Abstract
The Saccharomyces cerevisiae splicing factor Prp2 is an RNA-dependent ATPase required before the first transesterification reaction in pre-mRNA splicing. Prp2 binds to the spliceosome in the absence of ATP and is released following ATP hydrolysis. It contains three domains: a unique N-terminal domain, a helicase domain that is highly conserved in the DExD/H protein family, and a C-terminal domain that is conserved in spliceosomal DEAH proteins Prp2, Prp16, Prp22, and Prp43. We examined the role of each domain of Prp2 by deletion mutagenesis. Whereas deletions of either the helicase or C-terminal domain are lethal, deletions in the N-terminal domain have no detectable effect on Prp2 activity. Overexpression of the C-terminal domain of Prp2 exacerbates the temperature-sensitive phenotype of a prp2Ts strain, suggesting that the C-domain interferes with the activity of the Prp2Ts protein. A genetic approach was then taken to study interactions between Prp2 and the spliceosome. Previously, we isolated dominant negative mutants in the helicase domain of Prp2 that inhibit the activity of wild-type Prp2 when the mutant protein is overexpressed. We mutagenized one prp2 release mutant gene and screened for loss of dominant negative function. Several weak binding mutants were isolated and mapped to the C terminus of Prp2, further indicating the importance of the C terminus in spliceosome binding. This study is the first to indicate that amino acid substitutions outside the helicase domain can abolish spliceosome contact and splicing activity of a spliceosomal DEAH protein.
Keywords: Pre-mRNA splicing, DExD/H-box proteins, RNA helicase, RNA-dependent ATPase, Saccharomyces cerevisiae
INTRODUCTION
Pre-mRNA splicing occurs in the spliceosome, which is a dynamic RNA-protein complex. Spliceosomes contain the U1, U2, U4/U6, and U5 small ribonucleoprotein particles (snRNPs) as well as non-snRNP factors. Proteins in the DExD/H family are involved in splicing, both as integral components of snRNPs, such as Brr2 (Raghunathan and Guthrie 1998), and as extrinsic factors, such as Prp2 (Lin et al. 1987). Members of the spliceosomal DExD/H family show RNA-dependent ATPase activity and have a high degree of amino acid conservation in the ATPase, or helicase (H), domain. The ATPase domain can be subdivided into seven conserved motifs that are required for ATP binding, Mg2+ binding, and RNA binding (for review, see Hamm and Lamond 1998; Staley and Guthrie 1998; Tanner and Linder 2001).
Within the superfamily of DExD/H proteins, the yeast splicing factors Prp2, Prp16, Prp22, and Prp43, which have DEAH in motif II of the H-domain, form a group with conservation extending beyond the H-domain. The carboxyl (C) terminus is conserved whereas the amino (N) terminus is not highly conserved in this group of proteins. Each of these DEAH proteins has a different function in vivo and works in a sequential manner. Prp2 is required for the first transesterification reaction (King and Beggs 1990; Kim and Lin 1993; Plumpton et al. 1994), Prp16 is required for the second transesterification reaction (Schwer and Guthrie 1992), Prp22 acts both in the second step of pre-mRNA splicing and as a recycling factor (Schwer and Gross 1998; Wagner et al. 1998), and Prp43 is involved in spliceosome disassembly (Arenas and Abelson 1997; Martin et al. 2002).
The H-domain has been well studied in the DExD/H proteins, including in the DEAH subfamily (Schwer and Guthrie 1992; Plumpton et al. 1994; Hotz and Schwer 1998; Edwalds-Gilbert et al. 2000; Campodonico and Schwer 2002; Martin et al. 2002; Schneider et al. 2002), but much less is known about the roles of the N and C termini. Analysis of Prp16 suggests roles for the amino and carboxyl termini in protein function, including spliceosome binding (Hotz and Schwer 1998; Wang and Guthrie 1998). Studies of Prp22 show that portions of the N terminus as well as most of the C terminus are important for optimal protein function (Schneider and Schwer 2001). Most of the N terminus of Prp43 is not essential, whereas most of its C terminus is required in vivo (Martin et al. 2002).
In order to identify the regions of Prp2 that define spliceosome binding specificity, we performed deletion and mutational analyses of Prp2. We reason that mutations abolishing the ability of Prp2 to interact with the spliceosome would affect Prp2 function in vivo and even be detrimental to cell viability. Deletion of amino acids 6–206 in the N terminus did not affect Prp2 activity in vivo; deletions of amino acids 89–206 had no effect on Prp2 activity in vitro or in vivo. However, deletion of the C terminus ablated Prp2 activity in vivo and in vitro. Overexpression of the C terminus exacerbated the phenotype of a prp2Ts strain, indicating that the C terminus can interfere with the mutant Prp2 protein. We then took a genetic approach to identify residues in Prp2 important for spliceosome binding, by screening for loss of dominant negative function without a large truncation or a reversion. We isolated seven nonsense mutations and a double missense mutation, all of which mapped to the C terminus, further indicating the importance of the C terminus in spliceosome binding. This study is the first to indicate that amino acid substitutions outside the H-domain can abolish spliceosome contact and splicing activity of a spliceosomal DEAH protein.
RESULTS
Deletion mapping of Prp2
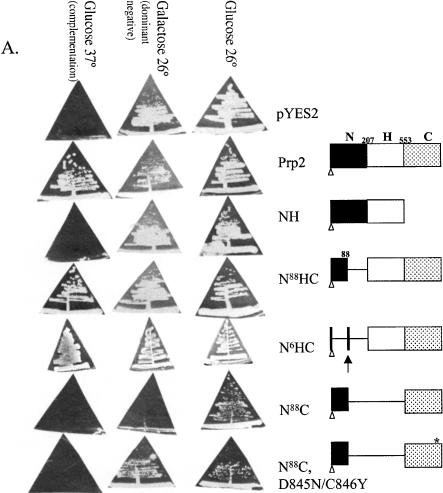
We first performed a deletion analysis of Prp2 to determine which regions are essential for its in vivo function. Prp2 is an 876 amino acid protein with three domains: the unique N-terminal domain, the highly conserved H-domain, and the C-terminal domain that is conserved within the DEAH-box subfamily of DExD/H proteins. To examine the function of each domain, we constructed pYES2-derived plasmids that could express, under the control of the GAL1 promoter, the N terminus alone (N207), the N terminus and H-domain (NH), the H-domain and C terminus (N88HC and N6HC), and the C terminus alone (N88C), including the putative nuclear localization signal (Fig. 1A ▶). The constructs were tested for function by transforming them into a temperature-sensitive prp2Ts strain (D12) and testing for growth on glucose (basal expression) and galactose (overexpression) at the permissive temperature and glucose at the nonpermissive temperature (Fig. 1A ▶). Basal expression of wild-type Prp2 rescued the Ts− phenotype of D12 at 37° (a complementation assay). Expression of a protein containing a deletion of the N terminus (N88HC or N6HC) complemented the Ts− phenotype of D12 (Fig. 1A ▶), which indicates that nearly all of the N terminus is dispensable for Prp2 activity. N6HC could also substitute for wild-type Prp2 in supporting cell viability of a prp2-deletion strain in a plasmid shuffle assay (data not shown). This result indicated that the first 206 amino acids of Prp2, perhaps with the exception of the putative nuclear localization signal at 83–88, are dispensable for life.
FIGURE 1.
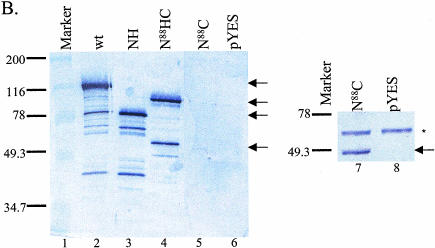
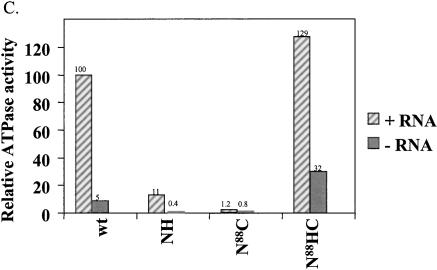
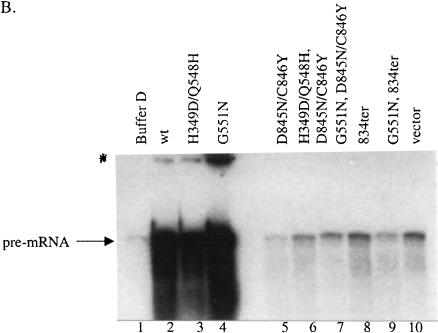
Characterization of Prp2 truncation mutants. (A) Map of Prp2 domain constructs and their in vivo phenotypes in temperature-sensitive prp2Ts strain D12. The domain or deletion constructs of PRP2 were expressed behind the GAL1 promoter in the 2μ plasmid pYES2.0 (Invitrogen). All constructs contain a six-histidine tag at the N terminus noted by a clear triangle. The putative nuclear localization signal, KKSSKY, from amino acids 83–88, is indicated by the arrow and was present in all constructs. The amino acid number 88, 207, or 553 indicates where the deletion occurs. The * indicates the D845N/C846Y double mutations (see Fig. 4A ▶). The indicated plasmids were transformed into the strain D12, and the Ura+ transformants were isolated and streaked onto SC-glucose or SC-galactose plates. Plates were photographed after 3 d (glucose) or 5 d (galactose) of growth at either 26°, the permissive temperature, or 37°, the nonpermissive temperature. (B) The expression of the mutant Prp2 proteins in D12. Five μL of the protein that eluted in 150 mM imidazole (Protocol A) were separated by 10% SDS-PAGE, transferred to PVDF membrane, and probed with rabbit anti-full-length Prp2 antibodies (lanes 1–6) or anti-histidine-tag antibodies (lanes 7,8). The samples shown on the right panel were concentrated by ammonium sulfate before loading on a nickel column. Arrows on the left panel, from top to bottom, indicate the bands corresponding to wild type, N88HC, NH, and N88C of Prp2. The arrow on the right panel indicates the N88C protein; * indicates a protein cross-reacted with the anti-histidine-tag antibodies. (C) The ATPase activity assay of the Prp2 mutant proteins. Estimated 15 ng of wild-type or mutant Prp2 proteins that eluted in 150 mM imidazole (Protocol A) were incubated with γ-[32P]-ATP with or without poly(U) RNA. Note that proteins used in this assay were purified in parallel, except for N88C. Activity is calculated relative to wild-type Prp2 in the presence of RNA, which is set at 100%. (D) The in vitro splicing and spliceosome binding of the Prp2 mutant proteins. Spliceosomes were assembled from heat-treated prp2Ts extract. Following depletion of ATP, the indicated purified proteins were added and incubated for 20 min at 23°C. An aliquot of the reaction was removed prior to immunoprecipitation to assess splicing (splicing—ATP). Anti-full-length Prp2 antibodies were added to half the reaction, which corresponds to immunoprecipitation—ATP. ATP was added to the remainder of the reaction and incubation continued at 23°C for 20 min. An aliquot of the reaction was removed to assess splicing (splicing+ATP), and the remainder was subjected to immunoprecipitation with anti-Prp2 antibodies (Immunoprecipitation+ATP). The positions of the pre-mRNA, mRNA, intron lariat-exon 2 (IVS*-E2), and intron lariat (IVS*) are indicated. D, Buffer D.
Deletion of the C terminus of Prp2 resulted in a protein, NH, which was not able to rescue the Ts− phenotype of D12 (Fig. 1A ▶). Another construct with a shorter deletion of the C-terminal 262 amino acid could not substitute for wild-type Prp2 in a plasmid shuffle assay (data not shown). Thus, the C terminus of Prp2 appears to be required for activity in vivo. N88C did not complement the Ts− phenotype of D12; however, overexpression of this C-terminal construct inhibited growth of D12 at the permissive temperature (Fig. 1A ▶, N88C in galactose at 26°). In addition, D12 cells expressing the C terminus could not grow in liquid media containing raffinose, an uninduced and unsuppressed condition (data not shown). Therefore, expression of the C terminus interferes with the mutant prp2Ts in the absence of most of the N terminus and the entire H-domain. PRP2 constructs expressing individual domains were also tested for their overexpression phenotypes in a wild-type PRP2 strain. None of the constructs showed a dominant negative phenotype when overexpressed in a wild-type strain (data not shown).
To test whether the yeast transformants expressed the deletion constructs, cell extracts were prepared in parallel and subjected to nickel affinity chromatography, because a histidine-tag is present in all constructs (Fig. 1A ▶). The proteins isolated were probed with anti-Prp2 antibodies in a Western blot (Fig. 1B ▶, lanes 1–6). Proteins for the NH and N88HC constructs were detected, and their expression levels were comparable to wild type (Fig. 1B ▶, lanes 3,4,2, respectively). The protein for N88C was expressed at a lower level (Fig. 1B ▶, lane 5) but its expression was confirmed in another preparation probed with anti-histidine-tag antibodies (lane 7). No protein was detected in the empty vector control (Fig. 1B ▶, lane 6 or 8). The protein for the N207 construct could not be detected (data not shown), so the construct was not further analyzed.
The purified NH, N88HC, and N88C proteins were tested for RNA-dependent ATPase (Fig. 1C ▶), splicing (Fig. 1D ▶, lanes 1–8), and spliceosome binding (Fig. 1D ▶, lanes 9–16) activities. Neither ATPase nor splicing activity was detected in the N88C protein, as expected (Fig. 1C,D ▶, lane 7). The NH protein showed a very small amount of ATPase activity (Fig. 1C ▶) but could not carry out splicing (data not shown). The N88HC protein had an RNA-dependent ATPase activity comparable to wild-type Prp2 (Fig. 1C ▶) and could complement prp2Ts mutant extracts (Fig. 1D ▶, lane 6). Immunoprecipitation of labeled actin pre-mRNA with anti-Prp2 antibody was used to measure spliceosome binding activity. In the absence of ATP, N88HC bound tightly to the spliceosome, as seen by immunoprecipitation of pre-mRNA (Fig. 1D ▶, lane 10); and after addition of ATP, N88HC released from the spliceosome (Fig. 1D ▶, lane 14). Attempts to immunoprecipitate purified NH and N88C with the spliceosome in the presence or absence of ATP were unsuccessful (Fig. 1D ▶, lanes 11,15; data not shown). Taken together, these results indicate that the N terminus of Prp2 is dispensable for all of its measurable activities, whereas the C terminus is important for an activity of Prp2 that needs to be further determined (see below).
Identification of a spliceosome binding region of Prp2
We took a genetic approach to investigate whether the dominant negative phenotype of the C terminus in the prp2Ts strain (Fig. 1A ▶) is due to the binding of the C terminus to the spliceosome, thereby interfering with the function of the temperature-sensitive prp2 protein. If the C terminus is important for binding, we should be able to isolate mutants in the C terminus that are impaired for spliceosome interaction. We previously isolated H-domain mutants of PRP2 that had a dominant negative phenotype when overexpressed behind the GAL1 promoter in vivo; the mutant proteins failed to release from the spliceosome in vitro (Edwalds-Gilbert et al. 2000). Here we used the GAL1-PRP2 system to identify potential spliceosome binding mutants of Prp2 that lost the dominant negative phenotype (Fig. 2A ▶).
FIGURE 2.
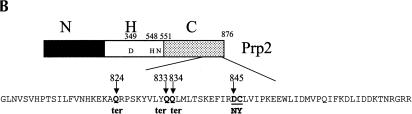
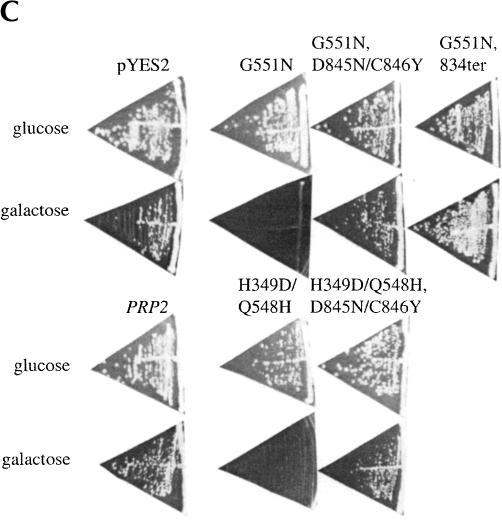
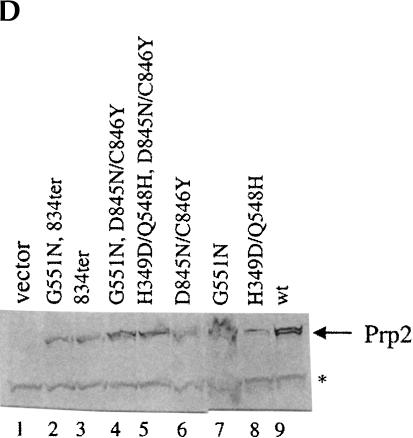
Identification and characterization of Prp2 spliceosome binding mutations. (A) Strategy for isolation of PRP2 spliceosome binding mutants. (B) Spliceosome binding mutants map to the C terminus of Prp2. The dominant negative H domain mutants used in this study are indicated. The final 73 amino acids are shown, with the locations of the mutations indicated. ter, termination codons. (C) C-terminal mutations permit growth of dominant negative mutants on galactose. The indicated plasmids were transformed into BJ168. Isolated colonies grown on SC-glucose were streaked onto SC-glucose or SC-galactose plates. Plates were photographed after 3 d (glucose) or 6 d (galactose) of growth at 30°C. (D) Mutant Prp2 proteins are expressed and stable in glass bead extracts. Five μL of the indicated glass bead extracts were separated by 10% SDS-PAGE and transferred to PVDF membrane. The membrane was incubated with rabbit anti-N-term Prp2 followed by alkaline-phosphatase-conjugated goat anti-rabbit IgG. The band corresponding to Prp2 is indicated. A cross-reacting band, marked *, is seen in glass bead extracts but is not present in purified Prp2 preparations (data not shown).
The dominant-negative GAL1-PRP2-G551N plasmid was treated with hydroxylamine and transformed into yeast. Transformants that could grow on galactose plates (Gal+) were isolated. To eliminate studying large truncation mutants, we estimated the size of the Prp2 protein expressed by immunoblotting extracts prepared from the Gal+ clones and selected mutants that expressed apparently full-length Prp2 protein (Fig. 2D ▶; data not shown). In order to avoid functional true or pseudo-revertants, cell extracts containing the apparently full-length Prp2 protein were added to heat-treated prp2Ts extract to assess the splicing activity of the mutant proteins. None of the mutants that we tested were able to complement splicing when the extract was diluted 1:20 (data not shown), which indicated that the Gal+ phenotype was not caused by reversion to wild type or pseudo wild type PRP2. A fragment-swap experiment was then performed to map the mutants. The GAL promoter and the region of DNA encoding the first 658 amino acids of Gal+ revertants were swapped with the same region from the G551N clone (that contained the G551N dominant negative mutation), and the Gal phenotype was again assayed to determine which region of Prp2 conferred the Gal+ phenotype (Fig. 2C ▶; data not shown). All clones tested remained Gal+ after the swap, indicating that mutations were localized to the C-terminal domain of Prp2. Of the nine mutations sequenced, seven had nonsense mutations (at amino acid residues 663, 824, 833, or 834; Fig. 2B ▶), and two of the clones had a double missense mutation (D845N/C846Y). Western analysis of the protein expressed in the mutants showed that the mutant proteins were stably expressed and of the expected size (Fig. 2D ▶). Thus, these C-terminal mutations eliminate the dominant negative function of the G551N mutation and are not active in splicing. These experiments support our observations indicating the importance of the C terminus in Prp2 activity.
Mutations in the C terminus of Prp2 confer a Gal+ phenotype on a second dominant negative H-domain mutant
The C-terminal mutations were identified by their ability to grow on galactose, presumably because the dominant negative mutant Prp2 protein could no longer bind tightly to the spliceosome. To test whether the D845N/C846Y mutation could confer a Gal+ phenotype to a different dominant negative Gal− H-domain mutant, site-directed mutagenesis was performed on pYES-PRP2-H349D/Q548H, whose gene product remains bound to the spliceosome in the presence of ATP and which has very low ATPase activity (Edwalds-Gilbert et al. 2000). The D845N/C846Y C-terminal mutations also conferred a Gal+ phenotype to the H349D/Q548H mutant (Fig. 2C ▶). When the D845N/C846Y mutations were introduced to the N88C plasmid, the mutated N88C protein could no longer exacerbate the prp2Ts phenotype when overexpressed (Fig. 1A ▶, N88C, D845N/C846Y). This indicates that the disruption caused by the D845N/C846Y mutation can affect binding function directly through the C terminus, rather than through potential interactions throughout a full-length protein.
We also tested the ability of Prp2 containing a wild-type H-domain and the D845N/C846Y mutation to complement a temperature-sensitive prp2Ts strain. The construct was made in 2μ-pYES2.0 vectors under a GAL1 promoter. No complementation was observed on glucose or galactose plates. Thus, the C-terminal mutation completely prevented the otherwise wild-type protein to complement even when overexpressed (data not shown). Therefore the D845 and C846 residues are critical to Prp2 activity in vivo.
C-terminal mutations relieve the block to in vitro splicing seen in the H-domain mutants
If the C-terminal mutants are spliceosome binding mutants, extracts prepared from strains expressing Prp2 with mutations in both the H- and C-terminal domains should not block endogenous Prp2 from acting in a splicing complementation assay. Glass bead extracts were prepared from cells grown in galactose-containing medium and were added to splicing mixtures containing heat-treated prp2Ts extract. We have shown that extracts containing Prp2 with a dominant negative H-domain mutation inhibit splicing by the endogenous Prp2 in the same extract (Fig. 3A ▶, lanes 2,3; Edwalds-Gilbert et al. 2000). The combination of the dominant negative H349D/Q548H H-domain mutations with the C-terminal mutations D845N/C846Y permits in vitro complementation (Fig. 3A ▶, lane 4). Complementation of heat-treated prp2Ts extract was also evident when glass bead extracts were added from the G551N, D845N/C846Y, or G551N;834ter mutants (Fig. 3A ▶, lanes 5,6). Extracts from C-terminal mutants with a wild-type H-domain also permit complementation of the heat-treated prp2Ts extract (Fig. 3A ▶, lane 7; data not shown). The mutant Prp2 proteins were all present to a similar extent in the glass bead extracts (Fig. 2D ▶), ruling out differential protein expression as the explanation for lack of inhibition. The amount of Prp2 in the vector alone extract is too low to detect by Western blots (Fig. 2D ▶, lane 1), but there is enough endogenous protein expressed from the chromosomal PRP2 gene to complement the splicing activity of heat-treated prp2Ts extract (Fig. 3A ▶, lane 1). These results do not rule out the possibility that C-terminal mutants with a wild-type H-domain are active; however, taken together with spliceosome binding data, these results are consistent with the notion that C-terminal mutant proteins do not stably bind to the spliceosome and are inactive for splicing.
FIGURE 3.
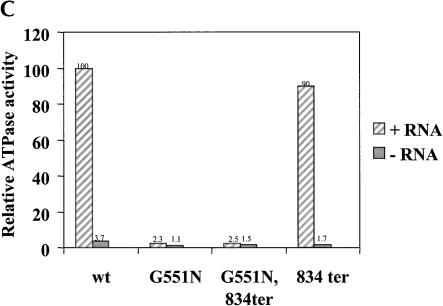
Mutations in the C terminus of Prp2 relieve the block to splicing seen in the H-domain mutants. (A) In vitro splicing. Glass bead extracts were prepared from the indicated strains and added to splicing mixtures containing heat-treated prp2Ts extract (Δprp2), 32P-labeled actin pre-mRNA, and ATP. Following incubation at 23° for 30 min, RNA was extracted and separated on a 7.5% polyacrylamide/7.5M urea gel. The positions of the pre-mRNA, mRNA, lariat intron-exon 2 (IVS*-E2), and intron lariat (IVS*) are indicated. (B) Prp2 proteins with mutations in the C terminus do not bind to the spliceosome in the absence of ATP. Splicing reactions were assembled in heat-treated prp2Ts extract with 32P-labeled actin pre-mRNA and ATP for 30 min at 23°. Glucose was added (5 mM final concentration), and incubation continued for 5 min to deplete ATP. Glass bead extracts prepared from BJ168 transformed with the mutants were added, and incubation continued for 10 min at 4°C. Half of the reaction mixture containing the indicated glass bead extracts were subjected to immunoprecipitation with anti-N-term Prp2 antibodies immobilized on Sepharose CL-4B beads. The band corresponding to actin pre-mRNA is indicated. The origin of the gel is marked *. (C) Prp2 containing a wild-type H-domain and a mutant C terminus has wild-type RNA-dependent ATPase activity. Estimated 15 ng of wild-type or mutant Prp2 proteins purified from poly(U)-Sepharose (Protocol B) were incubated with γ-[32P]-ATP with or without poly(U) RNA. Activity is shown relative to wild-type Prp2 in the presence of RNA, which is set at 100%. Note that proteins used in this assay were purified and their activities were measured in parallel. The measurements of wild-type and G551 proteins were shown (Edwalds-Gilbert et al. 2000).
Prp2 proteins with C-terminal mutations fail to bind to the spliceosome
The spliceosome binding activity of the C-terminal mutants was measured by immunoprecipitation of splicing reactions in the absence of ATP using polyclonal antiserum directed against the full-length Prp2 protein. Wild-type Prp2 and the H-domain mutants bind tightly to the spliceosome in the absence of ATP (Fig. 3B ▶, lanes 2–4). The binding activity in the C-terminal mutants is identical to that seen in vector alone (Fig. 3B ▶, cf. lanes 5–9 and lane 10), which indicates that the binding seen is due to the endogenous wild-type Prp2 expressed in the glass bead extracts from the chromosomal copy of PRP2 present in BJ168. In the presence of ATP, pre-mRNA is no longer immunoprecipitated from splicing reactions containing wild-type Prp2, whereas the H-domain mutant proteins remain bound (Edwalds-Gilbert et al. 2000). The C-terminal mutants did not bind to the spliceosome in the presence of ATP (data not shown). Therefore, Prp2 proteins with mutations in the C terminus do not bind to the spliceosome, regardless of ATP.
A Prp2 protein with a C-terminal mutation and a wild-type H-domain has no defect in RNA-dependent ATPase activity
To confirm that the C-terminal mutation did not affect the ATPase activity of the H-domain of the protein, wild-type and mutant Prp2 proteins were purified from yeast and assayed for RNA-dependent ATPase activity (Fig. 3C ▶). The G551N/834ter mutant, which has mutations in both the H-domain and the C terminus, has activity as low as the G551N mutant alone. The C-terminal mutant 834ter, which has a wild-type H-domain, has almost wild-type RNA-dependent ATPase activity (Fig. 3C ▶). Therefore, the mutant protein probably folds correctly in that it maintains the H-domain in an active conformation. Taken together, our results indicate that the C-terminal region of Prp2 is required for spliceosome interaction.
DISCUSSION
This study used deletion analysis and random mutagenesis to identify critical spliceosomal interacting regions in a transiently associated splicing ATPase, the yeast Prp2. Our data from the analysis of the domains of Prp2 and from the genetic screen indicate that (1) the C terminus of Prp2 is essential for interaction with the spliceosome, and (2) the N terminus of Prp2 is dispensable for activity. Although studies of other DEAH family members have evaluated the role of the N-terminal and C-terminal domains in splicing and cell viability, this is the first study that indicates that amino acid substitutions outside the H-domain can abolish spliceosome contact and splicing activity of a spliceosomal DEAH protein.
Our results define a function for the C terminus for spliceosome interaction. Overexpression of the C terminus in a temperature-sensitive prp2Ts strain exacerbates the Ts− phenotype, and deletion of the C terminus results in a nonfunctional Prp2 protein. Mutants selected for their ability to revert a dominant-negative release, mutant phenotype mapped to the C terminus of Prp2. Prp2 with a wild-type H-domain and mutations in the C terminus could not complement temperature-sensitive prp2Ts mutants, supporting the idea that the C terminus is required for interactions with the spliceosome. The C-terminal mutants show a spliceosome binding defect in vitro with no effect on RNA-dependent ATPase activity, which would indicate that the loss of binding function is not due to massive conformational changes within the protein. In contrast to the importance of the C terminus, Prp2 with a deletion of the N terminus can substitute for wild-type Prp2 in vivo, and has RNA-dependent ATPase and splicing activity in vitro.
We have looked for evidence that the C-terminal domain would by itself perform a Prp2-specific function such as spliceosome binding. We found that the N88C construct inhibited the growth of a prp2Ts strain when overexpressed, but not the growth of a wild-type PRP2 strain. This result suggests that the N88C protein could compete with a mutant Prp2 protein in vivo. The lack of competition in wild-type cells might be due to a relatively low expression of the truncated protein or a weak activity of the mutant protein. The purified N88C protein did not bind to the spliceosome as measured by immunoprecipitation of labeled pre-mRNA and did not inhibit splicing. Therefore, although the C terminus is required for interaction with the spliceosome, and can compete in vivo with mutant Prp2Ts protein, it does not bind sufficiently strong enough to account for wild-type spliceosome binding activity. It is likely that additional binding determinants reside within the H-domain of Prp2.
Figure 4A ▶ shows an alignment of amino acids 780–876 of Prp2 with related spliceosomal DEAH-box proteins Prp16, Prp22, and Prp43. Also indicated are results of deletion and mutation experiments with these proteins, which have shown the importance of the C terminus in viability and in in vitro activity (Hotz and Schwer 1998; Wang and Guthrie 1998; Schneider and Schwer 2001; Martin et al. 2002). Deletion of 42 amino acids from the C terminus of Prp2 results in a mutant that cannot complement a temperature-sensitive prp2 strain nor bind to the spliceosome in vitro. Deletion of the entire C-domain of Prp16 (amino acids 700–1024) is lethal (Wang and Guthrie 1998). In Prp22, deletion of the C-terminal 87 amino acids results in lethality, and a deletion of 55 amino acids from the C terminus of Prp43 shows lethality, further illustrating the importance of the C terminus (Schneider and Schwer 2001; Martin et al. 2002).
FIGURE 4.
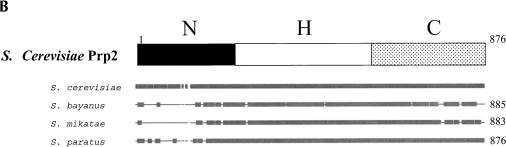
Conservation of Prp2. (A) Alignment of the C-terminal region of Prp2 with other DExD/H family members shows conservation surrounding the D845N/C846Y mutations. Amino acids 780–876 of Prp2 were aligned with Prp16, Prp22, and Prp43 using the BLASTP program. Amino acid identity with Prp2 is shown as a black background with white letters, and conserved residues are shown in a gray background with white letters. The site from which a deletion is known to be lethal is indicated above the sequence by >; #, deletion from here has a temperature-sensitive growth phenotype, but is lethal in combination with a shortened N terminus; *, deletion from here has a temperature-sensitive growth phenotype; ~, deletion has no effect on growth; STOP, stop codon; numbers in parentheses indicate the number of amino acids to the end of the protein. (B) Comparison of the Prp2 protein from S. cerevisiae to the ortholog from three closely related Saccharomyces species, S. bayanus, S. mikatae, and S. paratus. The alignment was performed using Align Plus 4, Version 4.1 (Scientific & Educational Software) with a similarity significance cutoff equal to or greater than 90%. Areas of gray represent windows of 10 amino acids in length where the protein is 90% identical to the cerevisiae Prp2. S. bayanus Prp2 is 81% matched with S. cerevisiae, S. mikatae is 82% matched, and S. paratus is 90% matched. Lines in the diagram indicate less than 90% identity; gaps indicate deletions in the alignment. A representative version of the domain boundaries of Prp2 indicating N (N-terminal domain), H (Helicase domain), and C (C-terminal domain) relative to the alignment is provided on the top.
The genetic screen for spliceosome binding mutants used the GAL1-PRP2 system to select for Gal+ revertants of a dominant negative, Gal− H-domain mutant after hydroxylamine mutagenesis. We recovered seven nonsense and two missense mutations, all of which mapped to the C terminus of Prp2. There is no indication that hydroxylamine mutagenesis would bias towards nonsense mutations. The intolerance of the C terminus to deletion combined with the dispensability of the N terminus suggests that the C terminus of Prp2 contains critical spliceosome binding determinants, which may be why of all the mutants we isolated map to the C terminus. The preponderance of nonsense mutations may be due to the sensitivity of the C terminus to truncation, most of which would show a Gal+ phenotype in the assay. Within the C terminus, perhaps only a specific region is most important in spliceosome interactions, again making Gal+ missense mutations less frequent than premature termination mutations.
The C termini of spliceosomal DEAH proteins Prp2, Prp16, Prp22, and Prp43 are highly conserved. The final 321 amino acids of Prp2 are 41% identical and 52% similar to amino acids 695–977 of Prp16, which are in the C terminus before the tail region. The extent of sequence identity with amino acids 813–1100 of Prp22 is 33% (48% similar) and that with amino acids 433–726 of Prp43 is 36% (49% similar). The conserved residues within the C terminus could play roles in domain formation, general spliceosome recognition, or intramolecular interaction. The proteins are required at different steps in the spliceosome assembly pathway and therefore must have a mechanism to distinguish their targets within the spliceosome. Prp2 binds to the pre-catalytic SS2 spliceosome (Kim and Lin 1993) and Prp16 does not (S.-H. Kim and R.-J. Lin, unpubl.). Prp16 is present in the SI spliceosome, which contains splicing intermediates formed after Prp2 has acted. Prp2 does not associate with the SI spliceosome or with the spliceosome after Prp16 has acted, suggesting that the docking site for these DEAH proteins within the spliceosome undergoes a conformational change after each interaction. Although the C termini of the spliceosomal DEAH proteins are highly conserved, being at least 50% different may be sufficient to provide for specificity determinants within the C-terminal domain that permit temporary docking of the proteins within the spliceosome. The nonconserved residues may mediate this interaction for Prp2, or be involved in intramolecular interactions with the H-domain. Interactions with distinct accessory factors may also provide specificity, such as Spp2 for Prp2 (Roy et al. 1995) or Isy1 for Prp16 (T. Villa and C. Guthrie, pers. comm.). The accessory factors may help target the DEAH proteins or modulate their activity, as has been noted for other members of the DExD/H family (for review, see Silverman et al. 2003).
In contrast to the importance of the C-terminal domain, a large portion of the N terminus of Prp2 can be deleted without apparent consequence in vivo, as measured by complementation assays. In vitro, amino acids 89–206 are dispensable. This phenomenon was also observed in deletion analyses of the N termini of Prp16 (Δ220), Prp22 (Δ260), and Prp43 (Δ90) Hotz and Schwer 1998; Wang and Guthrie 1998; Schneider and Schwer 2001; Martin et al. 2002). It is possible to delete the first 220 amino acids from the N terminus of Prp16 before affecting growth in vivo; in conjunction with deletions from the C terminus however, loss of the N-terminal 136–150 amino acids is lethal (Hotz and Schwer 1998; Wang and Guthrie 1998). In vitro, the N terminus of Prp16 is important for spliceosome binding, although the C terminus also plays a role (Wang and Guthrie 1998). We were unable to stably express the N terminus of Prp2; however, deletion of amino acids 89–206 in the N terminus did not affect Prp2 activity, including spliceosome binding, in vivo or in vitro. Because the N6HC protein is also fully functional in vivo, it appears that Prp2 does not rely on the N terminus for its activity. In a comparison of yeast genomes, Prp2 was shown to have extensive conservation in the H- and C-domains, with less conservation in the N terminus, up to amino acids 137 (Fig. 4B ▶). Interestingly, in Prp16, conservation is seen throughout the protein (http://db.yeastgenome.org/cgi-bin/FUNGI/nph-showAlign? locus=YKR086W; data not shown). This indicates that even among closely related yeast species, there has been a lack of selective pressure on the N terminus of Prp2 compared to the N terminus of Prp16. All of the Prp2 proteins contain the DC (845 and 846 in S. cerevisiae) amino acids, which are not found in other spliceosomal DEAH helicases. Accessory proteins such as Spp2 (Roy et al. 1995) and Prp44 (van Nues and Beggs 2001) could fulfill some of the role in spliceosome recruitment of Prp2 ascribed to the N termini of Prp16 and Prp22.
It will be interesting to determine whether Prp2, Prp16, Prp22, and Prp43 have a common spliceosome binding site and to unravel the in vivo substrates for their RNA-dependent ATPase activities. Further studies in Prp2 to define the borders of the spliceosome binding domain and the function of conserved and nonconserved residues in the C terminus will be important for identifying a potential general spliceosome binding motif, as well as regions responsible for binding specificity.
MATERIALS AND METHODS
Yeast strains and culture
Yeast genetic manipulations and growth conditions were performed as described in Rose et al. (1990). Yeast were transformed using the lithium acetate method as described by Rose et al. (1990) or Gietz and Woods (1994). Strains used were BJ168 (MATa, ura3, leu2, trp1, prb, pep4, prc), B4-7-2D (MATa, his3-Δ200, lys2-801, ura3-52), and D12 (MATa, ura3, leu2, his3, prp2-1).
Plasmid construction and mutagenesis
The N terminus of Prp2 is a HindIII fragment from pYES2.0-PRP2, which includes a six-histidine tag through amino acid 206. The NH, N88HC, and N88C constructs were made from pYES2.0-PRP2 by using overlap recombination PCR. N6HC was derived from N88HC by overlap recombination PCR. Deletions were confirmed by sequencing by the City of Hope DNA sequencing facility. Site-directed mutagenesis of plasmids pYES2.0-PRP2 and pYES2.0-H349D/Q548H was done using the QuikChange site-directed mutagenesis kit (Stratagene). Mutations were confirmed by sequencing. To isolate Gal+ revertants of G551N. the plasmid pYES2.0-PRP2(G551N) was mutagenized by hydroxylamine as described (Edwalds-Gilbert et al. 2000). After treatment with hydroxylamine, the DNA was precipitated and washed in 70% ethanol. The mutagenized DNA was transformed into strain B4-7-2D to select for Ura+ transformants on SC-glucose plates lacking uracil. Transformants were replica-plated onto SC-galactose lacking uracil. After incubation at 30° for 3 to 4 d, colonies that formed on the galactose plate (Gal+) were picked and streaked onto fresh plates for single colony isolation. The PvuII/PvuII fragment of the pYES2.0-PRP2(G551N) plasmid, which contained the G551N mutation in motif VI, was used to replace the corresponding fragment in each revertant able to grow on galactose. The chimeric DNA contained the PvuII/PvuII fragment from the unmutagenized G551N or wild-type DNA and the C-terminal region of PRP2 from the Gal+ revertants. Chimeric DNA was transformed into strain B4-7-2D and tested for growth on SC-galactose.
Preparation of yeast splicing extracts and glass bead extracts
Yeast whole-cell splicing extracts were prepared as described (Lin et al. 1987). For preparation of glass bead extracts, yeast strains containing the pYES2.0-PRP2 wild-type or mutant plasmids were cultured in growth medium containing 2% raffinose or 2% glucose (for dominant negative mutants) overnight. The following day, cultures were diluted 1:100 in SC-raffinose or SC-glucose and grown overnight at 30°C. Cultures grown in raffinose were induced by the addition of galactose to 2% and continued growth for 6–12 h; cultures grown in glucose were harvested and washed prior to resuspension in SC-galactose. After induction, cells were harvested, washed with cold water, and resuspended in 1/100 volume of cold buffer A (10 mM HEPES, pH 7.8, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT) containing proteinase inhibitors PMSF (1 mM), benzamidine (1 mM), leupeptin (1 μM), and pepstatin (1 μM). An equal volume of 0.5 mm glass beads was added, and cells were broken by vortexing three times for 1 min, with a 1-min incubation on ice after each vortex. The mixture was centrifuged at 3,000g for 5 min. The resulting supernatant was incubated with 0.1 volume of cold 2 M KCl on ice. The mixture was centrifuged at 105,000g for 20 min in a Beckman Optima tabletop ultracentrifuge using a TLA 100.4 rotor. The supernatant was used as a glass bead extract and to purify the histidine-tagged Prp2 proteins as described below.
Purification of histidine-tagged proteins
Histidine-tagged full-length or truncated Prp2 proteins were purified by Ni-affinity chromatography by one of the following protocols. Protocol A was performed according to Tang et al. (1998). In some cases as specified, yeast extracts were first fractionated with 30%–60% of ammonium sulfate, dialyzed, and then loaded onto a nickel column. Protocol B was modified from Plumpton et al. (1994) and is described here. Glass bead extracts were mixed with an equal volume a buffer containing 20% glycerol, 20 mM Tris-HCl, pH 7.9, 1 M NaCl, and 2 mM imidazole. Samples were loaded twice onto a 0.4-mL Ni column prepared according to the manufacturer’s instructions (Novagen) and equilibrated in BC buffer (10% glycerol, 20 mM Tris, pH 7.9) containing 500 mM KCl (BC500). Columns were washed with 30 column volumes of BC500 containing 20 mM imidazole, and then washed with 20 column volumes of BC buffer containing 100 mM KCl (BC100) and 50 mM imidazole. Prp2 protein was eluted stepwise with six column volumes of BC100 containing 50 mM, 100 mM, 150 mM, 200 mM, or 250 mM imidazole. Peak Prp2-containing fractions were further purified on a poly(U)-Sepharose column equilibrated with buffer BP50 (20 mM HEPES, pH 7.9, 0.5 mM EDTA, 0.01% NP-40, 10% glycerol, 50 mM KCl). Columns were washed with six column volumes of BP50, and Prp2 protein was eluted stepwise with three column volumes each of buffer BP containing 100 mM, 150 mM, 200 mM, 250 mM, or 300 mM KCl. Protein concentrations were determined by Bradford assay (Bio-Rad) or estimated by Coomassie or silver staining gels.
In vitro splicing and ATPase assays
In vitro splicing was performed as described (Lin et al. 1987; Kim et al. 1992; Kim and Lin 1996). Spliceosome binding assays and immunoprecipitation of splicing reactions with or without ATP were performed as described (Kim and Lin 1996; Edwalds-Gilbert et al. 2000).
The 32P-labeled RNAs on gels were quantified using a Molecular Dynamics Phosphorimager. ATPase assays of purified Prp2 proteins were performed as described (Kim and Lin 1993) using 15 ng of protein with or without 0.5 μg/mL poly(U) RNA.
Western analysis
Glass bead extract or protein samples was separated by SDS-PAGE and electrophoretically transferred to PVDF membrane (Millipore) in Tris-Glycine transfer buffer (25 mM Tris, pH 8.3, 192 mM glycine) according to the manufacturer’s instructions (Bio-Rad). In some cases, following protein transfer, the filter was stained for 1 min with 50 mL of an aqueous solution containing 0.25 g Ponceau S and 0.5 mL acetic acid and then destained for 2 min in water. The filter was photographed and then further destained prior to incubation in Blotto (5% nonfat dry milk in PBS) for 2 h at room temperature. Further manipulation was done as described (Kim et al. 1992). N-terminal-specific anti-Prp2 polyclonal antisera were used at 1:500–1:2,000 dilution as described (Kim and Lin 1996). Rabbit polyclonal antisera were generated against histidine-tagged, full-length Prp2 purified from yeast. Anti-full length Prp2 antisera were used at 1:1,000 dilutions. Anti-tetra His antibodies (QIAGEN) were used at 1:1000 dilutions. Alkaline-phosphatase-conjugated goat anti-rabbit IgG was used as the secondary antibody for detection.
Acknowledgments
We thank Carol Lee for the preparation of the antibody against full-length Prp2 protein, Ying Yu for construction of pYES2-N88C, Cindy Cheng for construction of pYES2-N6HC, and Marcy Diaz for the N6HC plasmid shuffle assay. We also thank Glenn Manthey for review of the manuscript. This work was supported by NIH GM 40639. The Molecular Dynamics Phosphorimager was purchased through grant BIR-9220534 from the National Science Foundation. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5151404.
REFERENCES
- Arenas, J.E. and Abelson, J.N. 1997. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. 94: 11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campodonico, E. and Schwer, B. 2002. ATP-dependent remodeling of the spliceosome: Intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22. Genetics 160: 407–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert, G., Kim, D.H., Kim, S.H., Tseng, Y.H., Yu, Y., and Lin, R.J. 2000. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA 6: 1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D. and Woods, R.A. 1994. High efficiency transformation in yeast. Molecular genetics of yeast: Practical approaches (ed. J.A Johnston), pp. 121–134. Oxford University Press, Oxford, UK.
- Hamm, J. and Lamond, A.I. 1998. Spliceosome assembly: The unwinding role of DEAD-box proteins. Curr. Biol. 8: R532–534. [DOI] [PubMed] [Google Scholar]
- Hotz, H.R. and Schwer, B. 1998. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics 149: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. and Lin, R.J. 1993. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. 90: 888–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. and Lin, R.J. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell Biol. 16: 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Smith, J., Claude, A., and Lin, R.J. 1992. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11: 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, D.S. and Beggs, J.D. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18: 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R.J., Lustig, A.J., and Abelson, J. 1987. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes & Dev. 1: 7–18. [DOI] [PubMed] [Google Scholar]
- Martin, A., Schneider, S., and Schwer, B. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277: 17743–17750. [DOI] [PubMed] [Google Scholar]
- Plumpton, M., McGarvey, M., and Beggs, J.D. 1994. A dominant negative mutation in the conserved RNA helicase motif “SAT” causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan, P.L. and Guthrie, C. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8: 847–855. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. 1990. Methods in yeast genetics: A laboratory course. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Roy, J., Kim, K., Maddock, J.R., Anthony, J.G., and Woolford Jr., J.L. 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1: 375–390. [PMC free article] [PubMed] [Google Scholar]
- Schneider, S. and Schwer, B. 2001. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 276: 21184–21191. [DOI] [PubMed] [Google Scholar]
- Schneider, S., Hotz, H.R., and Schwer, B. 2002. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 277: 15452–15458. [DOI] [PubMed] [Google Scholar]
- Schwer, B. and Gross, C.H. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17: 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B. and Guthrie, C. 1992. A dominant negative mutation in a spliceosomal ATPase affects ATP hydrolysis but not binding to the spliceosome. Mol. Cell. Biol. 12: 3540–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, E., Edwalds-Gilbert, G., and Lin, R.J. 2003. DExD/H box proteins and their partners: Helping RNA helicases unwind. Gene 312: 1–16. [DOI] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Tang, Z., Yanagida, M., and Lin, R.J. 1998. Fission yeast mitotic regulator Dsk1 is an SR protein-specific kinase. J. Biol. Chem. 273: 5963–5969. [DOI] [PubMed] [Google Scholar]
- Tanner, N.K. and Linder, P. 2001. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell 8: 251–262. [DOI] [PubMed] [Google Scholar]
- van Nues, R.W. and Beggs, J.D. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, J.D., Jankowsky, E., Company, M., Pyle, A.M., and Abelson, J.N. 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17: 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Guthrie, C. 1998. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA 4: 1216–1229. [PMC free article] [PubMed] [Google Scholar]