Abstract
The expression of some genes requires a high proportion of ribosomes to shift at a specific site into one of the two alternative frames. This utilized frameshifting provides a unique tool for studying reading frame control. Peptidyl-tRNA slippage has been invoked to explain many cases of programmed frameshifting. The present work extends this to other cases. When the A-site is unoccupied, the P-site tRNA can be repositioned forward with respect to mRNA (although repositioning in the minus direction is also possible). A kinetic model is presented for the influence of both, the cognate tRNAs competing for overlapping codons in A-site, and the stabilities of P-site tRNA:mRNA complexes in the initial and new frames. When the A-site is occupied, the P-site tRNA can be repositioned backward. Whether frameshifting will happen depends on the ability of the A-site tRNA to subsequently be repositioned to maintain physical proximity of the tRNAs. This model offers an alternative explanation to previously published mechanisms of programmed frameshifting, such as out-of-frame tRNA binding, and a different perspective on simultaneous tandem tRNA slippage.
Keywords: translation, recoding, kinetic model, frameshifting, ribosome
INTRODUCTION
A primary feature of the genetic code is its triplet character. During standard translation, codons do not overlap and are not separated by any nucleotides. This feature provides the basis for predicting coding sequences in mRNA. A sequence of triplets in mRNA not interrupted by a stop codon is defined as an open reading frame (ORF). Generally, a single ORF encodes a single protein. However, during translation of certain mRNAs, a proportion of ribosomes shift frame at a particular site, and a second product is synthesized. This product can have more or fewer amino acid residues than the product of standard decoding, depending on the existence of a partially overlapping ORF and its disposition. ORF fusion is widely used during the expression of viral genes and mobile elements, and several examples in decoding nonmobile chromosomal genes have been studied in detail (for reviews, see Brierley and Pennell 2001; Baranov et al. 2002a; Stahl et al. 2002; Baranov et al. 2003).
Programmed ribosomal frameshifting is an extension of genetic decoding that expands standard gene readout. More than one product can be synthesized from the same gene. For example, in Escherichia coli dnaX decoding, two proteins are synthesized from the same mRNA in an equimolar ratio, one by standard translation and the other one via frameshifting (Blinkowa and Walker 1990; Flower and McHenry 1990; Tsuchihashi and Kornberg 1990). Frameshifting also offers the prospect of conditional regulation of gene decoding: with bacterial release factor 2 (RF2), its own cellular concentration affects the efficiency of the frameshifting that is required for its biosynthesis (Craigen and Caskey 1986); whereas with mammalian antizyme, the efficiency of obligatory frameshifting depends on the concentration of polyamines whose synthesis is negatively regulated by antizyme (Matsufuji et al. 1995).
In addition to intrinsic interest, genes utilizing frameshifting for their expression provide an opportunity to understand the mechanism of open reading frame control. There have been several models dealing with the phase of the ribosomal cycle at which frameshifting occurs (Jacks et al. 1988a; Weiss et al. 1989; Horsfield et al. 1995; Farabaugh and Björk 1999; Harger et al. 2002) and others dealing with the influence of codon–anticodon interactions on frameshifting (Brierley et al. 1991; Tsuchihashi and Brown 1992; Curran 1993; Kolor et al. 1993; Sundararajan et al. 1999; Atkins et al. 2000). The relationship of these models to the present one, will be considered below.
Reading frame switching due to repositioning of tRNA in the P-site
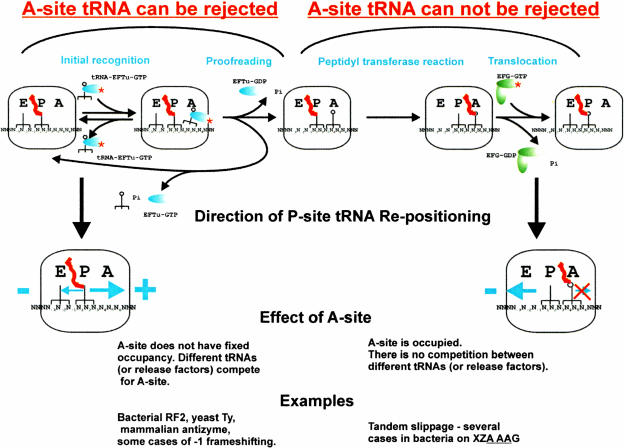
During translation, the P-site tRNA helps to define a boundary for the A-site tRNA. If the A-site tRNA arrives out-of-frame, it should be rejected as a noncognate tRNA, because no base pairs are formed in the first position of the A-site codon. If this is true, and supportive evidence has been presented by Hansen et al. (2003), the incoming tRNA can be incorporated into the ribosome only after P-site tRNA re-pairing to mRNA at a codon in a new frame. There are two distinct elongation phases where repositioning of the P-site tRNA relative to mRNA can occur (Fig. 1 ▶). Repositioning involves codon:anticodon dissociation, relative movement of the anticodon to mRNA, and re-pairing to mRNA at a new codon.
FIGURE 1.
Simplified scheme of the translation elongation cycle. Two main types of ribosomal frameshifting are distinguished depending on the phase of the elongation cycle where tRNA repositioning occurs.
The first occurs prior to the final (irreversible) acceptance of the A-site tRNA, that is, during the initial recognition of the A-site tRNA and/or the proofreading step (Fig. 1 ▶). If P-site tRNA repositioning occurs at this phase, then the next incorporated amino acid will be specified by the codon in the new frame. Repositioning can occur in both plus and minus directions, although in the case of the minus direction, the tRNA in the E-site could interfere (Horsfield et al. 1995; Baranov et al 2002b; K.H. Nierhaus, pers. comm.). If the E-site plays such a role, E-site tRNA slippage or its relative dislocation should precede P-site tRNA repositioning. If the P-site tRNA can dissociate from its codon in the initial frame and bind to a codon in a new frame, then it may also be able to shift back to the initial frame, before the A-site tRNA is irreversibly incorporated into the ribosome. This would mean that the success of frameshifting largely depends on the availability of tRNAs or release factors competing for available A-site codons in different frames. This point is considered further below.
The second elongation phase to consider is after final acceptance of the A-site tRNA into the ribosome, but before translocation is complete (Fig. 1 ▶). In this case, the identity of the A-site tRNA corresponds to that specified by the zero frame codon. Because an irreversible choice of A-site tRNA has already been made, the concentration of potential competing tRNAs for the A-site is irrelevant for frameshift efficiency. Productive forward repositioning of the A-site tRNA is assumed to be unlikely because no base pairs would be formed with the first A-site codon base. In this scenario, P-site repositioning can occur only in the minus direction, because nucleotides 3′ of the P-site codon are occupied by A-site tRNA that cannot be rejected. A substantial refinement of the presently accepted hypothesis of simultaneous slippage of two tRNAs suggested for this elongation phase by Jacks et al. (1988a) is provided below.
Effect of stability of the P-site codon:anticodon complex on reading frame maintenance
Selection of tRNA in the A-site is efficiently monitored by the ribosome. The geometry of base pairs in the first two positions of the A-site codon:anticodon duplex is controlled more precisely than the geometry of the base pair in the third position (for review, see Ramakrishnan 2002). This allows the ribosome to discriminate cognate tRNAs from near-cognate tRNAs that could form energetically stable complexes with the same codon in mRNA (i.e., although the codon UUU can form base pairs with both the AAG and GAI anticodons and energetically both complexes are nearly equal, only AAG is a cognate anticodon for UUU). Such discrimination of tRNAs in the A-site is vital for accurate translation. However, there is no need to control the geometry of the P-site tRNA codon:anticodon complex in the same way. First, the P-site tRNA correctness has already been monitored while it was in the A-site. If the identity of the P-site tRNA were controlled, its rejection would terminate translation because the tRNA is already attached to a growing polypeptide chain. (Indeed, studies by Potapov et al. [1995] have shown that DNA matrices may be used instead of mRNA for the P-site tRNA binding, but not for the A-site tRNA binding.) The only case in which recognition of mRNA occurs in the P-site is during translational initiation. In bacteria there are several other initiator codons that can be used instead of the standard AUG, for example, GUG, UUG, and in rare cases AUU. These codons are recognized by the same tRNAfMet (whereas elongation tRNAMet recognizes internal AUG codons exclusively). This leads to the conclusion that ribosomes do not discriminate the positions of a codon:anticodon duplex in the P-site in the same way as they do in the A-site (e.g., tRNAs with the anticodons AAG and GAA may be equally stable in complex with the codon UUU).
If we consider the P-site tRNA repositioning as a chemical reaction, its rate may be expressed by the following equation:
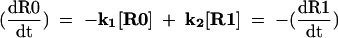 |
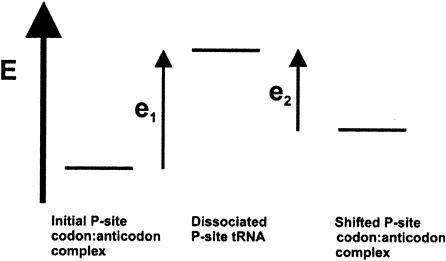
where [R0] is the concentration of ribosomes with P-site tRNA in the complex with the codon in the zero frame, [R1] is the concentration of ribosomes with the complex in a new frame, k1 is the rate constant of the forward reaction, and k2 is the rate constant of the reverse reaction. The stage when P-site tRNA is entirely dissociated from the mRNA can be considered as the activation complex for this reaction. In this case, the value of k1 and k2 would depend primarily on the energetic barriers between the corresponding codon:anticodon complexes and dissociated complexes (Fig. 2 ▶). This means that the efficiency of frameshifting depends not only on the stability of the complex in a new frame, but also on the stability of the complex in the initial, or zero frame.
FIGURE 2.
Energetic profile of P-site codon:anticodon complexes. P-site tRNA dissociated from mRNA is considered as an activation complex for P-site tRNA repositioning. Transition from a tRNA: mRNA complex with the codon in the initial frame to where the codon is in an alternative frame, can be achieved in two ways: (1) by increasing the rate constant of the forward reaction (higher energy of the initial complex); and (2) by decreasing the rate constant of the reverse reaction (lower energy of the new complex).
The importance of the stability of the codon:anticodon complex in a new frame has been noted before (Jacks et al. 1988a; Weiss et al. 1990a; Tsuchihashi and Brown 1992). Curran (1993) performed a systematic analysis of frameshifting efficiency in a derivative of the release factor 2 (RF2) frameshifting cassette CUU UAG C with 32 different codons preceding the stop codon (the wild-type UGA stop codon was substituted with UAG). (Spaces are used to separate codons in the initial ORF, underlining shows the new frame.) He found that the most frameshift-prone codons are CUU > CCC > UUU > GUU > CCU, where CUU is the naturally occurring shifty codon in all bacterial genes encoding RF2 that use frameshifting for their expression. Curran (1993) was able to build an exponential correlation between frameshifting efficiency on these codons and the stability of the P-site tRNA in a complex with an overlapping +1 frame codon using simplified calculations of the energies of the codon:anticodon complex. However, in some cases no correlation was found. For instance, tRNAPhe(3′-AAG-5′) should form a more stable complex with the +1 codon UUU in the sequence U UUU than would tRNALeu(3′-GAG-5′) in the sequence C UUU. Nevertheless, the observed frameshifting efficiency at the sequence U UUU was lower. Thus, it is not sufficient to estimate the stability of complexes relying solely on the energies of the base pairs involved. Certain mutations in tRNAs outside of the anticodon loop can affect the rate of P-site tRNA dissociation (e.g., Herr et al. 1999).
The effect of stability of the initial P-site tRNA:mRNA complex on frameshifting has been observed (e.g., Farabaugh et al. 1993) and experimentally studied in detail especially using the efficient ribosomal frameshifting that occurs during decoding of yeast Ty transposable element mRNAs (Vimaladithan and Farabaugh 1994; Pande et al. 1995; Sundararajan et al. 1999). (A similar effect of the stability of the initial tRNA–mRNA complex in the A-site had been noticed earlier [Tsuchihashi and Brown 1992].) In Ty1, the frameshifting site is CUU AGG C (Belcourt and Farabaugh 1990) and in Ty3 it is GCG AGU U (Farabaugh et al. 1993). If the tRNAAla(3′-CGU-5′) that recognizes GCG is repositioned to the overlapping +1 CGA codon, it can form only one canonical base pair. Vimaladithan and Farabaugh (1994) checked frameshifting efficiency with all 61 possible codons preceding AGG C or AGU U in a yeast strain lacking tRNAArg(3′-UCC-5′). They found that the most efficient frameshifting (>40%) occurs on those codons (CUU, CCG, GCG, and CUG) that do not have cognate tRNAs that could form standard codon:anticodon interactions (e.g., canonical Watson-Crick base pairs in all three positions). In addition, frameshifting efficiency on those codons does not directly correlate with the ability of their corresponding tRNAs to re-pair with the mRNA in the +1 frame. This led to the proposal that there are special features in those tRNAs that promote frameshifting by interfering with the binding of the incoming A-site tRNA (Vimaladithan and Farabaugh 1994). In this model, A-site tRNA recognition occurs in the alternative frame without a shift of the P-site tRNA. In their latest review, this group even considered the more radical hypothesis that in yeast, frameshifting occurs only caused by the out-of-frame binding of tRNA in the A-site (Stahl et al. 2001). However, if the initial codon:anticodon complex is weak, P-site tRNA repositioning to a codon in a new frame may be efficient even if the stability of the new complex is also weak (Fig. 2 ▶; kinetic model below); in this case, there would be a correlation between the propensity for re-pairing in the new frame and frameshifting efficiency, but it is not direct. The idea that near-cognate tRNAs are prone to slippage was also proposed by Gallant et al. (2000). Furthermore, recent work (A.J. Herr, N.M. Wills, C.C. Nelson, R.F. Gesteland, and J.F. Atkins, in prep.) has shown that in a case of “extreme” P-site repositioning—hopping—ribosomes carrying P-site tRNA can, in certain circumstances, re-engage with mRNA, in positions where no Watson-Crick base pairs can be formed between the codon and anticodon. Taken together, these several considerations indicate that there is no need to invoke structural obstacles to explain frameshifting on such codons. Based on experiments with mutated mammalian antizyme +1 frameshifting cassettes, Ivanov et al. (2003) proposed that <2 bp in the new, shifted codon:anticodon complex may be sufficient for the efficient frameshifting. The wild-type frameshifting site in mammalian antizyme 1 is UCC UGA U; however, several mutations in the UCC codon that diminish base-pairing of tRNA anticodon with mRNA codon in the +1 frame do not eliminate frameshifting (Matsufuji et al. 1995).
In summary, the stability of P-site codon:anticodon complex formation in the initial and new frames should greatly influence to the efficiency of frameshifting.
Effect of competitors in the A-site
As considered above, when P-site tRNA repositioning occurs during tRNA recognition before the peptidyl transferase reaction, the efficiency of A-site tRNA acceptance should depend on the concentration of tRNAs or release factors corresponding to A-site codons in the different frames. Despite the potential ability of P-site tRNA to be repositioned on certain overlapping codons, realization of this potential depends on several features, among which relative tRNA concentration and affinity for mRNA are prominent. The kinetic model described below indicates how these features affect P-site tRNA repositioning and consequently frameshifting.
Let us consider a sequence of the general type 000 111 2, where the tRNA that recognizes codon 000 is capable of being repositioned to codon 001 when it is in the P-site. These processes can be considered as a combination of five reactions:
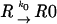 |
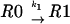 |
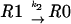 |
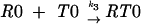 |
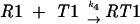 |
where R is the ribosome incoming into this particular sequence; R0 is the ribosome with its P-site tRNA base-paired to the 000 codon; R1 is the ribosome with its P-site tRNA base-paired to the 001 codon; T0 is a tRNA corresponding to the A-site codon 111; T1 is the A-site tRNA corresponding to the 112 codon; RT0 is a complex of the ribosome with irreversibly bound P-site and A-site tRNAs in frame 000 111; and RT1 is a complex where P-site and A-site tRNAs are irreversibly bound in the +1 frame 001 112. The rates of all reactions can be expressed as follows:
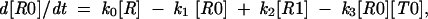 |
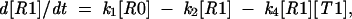 |
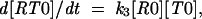 |
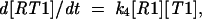 |
where we assume that the concentration of the incoming ribosomes [R] is large and does not change significantly in the course of this reaction, so that ribosomes arrive at a given sequence with a constant speed k0[R]. In the stationary regime, from
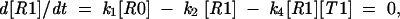 |
we get a value for [R0]/[R1], which is
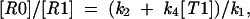 |
where we also assume that the concentration of ribosomes at this particular sequence is significantly lower than of tRNAs, so that [T0] and [T1] can also be considered as constants.
Next, from the constant [R0] and [R1] of the stationary regime, we obtain
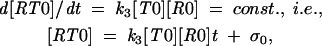 |
and
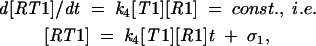 |
where σ1, σ2 are integration constants representing the concentrations [RT0] and [RT1] achieved during the initial equilibration stage of the reaction. After some time, these constant terms become negligible as compared with the terms linear in time, t, and then we can write for this later stage,
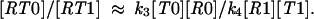 |
Substituting the stationary value of [R0]/[R1], we finally obtain
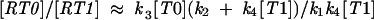 |
or
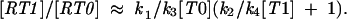 |
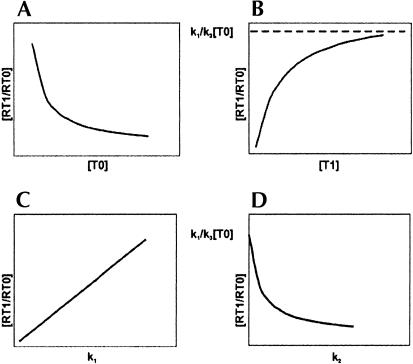
This equation illustrates the dependence of frameshifting efficiency [RT1/RT0] on the concentration of “in-frame” and “out-of-frame” A-site tRNAs. The graphical representation of this dependence is shown in Figure 3 ▶.
FIGURE 3.
Graphical representation of the affect of several parameters on frameshifting efficiency in Ty1. (A) The effect of concentration of “in-frame” A-site tRNA. (B) The effect of concentration of “out-of-frame” tRNA. (C,D) The effects of the rate constants for the repositioning of P-site tRNA, k1 and k2, depend on “instability” of the initial P-site codon:anticodon complex and “instability” of the P-site tRNA complex with the +1-frame codon, respectively.
It is clear that the effect of A-site tRNA corresponding to the +1 frame codon and the effect of stability of P-site tRNA in a shifted complex are limited. As seen in Figure 3B ▶, a low concentration of this tRNA would favor standard triplet translation. A high concentration should increase the efficiency of frameshifting; however, this effect is limited so that the efficiency cannot exceed the limit k1/k3[T0]. On the contrary, the concentration of tRNA corresponding to the in-frame A-site codon (Fig. 3A ▶) plays a more significant role. Its high concentration favors triplet decoding, its lower concentration favors frameshifting. Depletion of this tRNA should bring frameshifting efficiency to virtually 100%. Figure 3 ▶, C and D, illustrates the effect of forward and reverse P-site tRNA repositioning rates on frameshifting. k1 is the rate of forward repositioning from the zero-frame codon to the plus-frame codon. This rate depends on energetic differences between the zero-frame complex and the dissociated complex (Fig. 2 ▶). Decreasing its stability, for example, by incorporating a near-cognate tRNA in the P-site, should increase frameshifting efficiency (Fig. 3C ▶). k2 corresponds to the rate of reverse P-site tRNA repositioning. It depends on energetic differences between the complex of P-site tRNA with the plus-frame codon and the dissociated complex (Fig. 2 ▶). Low affinity of the P-site tRNA to the plus-frame codon favors triplet decoding, whereas its high affinity favors frameshifting. However, even if P-site tRNA forms a perfect complex with the plus-frame codon, frameshifting efficiency cannot exceed the k1/k3[T0] value (Fig. 3D ▶). These considerations illustrate the parameters, cellular tRNA concentration and their codon affinity, that are necessary for standard triplet decoding. If these requirements are not fulfilled for tRNAs involved in decoding a particular combination of codons, this combination of codons would be shift-prone. A later paragraph deals with such examples.
There are several cases of experimentally analyzed +1 frameshifting that support this kinetic model. The Ty1 and Ty3 frameshifting sites are CUU AGG C (Belcourt and Farabaugh 1990) and GCG AGU U (Farabaugh et al. 1993), respectively. In both cases, the frameshifting rate should depend on the concentration of tRNAs that recognizes the zero- and +1-frame A-site codons. Under normal conditions in yeast, the cellular concentration of tRNAs depends on their gene copy numbers (Percudani et al. 1997). With Ty1, the zero-frame codon is recognized by tRNAArg(3′-UCC-5′), which is encoded by only a single gene. The +1-frame codon is recognized by tRNAGly(3′-CCG-5′), which is encoded by 16 identical genes. In decoding Ty3 mRNA, the zero-frame codon is recognized by tRNASer(3′-UCG-5′) encoded by two genes, and the +1-frame codon is recognized by tRNAVal(3′-CAI-5′), which is encoded by 14 gene copies. In both cases, misbalance between the concentrations of the corresponding tRNAs is exceptional, and favors frameshifting (Sundararajan et al. 1999), as it should according to Figure 3, A and B ▶. Indeed, following the finding of earlier experiments (Atkins et al. 1979; Weiss and Gallant 1983), manipulations of the concentration of the corresponding tRNAs should affect frameshifting efficiency. Belcourt and Farabaugh (1990) have shown that overexpression of tRNAArg(3′-UCC-5′), corresponding to the zero-frame codon, reduces Ty1 frameshifting about 40-fold (Fig. 3A ▶). Deletion of the only gene encoding this tRNA increases frameshifting and as a result decreases transposition of Ty1 (Kawakami et al. 1993). Furthermore, overexpression of tRNAs corresponding to the +1-frame codon can increase frameshifting efficiency (Pande et al. 1995) as it should according to Figure 3B ▶. Both these effects are evident in Figure 3, A and B ▶. At the same time, mutations of the last C in the Ty1 frameshifting site (CUU AGG C) to any other nucleotide reduces frameshifting efficiency ~40-fold (Belcourt and Farabaugh 1990). The tRNA that normally recognizes the potential +1-frame codons GGA or GGG, is significantly less abundant than tRNAGly(3′-CCG-5′), which decodes the wild-type +1-frame codon GGC. The codon GGU is presumably recognized by the same tRNAGly (3′-CCG-5′), but it may have lower affinity because of the G:U pair at the third position of the codon.
This model is also valid for the frameshifting observed during expression of Escherichia coli release factor 2 (RF2), where the efficiency of frameshifting is highly dependent on the concentration of RF2 itself competing for the A-site, UGA, in the zero frame [shift site CUU U(GA)]. However, its precise description in the model shown above is complicated, because the efficiency of frameshifting depends on the rate of Shine Dalgarno/anti-Shine Dalgarno complex formation. This complex formation is important for destabilization of P-site tRNA interactions with the zero-frame codon.
This model indicates that there are certain concentrations of tRNAs that are necessary for the maintenance of accurate triplet decoding.
Caution about the phrase “simultaneous slippage” for tandem A-site and P-site anticodon repositioning
All the cases considered so far have involved P-site tRNA repositioning before the A-site tRNA has been irreversibly incorporated into the ribosome. When the A-site tRNA cannot be rejected, a different consideration applies (this arises after the last step of proofreading, during the peptidyl transferase reaction or during translocation as suggested earlier [Weiss et al. 1989], but certainly before translocation is complete). The cellular concentration of tRNA corresponding to the codon in the A-site does not play any role. The success of frameshifting now depends on the ability of the A-site tRNA to be repositioned to a new frame. This kind of frameshifting can occur only in the minus direction, because, as discussed above, the P-site tRNA cannot pair with the codon, 1 nt of which is already occupied by the A-site tRNA. The ability of P-site tRNA to pair with the new codon most probably depends primarily on the energies of the corresponding codon:anticodon complexes in the same manner as during recognition of tRNA in the A-site. However, for A-site tRNA repositioning, the effect of stability of the corresponding complexes is more difficult to estimate. The anticodon of this tRNA is still located in the A-site, where ribosomal monitoring of the correct geometry of the codon:anticodon duplex may still occur. This implies that correct interactions at the first two positions in the codon: anticodon duplex play a bigger role than the correct interactions at the third position. On the other hand, because normally there are no unoccupied nucleotides in the mRNA between the A-site and P-site codons, positioning an A-site tRNA should be more favorable at a codon immediately adjacent to the P-site codon. P-site tRNA repositioning in this case does not necessarily lead to frameshifting. Whether a ribosome will shift depends also on the efficiency of A-site tRNA repositioning.
The classical and important example of such frameshifting is tandem repositioning on the sequence X XXY YYZ. The P-site tRNA that forms the initial complex with XXY can also form a complex with XXX, and the A-site tRNA that recognizes YYZ can also pair with YYY (underlined). Jacks et al (1988a) suggested that this kind of frameshifting is caused by simultaneous slippage of two tRNAs. This has been interpreted by many to mean relative movement of two tRNAs as a single unit with respect to mRNA. If so, one would expect such movement in the plus direction by the same mechanism, for example, XXX YYY Z. However, no examples of this have been found so far, whereas the number of the examples of minus frameshifting on such sequences is increasing. The hypothesis of simultaneous tandem slippage does not explain the perceived directional selectivity.
In addition, simultaneous slippage of tRNAs presumes something that will coordinate their simultaneous movement with respect to mRNA (and perhaps interactions between these tRNAs). Otherwise, it is not clear what would simultaneously dissociate two codon:anticodon complexes. (Although 3′ stimulatory elements can greatly elevate the level of frameshifting, shifting can occur at a significant level in their absence.) Ironically, before structural data emerged, simultaneous slippage was invoked as an argument in support of the ‘R’-configuration of A- and P-site tRNAs inside the ribosomes, which would allow such interactions between codon–anticodon duplexes of two tRNAs in contrast to the ‘S’-configuration (Lim 1997). It is well known now that the mutual orientation of tRNAs is closer to the ‘S’-configuration (e.g., Stark et al. 1997; Cate et al. 1999). Therefore, contemporary structural data do not provide any information about possible interactions between A- and P-site tRNA that might be responsible for coordinated dissociation of these tRNAs from mRNA. Although it is possible that the ribosome is responsible for coordinated and simultaneous movement of two tRNAs, it is not obvious why such a feature would have evolved in the ribosome. Moreover, there is at present no data supporting such a hypothesis.
One mechanism of frameshifting versus two
Why do some cases of frameshifting involve P-site tRNA repositioning that takes place during recognition of a new tRNA in the A-site and other cases involve the repositioning of P-site tRNA at the stage when A-site tRNA is irreversibly incorporated? Is there anything special that determines at what elongation phase repositioning should take place?
The site of frameshifting required for synthesis of the HIV-1 GagPol precursor polyprotein is U UUU UUA. Translation studies in reticulocyte lysates show that there are two products. In ~70% of the synthesized polypeptide, the amino acid incorporated after the UUU-encoded Phe is Leu, and this is explicable by tandem repositioning. In the remaining 30% of the synthesized product, there is tandem Phe (Jacks et al. 1988b), and this corresponds to single P-site tRNA repositioning in the posttranslocated complex. (Studies in heterologous translation systems provided further evidence for two different products; Yelverton et al. 1994.) Horsfield et al. (1995) proposed that E- and P-site slippage take place in this case. If E-site slippage indeed occurs in this situation, than it should precede P-site repositioning. Thus, it is likely that repositioning of P-site tRNA in the HIV frameshifting cassette occurs in both elongation phases.
The discrimination between these two phases becomes even more problematic at a very shift-prone heptamer in E. coli, A AAA AAG. The explanation for the high frameshifting activity is dependent on E. coli lacking a tRNALys(3′-UUC-5′) that would exclusively recognize AAG. Uridine in the wobble position of tRNALys is modified to 5-methylaminomethyl-2-thiouridine (3′-UUmnm5s2U-5′), which pairs poorly with the codon’s third base, G (Sundaram et al. 2000 and references therein). Consequently, the affinity of this tRNA for AAA codons is significantly higher than for AAG. Based on its sequence analogy to X XXY YYZ, this kind of frameshifting has been considered to be caused by tandem tRNA slippage. This means that the A-site tRNA that recognizes AAG repositions to the −1-frame codon AAA. However, this is virtually impossible to verify experimentally, because tRNALys(3′-UUmnm5s2U-5′) normally recognizes both AAA and AAG codons. Alternatively, this frameshifting could be single −1 P-site frameshifting, where P-site tRNALys is repositioned in the minus direction and is stimulated by the competition of tRNALys for two different codons, AAA and AAG. It is also possible that the frameshifting occurs by both mechanisms: (1) During recognition of tRNA in the A-site, P-site tRNA is repositioned in the −1 direction with stimulation provided by the higher affinity of incoming A-site tRNALys for AAA than for AAG. (2) When A-site tRNA has been irreversibly incorporated into the ribosome, P-site tRNA is repositioned from the zero-frame AAA to the −1 frame AAA, and consequently the A-site tRNA is repositioned from AAG to AAA. The high activity of the frameshifting at A AAA AAG in E. coli may be due to the ability of the ribosome to shift at different phases of elongation.
In decoding the Bacillus subtilis cdd gene, efficient −1 frameshifting takes place at the sequence CGA AAG (Mejlhede et al. 1999). Mejlhede et al. (1999) speculated that this frameshifting is possible because of the weak base-pairing between inosine at position 34 of tRNAArg and the adenosine at the wobble position of the P-site codon. Inosine can detach from adenosine in the CGA codon, making it available for pairing in the −1 frame by tRNALys. Alternatively, this could happen because of the relative instability of the tRNA:codon complex in the P-site, with only minimal or no requirement for re-pairing in the P-site. As shown recently (Licznar et al. 2003), this frameshifting is efficient at other P-site codons. Some of these codons are decoded by tRNAs with nucleotides other than inosine at tRNA position 34. Independently of which of these two possibilities correspond to reality, they both can be described in terms of the scheme illustrated in Figure 1 ▶. In both cases, rearrangements within the P-site codon:anticodon complex precedes A-site tRNA repositioning. The only difference is that in the case of the “single A-site tRNA slippage hypothesis” (Mejlhede et al. 1999), P-site tRNA is required to rearrange the form of the complex with the same initial codon, but in a different manner.
As originally interpreted, one unusual case of noncognate tRNA-mediated −1 frameshifting is an exception to the model proposed. In E. coli cell-free protein synthesis with the normal balance of tRNAs, phage MS2 RNA directs the synthesis of two forms of its synthetase (viral-encoded replicase component). Synthesis of the longer form is due to tRNAThr(3′-UGG-5′) reading a CCA or CCG proline codon and causing a shift to the −1 frame by making the third codon base available for pairing with anticodon base 34 of the following tRNA (Bruce et al. 1986; Dayhuff et al. 1986). The original model for this frameshifting was that anticodon bases 34 and 35 pair with the second and first codon bases, respectively, by implication in the ribosomal A-site (Atkins et al. 1979). However, this explanation seems unlikely. When in the A-site, the third base of the proline codon is likely occluded by tRNAThr(3′-UGG-5′) or involved in pairing with anticodon base 33, so that it is unavailable for pairing with anticodon base 34 of the next incoming tRNA. Rearrangement of tRNAThr(3′-UGG-5′) in the P-site likely permits pairing by the next tRNA with the third codon base of the “proline” codon (Atkins et al. 2000). Thus, this case is unlikely to be an exception to the model proposed here.
All known cases of frameshifting can be explained within the framework described here. In all cases, rearrangements in the P-site precede slippage of A-site tRNA or its incorporation in a new frame. Rearrangements in the P-site include standard repositioning (but not necessarily completed), dissociation of initial complex, or conformational changes within tRNA. The identity of the incorporated amino acids, direction of the frameshifting, and the effect of tRNA concentrations depend on the elongation stage, where the P-site tRNA rearrangements occur.
There are several different stimulatory signals that can stimulate frameshifting through direct or indirect influence on one or several parameters (stability of P-site tRNAs in the ribosome, affinity of A-site ligands to a codon in the A-site, etc.) described in the present model. These stimulatory signals are highly diverse and include secondary RNA structures, such as pseudoknots (for review, see Brierley and Pennell 2001), stem–loops (Kollmus et al. 1994; Larsen et al. 1997) and RNA three-way junctions (Rettberg et al. 1999). Some mRNA elements act through interactions with the ribosome (Weiss et al. 1988; Larsen et al. 1994; Li et al. 2001; Stahl et al. 2001). Particular sequence elements flanking the “shifty” codon affect the affinity of incorporated tRNAs (Bertrand et al. 2002) or release factors (Tate et al. 1995). Among other elements are sequences that influence frameshifting (in this case translational bypass) via the encoded growing nascent peptide chain (Weiss et al. 1990b) and sequences located ~4000 nt downstream of the frameshifting site (Barry and Miller 2002). There are some cis-elements whose nature is unknown (Ivanov et al. 2000), and it is very likely that more new elements will emerge in the future.
CONCLUSIONS
The analysis presented results in several predictions:
For frameshifting, tRNA repositioning (or other rearrangements between codon and anticodon) in the P-site always paves the way to repositioning of A-site tRNA or to A-site tRNA binding in a new frame. Therefore, (a) out-of-frame binding of A-site tRNA does not occur without a preceding event in the P-site. (b) Tandem A- and P-site tRNA repositioning is unlikely to occur simultaneously. P-site tRNA rearrangement relative to mRNA should first provide a space for A-site tRNA repositioning.
The direction of frameshifting and its sensitivity to the intracellular concentration of tRNAs depends on the elongation phase, in which P-site tRNA repositions (or rearranges with mRNA in another manner).
The correct reading frame is controlled by an interplay of the ribosome, tRNAs and mRNAs. Ribosome and tRNAs maintain stable tRNA–mRNA interactions in the P-site. According to the kinetic model in the present work, proper cellular tRNA concentrations in combination with optimal codon usage in translated mRNAs are necessary for reading frame maintenance. Moreover, the effects of the relative concentrations of tRNAs competing for the initial, or new frame, A-site codons are not equivalent according to the kinetic model. Alterations in tRNA concentration of a given organism or expression of heterologous mRNA (with a very different codon bias) should result in elevation of frameshift errors.
Acknowledgments
We are grateful to Olivier Fayet, Olga Gurvich, Michael Howard, Ivaylo Ivanov, Mikhail Khoudiakov, Knud Nierhaus, Marina Rodnina, Petr Sergiev, Andrey Sivachenko, Daniel Wilson, and particularly to Robert Thompson and Mike Yarus for discussion of the manuscript. This work benefited enormously from the stimulatory effect of others in the field, especially Phil Farabaugh, who are driving the evolution of ideas about the role of P-site tRNAs in framing. This work was supported by NIH (grant R01-GM61200 to R.F.G. and R01-GM48152 to J.F.A) and DOE (Grant DE-FG03-O1ER63132 to R.F.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5122604.
REFERENCES
- Atkins, J.F., Gesteland, R.F., Reid, B.R., and Anderson, C.W. 1979. Normal tRNAs promote ribosomal frameshifting. Cell 18: 1119–1131. [DOI] [PubMed] [Google Scholar]
- Atkins, J.F., Herr, A.J., Massire, C., O’Connor, M., Ivanov, I., and Gesteland, R.F. 2000. Poking a hole in the sanctity of the triplet code: Inferences for framing. In The ribosome: Structure, function, antibiotics and cellular interactions (eds. R.A. Garrett et al.), pp. 369–383. ASM Press, Washington, DC.
- Baranov, P.V., Gesteland, R.F., and Atkins, J.F. 2002a. Recoding: Translational bifurcations in gene expression. Gene 286: 187–201. [DOI] [PubMed] [Google Scholar]
- ———. 2002b. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 3: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov, P.V., Gurvich, O.L., Hammer, A.W., Gesteland, R.F., and Atkins, J.F. 2003. Recode 2003. Nucleic Acids Res. 31: 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, J.K. and Miller, W.A. 2002. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. 99: 11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt, M.F. and Farabaugh, P.J. 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62: 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, C., Prére, M.F., Gesteland, R.F., Atkins, J.F., and Fayet, O. 2002. Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA 8: 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkowa, A.L. and Walker, J.R. 1990. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 18: 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I. and Pennell, S. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 66: 233–248. [DOI] [PubMed] [Google Scholar]
- Brierley, I., Rolley, N.J., Jenner, A.J., and Inglis, S.C. 1991. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 220: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, A.G., Atkins, J.F., and Gesteland, R.F. 1986. tRNA anticodon replacement experiments show that ribosomal frameshifting can be caused by doublet decoding. Proc. Natl. Acad. Sci. 83: 5062–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N., and Noller, H.F. 1999. X-Ray crystal structures of 70S ribosome functional complexes. Science 285: 2095–2104. [DOI] [PubMed] [Google Scholar]
- Craigen, W.J. and Caskey, C.T. 1986. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322: 273–275. [DOI] [PubMed] [Google Scholar]
- Curran, J.F. 1993. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 21: 1837–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhuff, T.J., Atkins, J.F., and Gesteland, R.F. 1986. Characterization of ribosomal frameshift events by protein sequence analysis. J. Biol. Chem. 261: 7491–7500. [PubMed] [Google Scholar]
- Farabaugh, P.J. and Björk, G.R. 1999. How translational accuracy influences reading frame maintenance. EMBO J. 18: 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P.J., Zhao, H., and Vimaladithan, A. 1993. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: Frameshifting without tRNA slippage. Cell 74: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower, A.M. and McHenry, C.S. 1990. The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. 87: 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant, J., Lindsley, D., and Massucci, J. 2000. The unbearable lightness of peptidyl-tRNA. The ribosome: Structure, function, antibiotics and cellular interactions (eds. R.A. Garrett et al.), pp. 385–396. ASM Press, Washington, DC.
- Hansen, T.M., Baranov, P.V., Ivanov, I.P., Gesteland, R.F., and Atkins, J.F. 2003. Maintenance of the correct open reading frame by the ribosome. EMBO Rep. 4: 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger, J.W., Meskauskas, A., and Dinman, J.D. 2002. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 27: 448–454. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Atkins, J.F., and Gesteland, R.F. 1999. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J. 18: 2886–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield, J.A., Wilson, D.N., Mannering, S.A., Adamski, F.M., and Tate, W.P. 1995. Prokaryotic ribosomes recode the HIV-1 gag-pol-1 frameshift sequence by an E/P site post-translocation simultaneous slippage mechanism. Nucleic Acids Res. 11: 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, I.P., Gesteland, R.F., and Atkins, J.F. 2000. Antizyme expression: A subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 28: 3185–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, I.P., Gurvich, O.L., Gesteland, R.F., and Atkins, J.F. 2003. Recoding: Site- or mRNA-specific alteration of genetic readout utilized for gene expression. In Translation mechanisms (eds. J. Lapointe and L. Braker-Gingras), pp. 354–369. Landes Bioscience, Austin, TX.
- Kollmus, H., Honigman, A., Panet, A., and Hauser, H. 1994. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T-cell leukemia virus type II in vivo. J. Virol. 68: 6087–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolor, K., Lindsley, D., and Gallant, J.A. 1993. On the role of the P-site in leftward ribosome frameshifting at a hungry codon. J. Mol. Biol. 230: 1–5. [DOI] [PubMed] [Google Scholar]
- Jacks, T., Madhani, H.D., Masiarz, F.R., and Varmus, H.E. 1988a. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks, T., Power, M.D., Masiarz, F.R., Luciw, P.A., Barr, P.J., and Varmus, H.E. 1988b. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331: 280–283. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., Pande, S., Faiola, B., Moore, D.P., Boeke, J.D., Farabaugh, P.J., Strathern, J.N., Nakamura, Y., and Garfinkel, D.J. 1993. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, B., Wills, N.M., Gesteland, R.F., and Atkins, J.F. 1994. rRNA–mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176: 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, B., Gesteland, R.F., and Atkins, J.F. 1997. Structural probing and mutagenic analysis of the stem–loop required for Escherichia coli dnaX ribosomal frameshifting: Programmed efficiency of 50%. J. Mol. Biol. 271: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Stahl, G., and Farabaugh, P.J. 2001. Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA 7: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznar, P., Mejlhede, N., Prére, M-F., Wills, N., Gesteland, R.F., Atkins, J.F., and Fayet, O. 2003. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 22: 4770–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, V.I. 1997. Analysis of interactions between the codon–anticodon duplexes within the ribosome: Their role in translation. J. Mol. Biol. 266: 877–890. [DOI] [PubMed] [Google Scholar]
- Matsufuji, S., Matsufuji, T., Miyazaki, Y., Murakami, Y., Atkins, J.F., and Gesteland, R.F., and Hayashi, S. 1995. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlhede, N., Atkins, J.F., and Neuhard, J. 1999. Ribosomal −1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J. Bacteriol. 181: 2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande, S., Vimaladithan, A., Zhao, H., and Farabaugh, P.J. 1995. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol. Cell. Biol. 15: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani. R., Pavesi, A., and Ottonello, S. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268: 322–330. [DOI] [PubMed] [Google Scholar]
- Potapov, A.P., Triana-Alonso, F.J., and Nierhaus, K.H. 1995. Ribosomal decoding processes at codons in the A or P sites depend differently on 2′-OH groups. J. Biol. Chem. 270: 17680–17684. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell 108: 557–572. [DOI] [PubMed] [Google Scholar]
- Rettberg, C.C., Prére, M.F., Gesteland, R.F., Atkins, J.F., and Fayet, O. 1999. A three-way junction and constituent stem–loops as the stimulator for programmed −1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 12: 1365–1378. [DOI] [PubMed] [Google Scholar]
- Stahl, G., Ben Salem, S., Li, Z., McCarty, G., Raman, A., Shah, M., and Farabaugh, P.J. 2001. Programmed +1 translational frameshifting in the yeast Saccharomyces cerevisiae results from disruption of translational error correction. Cold Spring Harb. Symp. Quant. Biol. 66: 249–258. [DOI] [PubMed] [Google Scholar]
- Stahl, G., McCarty, G.P., and Farabaugh, P.J. 2002. Ribosome structure: Revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 27: 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, H., Orlova, E.V., Rinke-Appel, J., Junke, N., Mueller, F., Rodnina, M., Wintermeyer, W., Brimacombe, R., and van Heel, M. 1997. Arrangement of tRNAs in pre- and posttranslocational ribosomes revealed by electron cryomicroscopy. Cell 88: 19–28. [DOI] [PubMed] [Google Scholar]
- Sundaram, M., Durant, P.C., and Davis, D.R. 2000. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39: 12575–12584. [DOI] [PubMed] [Google Scholar]
- Sundararajan, A., Michaud, W.A., Qian, Q., Stahl, G., and Farabaugh, P.J. 1999. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell 4: 1005–1015. [DOI] [PubMed] [Google Scholar]
- Tate, W.P., Poole, E.S., Horsfield, J.A., Mannering, S.A., Brown, C.M., Moffat, J.G., Dalphin, M.E., McCaughan, K.K., Major, L.L., and Wilson, D.N. 1995. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem. Cell Biol. 73: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi, Z. and Brown, P.O. 1992. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNALys and an AAG lysine codon. Genes & Dev. 6: 511–519. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi, Z. and Kornberg, A. 1990. Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. 87: 2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimaladithan, A. and Farabaugh, P.J. 1994. Special peptidyl-tRNA molecules can promote translational frameshifting without slippage. Mol. Cell. Biol. 14: 8107–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, R. and Gallant, J. 1983. Mechanism of ribosome frameshifting during translation of the genetic code. Nature 302: 389–393. [DOI] [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Dahlberg, A.E., Atkins, J.F., and Gesteland, R.F. 1988. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 7: 1503–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Shuh, M., Atkins, J.F., and Gesteland, R.F. 1989. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1: 159–169. [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Atkins, J.F., and Gesteland, R.F. 1990a. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 39: 159–183. [DOI] [PubMed] [Google Scholar]
- Weiss, R.B., Huang, W.M., and Dunn, D.M. 1990b. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell 62: 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelverton, E., Lindsley, D., Yamauchi, P., and Gallant, J.A. 1994. The function of a ribosomal frameshift signal from human immunodeficiency virus-1 in Escherichia coli. Mol. Microbiol. 11: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]