the RNA precipitated with ethanol as described above; the redissolved RNA was desalted using a Microcon-3 ultrafiltration device. Aliquots of the final RNA solution (5 μL) were added to matrix solution (5 μL), which is aqueous acetonitrile (50% vol/vol) containing 6-aza-2-thiothymine (10 mg/mL) and dibasic ammonium citrate (20 mM). The samples were spotted onto a MALDI sample plate, air dried, and then analyzed by MALDI-TOF MS in an OmniFLEX instrument (Bruker Daltonics); the detector was used in linear mode with negative ion detection. RNA oligonucleotides (Dharmacon) were used for mass calibration: CUGUGUf5UCGAUCCACAG, 5382.3 Mr; GGGGAUUGAAAAUCCCC, 5450.4 Mr; CCGCUAAUGUU GAAAAAUUAGCGG, 7701.7 Mr. Data was acquired with 300 laser shots at a sampling rate of 1 nsec, and 15-point smoothing was applied using the program XMASS, version 5.0 (Bruker Daltonics).
Acknowledgments
This work was supported by the National Institutes of Health (GM59636 to E.G.M). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5100104.
Footnotes
Dyskerin is part of the telomerase complex (Mitchell et al. 1999), and the autosomal dominant form of the dyskeratosis congenita stems from mutations in the RNA component of telomerase (Vulliamy et al. 2001), leaving the cause of the disease as a telomerase deficiency, a pseudouridylation deficiency, or both. Recent work with knock-out mice shows that the absence of the dyskerin homolog imparts disease symptoms in early generations even though telomere shortening is not evident until later generations, which strongly suggests that the disease results at least partially from an effect other than reduced telomerase activity—most likely a pseudouridylation deficiency (Ruggero et al. 2003).
Although inhibition at very low concentrations (0.1–100 nM) of [f5U]RNA is unambiguous, the use of nonhomogenous enzyme preparations and single time points frustrate a precise analysis of the stoichiometry and extent of inhibition for the yeast Ψ synthases (Samuelsson 1991). Assuming that the single time points were in the linear range, the value of Ki for the TruB homolog Pus4p (PS 55 is the older name) is on the order of 0.1 nM.
When only Laemmli sample buffer was added, the shifted band ran faster than RluA alone, likely due to incomplete denaturation (supplementary material can be found at http://www.udel.edu/chem/mueller/Spedalieresupp.pdf).
REFERENCES
- Becker, H.F., Motorin, Y., Planta, R.J., and Grosjean, H. 1997. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 25: 4493–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, M., Connick, M., and Ames, B.N. 1983. Complete analysis of tRNA modified nucleosides by high-performance liquid chromatography: The 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal. Biochem. 129: 1–13. [DOI] [PubMed] [Google Scholar]
- Collier, A.K., Arnold, J.R.P., and Fisher, J. 1996. NMR investigations of fluorine-labelled RNA: Application of two-dimensional heteronuclear experiments to study single-strand conformation. Magn. Reson. Chem. 34: 191–196. [Google Scholar]
- Foster, P.G., Huang, L., Santi, D.V., and Stroud, R.M. 2000. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat. Struct. Biol. 7: 23–27. [DOI] [PubMed] [Google Scholar]
- Gehrke, C.W., Kuo, K.C., McCune, R.A., Gerhardt, K.O., and Agris, P.F. 1982. Quantitative enzymatic hydrolysis of tRNAs: Reversed-phase high-performance liquid chromatography of tRNA nucleosides. J. Chromatogr. 230: 297–308. [PubMed] [Google Scholar]
- Giordano, E., Peluso, I., Senger, S., and Furia, M. 1999. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 144: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, H. and Benne, R. 1998. Modification and editing of RNA. ASM Press, Washington, DC.
- Gu, X.R., Yu, M., Ivanetich, K.M., and Santi, D.V. 1998. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37: 339–343. [DOI] [PubMed] [Google Scholar]
- Gu, X.R., Liu, Y.Q., and Santi, D.V. 1999. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. 96: 14270–14275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, C., Reid, R., Greene, P.J., and Santi, D.V. 1996. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 24: 3756–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss, N.S., Knight, S.W., Vulliamy, T.J., Klauck, S.M., Wiemann, S., Mason, P.J., Poustka, A., and Dokal, I. 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19: 32–38. [DOI] [PubMed] [Google Scholar]
- Hoang, C. and Ferre-D’Amare, A.R. 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939. [DOI] [PubMed] [Google Scholar]
- Horne, D.A., Rood, K., Levine, E., and Ofengand, J. 1997. RNA substrate recognition by E. coli pseudouridine synthase (RluA) with dual substrate specificity. Abstr. Pap. Am. Chem. Soc. 213: 308. [Google Scholar]
- Huang, L.X., Pookanjanatavip, M., Gu, X.G., and Santi, D.V. 1998. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 37: 344–351. [DOI] [PubMed] [Google Scholar]
- Kaya, Y. and Ofengand, J. 2003. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 9: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V. 1996. Pseudouridine synthases: Four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 24: 2411– 2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz, H.J., Manno, D.J., Danenberg, K.D., and Danenberg, P.V. 1994. Incorporation of 5-fluorouracil into U2 and U6 snRNA inhibits mRNA precursor splicing. J. Biol. Chem 269: 31962–31968. [PubMed] [Google Scholar]
- Ma, X.J., Zhao, X.L., and Yu, Y.T. 2003. Pseudouridylation of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., Wood, E., and Collins, K. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555. [DOI] [PubMed] [Google Scholar]
- Mueller, E.G. 2002. Chips off the old block. Nat. Struct. Biol. 9: 320–322. [DOI] [PubMed] [Google Scholar]
- Mueller, E.G., Buck, C.J., Palenchar, P.M., Barnhart, L.E., and Paulson, J.L. 1998. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 26: 2606–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, B., Billin, A.N., Cadwell, C., Buchholz, R., Erickson, C., Merriam, J.R., Carbon, J., and Poole, S.J. 1998. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol. Gen. Genet. 260: 20–29. [DOI] [PubMed] [Google Scholar]
- Ramamurthy, V., Swann, S.L., Paulson, J.L., Spedaliere, C.J., and Mueller, E.G. 1999a. Critical aspartic acid residues in pseudouridine syntheses. J. Biol. Chem 274: 22225–22230. [DOI] [PubMed] [Google Scholar]
- Ramamurthy, V., Swann, S.L., Spedaliere, C.J., and Mueller, E.G. 1999b. Role of cysteine residues in pseudouridine synthases of different families. Biochemistry 38: 13106–13111. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S., Conrad, J., Hall, B.G., and Ofengand, J. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D., Grisendi, S., Piazza, F., Rego, E., Mari, F., Rao, P.H., Cordon-Cardo, C., and Pandolfi, P.P. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299: 259–262. [DOI] [PubMed] [Google Scholar]
- Samuelsson, T. 1991. Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Res. 19: 6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaraman, J., Sauvé, V., Larocque, R., Stura, E.R., Schrag, J.D., Cygler, M., and Matte, A. 2002. Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP. Nat. Struct. Biol. 9: 353–358. [DOI] [PubMed] [Google Scholar]
- Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P.J., and Dokal, I. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435. [DOI] [PubMed] [Google Scholar]
- Wrzesinski, J., Nurse, K., Bakin, A., Lane, B.G., and Ofengand, J. 1995. A dual-specificity pseudouridine synthase: An Escherichia coli synthase purified and cloned on the basis of its specificity for Ψ746 in 23S RNA is also specific for Ψ32 in tRNAPhe. RNA 1: 437–448. [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., and Steitz, J.A. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian, Y., King, T., Fournier, M.J., Clarke, L., and Carbon, J. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19: 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]