Abstract
The kink-turn (K-turn) is a new motif in RNA structure that was identified by examination of the crystal structures of the ribosome. We examined the structural and dynamic properties of this element in free solution. The K-turn RNA exists in a dynamic equilibrium between a tightly kinked conformation and a more open structure similar to a simple bulge bend. The highly kinked form is stabilized by the noncooperative binding of metal ions, but a significant population of the less-kinked form is present even in the presence of relatively high concentrations of divalent metal ions. The conformation of the tightly kinked population is in excellent agreement with that of the K-turn structures observed in the ribosome by crystallography. The end-to-end FRET efficiency of this species agrees closely with that of the ribosomal K-turn, and the direction of the bend measured by comparative gel electrophoresis also corresponds very well. These results show that the tightly kinked conformation of the K-turn requires stabilization by other factors, possibly by protein binding, for example. The K-turn is therefore unlikely to be of itself a primary organizing feature in RNA.
Keywords: K-turn motif, RNA structure, RNA folding, metal ions, FRET
INTRODUCTION
Recent success in solving RNA structures has provided a large database of conformational information, from which some general architectural principles can be extracted (Leontis and Westhof 2003). These include a variety of ribozyme structures (Pley et al. 1994; Scott et al. 1995; Cate et al. 1996a; Ferré-d’Amaré et al. 1998; Golden et al. 1998; Rupert and Ferré-d’Amaré 2001; Krasilnikov et al. 2003), and most notably the ribosomal subunits (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000). A number of new structural motifs have been identified by examination of these molecules, including terminal loops, the A-minor motifs (Cate et al. 1996b; Nissen et al. 2001), the kink-turn (K-turn; Klein et al. 2001), and the loop-E motif (Correll et al. 1997).
Most of these structural motifs have been identified in crystal structures, usually as part of a much larger RNA structure and frequently in the presence of bound proteins. As a consequence, there is little information about the solution properties of these elements in most cases, such as their capacity for independent existence, their folding properties, and the requirement for other factors such as metal ions. There is therefore a need to take these features in isolation, and to study their solution properties by spectroscopic and other means.
The K-turn motif comprises an internal loop in duplex RNA that introduces a very tight kink into the helical axis (Klein et al. 2001). It occurs six times in the 23S rRNA of Haloarcula marismortui (Ban et al. 2000), and twice in the 16S rRNA of Thermus thermophilus (Schluenzen et al. 2000). The overall geometries of these RNA structures are closely similar; for example, the six K-turns of the 50S ribosomal subunit superimpose with an rmsd of 1.7 Å. The general sequence features of the K-turn are a three-nucleotide (3-nt) bulge flanked by a normal duplex on its 5′ side (termed the C-stem) and two or three G•A mispairs on the 3′ side (termed the NC-stem). In addition to the examples found in the ribosome, RNA sequences conforming to a consensus K-turn have been noted in a variety of nonribosomal RNA species (Klein et al. 2001) such as the U4 snRNA (Vidovic et al. 2000), and the motif is likely to be of general importance. Most of the examples of the K-turn in the ribosome are involved in protein binding, and thus the K-turn is an important protein recognition element.
The K-turn is typified by Kt-7, found in helix 7 of the 23S rRNA (Klein et al. 2001). The sequence and structure of Kt-7 as it exists in the ribosomal subunit is shown in Figure 1 ▶. Globally, the RNA is strongly bent by the presence of the Kt-7 element—the axes of the C and NC-stems subtend an included angle of 60°. There are three sheared G•A mismatches on the 3′-side of a bulge comprising the sequence GAA. G94 (at the 5′ end of the bulge) is stacked on G97 at the end of the NC stem, and A95 is stacked onto the end of the C stem, with an angle of ~ 90° between their two planes. A96 points away from the RNA, making no further interactions. The minor groove edges of the adenines of the G•A mismatches (A80, A98, and A99) are in intimate contact with the minor groove of the C-stem helix, forming a type I A-minor interaction (Nissen et al. 2001) that appears to be the principle stabilization of the sharp bend of the K-turn.
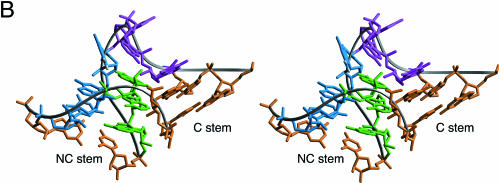
FIGURE 1.
The sequence and structure of a kink-turn motif. (A) The consensus sequence of the K-turn motif, and the sequence of the Kt-7 element found in the 23S rRNA. The conventional numbering is indicated for the Kt-7 sequence. (B) Parallel-eye stereoscopic view of the high-resolution structure of the Kt-7 motif from the crystal structure of the 50S ribosomal subunit (Klein et al. 2001). The color coding corresponds to the sequence shown in (A). The two helical stems are called C and NC. The two G•A mismatches proximal to the bulged nucleotides (purple) in the NC stem orient their adenine bases (green) into the minor groove of the C stem, generating a very tight kink with an included angle of 60°.
In this work we studied the global structure of the prototypical K-turn Kt-7 in an isolated RNA duplex in solution. We found that the structure is profoundly bent in the presence of magnesium ions, and in good agreement with its conformation in the ribosome. Perhaps surprisingly however, it transpires that the structure is significantly dynamic in solution, in a two-state equilibrium. In the absence of added metal ions, the Kt-7 RNA behaves much more like a simple three-purine base bulge, but some population of this conformation is present under all conditions explored. The transition between the two conformations involves a major change in both bend angle and direction, and is induced by the noncooperative binding of metal ions.
RESULTS
In these studies we analyzed the global conformation and transitions of RNA duplexes containing a central K-turn sequence. We chose to study the Kt-7 sequence, because this is closest to the consensus sequence for the various K-turn sequences found in the ribosomal 50S and 30S subunits (Fig. 1 ▶; Klein et al. 2001). We also made derivative sequences in which the three G•A mismatches on the 3′ side of the unpaired nucleotides are converted into Watson-Crick base pairs, effectively changing the K-turn into a simple 3-nt bulge.
Highly retarded electrophoretic mobility of RNA containing a K-turn
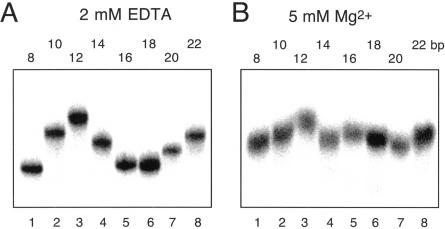
Gel electrophoresis is a sensitive means for examining deformations in double-helical RNA (Bhattacharyya et al. 1990; Tang and Draper 1990; Luebke and Tinoco 1996) or DNA (Marini et al. 1982; Diekmann and Wang 1985; Bhattacharyya and Lilley 1989; Hsieh and Griffith 1989) leading to a change in global structure. If the solution structure of the K-turn is the same as that observed in the crystal (Klein et al. 2001), RNA containing such a feature would be expected to exhibit severely retarded electrophoretic mobility in a polyacrylamide gel. We therefore prepared a series of duplex RNA molecules of 120 bp, containing a central sequence that should result in a bend in the helical axis (Fig. 2A ▶). We used a series of oligoadenine (An) bulges, which generate a kinking of the axis, and compared the mobility of these species with an equivalent duplex where the An bulge is replaced by the Kt-7 sequence. The relative mobilities were compared in a 12% polyacrylamide gel, electrophoresed at 20°C in 90 mM Tris-borate, pH 8.3 (TB buffer) containing 5 mM magnesium ions. The RNA was radioactively [α′-32P]-labeled, and the mobilities of the different species were compared by phosphorimaging (Fig. 2B ▶). The series of An bulged RNA species exhibit the expected progressive reduction of mobility with bulge size (n), which is known to result from an axial kinking that becomes greater as the number of unpaired bases increases (Gohlke et al. 1994; Zacharias and Hagerman 1995). The RNA containing the Kt-7 K-turn has a markedly impaired mobility (Fig. 2B ▶, lane 1), similar to that of the A7 bulged RNA. This is clearly consistent with a major deformation of the helix axis, in qualitative agreement with the structure of the RNA observed in the crystal of the 50S ribosomal subunit (Klein et al. 2001). The equivalent RNA containing the converted K-turn sequence (i.e., a simple three-base bulge) was also electrophoresed on the same gel (Fig. 2B ▶, lane 2). It has a much greater electrophoretic mobility than the complete K-turn, similar to that of the A3 bulge, as expected. Clearly some aspect of the G•A mismatches is essential for generating the severe kinking of the complete K-turn sequence; this is explored further below.
FIGURE 2.
Anomalous electrophoretic migration of K-turn RNA is dependent on the presence of magnesium ions. Comparison of electrophoretic mobility of 120-bp RNA duplexes containing a central Kt-7 sequence or variant with duplexes kinked by virtue of a central oligoadenine bulge. (A) Central sequences of the RNA species employed. Each is a 120-bp duplex with either the Kt-7 sequence (K, top), a derivative in which the three G•A mismatches have been each converted into a Watson-Crick base pair (K′, middle), or an oligoadenine bulge (An, bottom). (B,C) Electrophoretic mobilities in the presence of 5 mM magnesium ions (B) or 2 mM EDTA (C). Note that the Kt-7 sequence migrates very slowly in magnesium ions, indicating a highly kinked structure, comparable to the A7 bulged species. By comparison, the same species migrates much faster in EDTA, similar to an A3 bulged duplex. Lane 1, Kt-7; 2, K′ sequence, lacking the G•A mismatches; 3–8, oligoadenine An bulge-containing species, where n = 0 (i.e., perfect duplex), 1, 3, 4, 5, or 7, respectively.
The same RNA samples were applied to a polyacrylamide gel and electrophoresed in TB buffer containing 2 mM EDTA in place of the divalent metal ions (Fig. 2C ▶). Once again the An bulged-RNA species migrate progressively more slowly as the bulge size increases. However, the RNA containing the Kt-7 sequence has a considerably greater relative mobility under these conditions, and is now similar to the A3 bulged RNA, and to the converted K-turn sequence that has lost the three G•A mismatches.
The K-turn has a defined direction
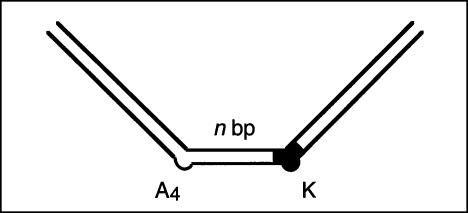
Axial kinks resulting from base bulges in RNA have a defined direction in solution. This can be demonstrated by constructing a series of duplexes containing two such bulges (Bhattacharyya et al. 1990; Tang and Draper 1990; Riordan et al. 1992), in which the spacing between bulges is varied systematically while keeping the overall length of the duplex constant. Electrophoresis of the series results in a sinusoidal modulation of mobility as the dihedral angle between the outer arms varies between 0° (greatest retardation) and 180° (maximum mobility). When two identical bends are used, the species of minimum mobility will be those where the two bends are separated by an exact helical turn (zero phase difference). However, if the two bending loci have different directions relative to the RNA structure, this will result in a phase shift that is directly related to the difference in the directions of the two bends. When the direction of one bend is known, the phase shift can be used to determine the direction of the unknown bend. Alternatively, the relative bending direction of two bends can be determined by comparison with a constant bend, such as that generated by an An bulge.
We performed this experiment using a series of 88-nt RNA duplexes containing an A4 bulge and the K-turn sequence. The two bent sequences were symmetrically located about the center of the RNA, and separated by a spacer region between 8 and 22 bp-long. The radioactively [α-32P]-labeled species were electrophoresed at 20°C in a 10% polyacrylamide gel in TB buffer containing either 2 mM EDTA or 5 mM magnesium ions. Phosphorimages of the gels are shown in Figure 3 ▶.
FIGURE 3.
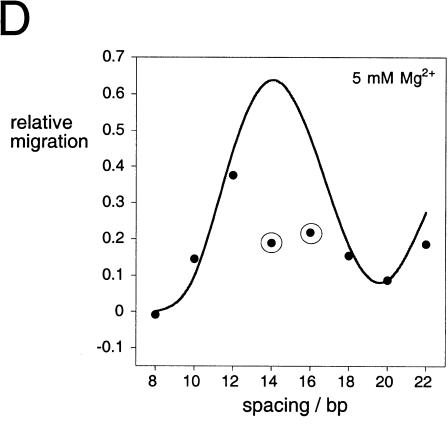
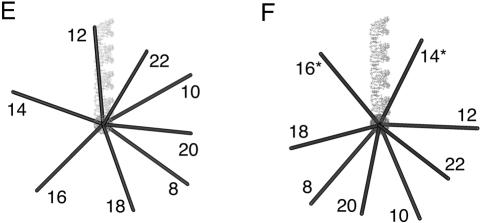
The direction of bending at the K-turn locus depends on the presence of metal ions. A series of 88-nt RNA duplexes containing an A4 bulge and the K-turn sequence symmetrically located about the center and separated by a spacer region of n bp (n = 8–22) were analyzed. (A,B) The radioactively [α′-32P]-labeled RNA was electrophoresed at 20°C in a 10% polyacrylamide gel in the presence of either 2 mM EDTA (A) or 5 mM magnesium ions (B). The RNA was visualized by phosphorimaging, and the migration of each species measured. (C,D) The experimentally determined relative inverse migrations in 2 mM EDTA or 5 mM magnesium ions are plotted as a function of spacing in C and D, respectively. The lines are smooth interpolations of the model derived migration factors that best fit the experimental data. (E,F) The data were analyzed using molecular graphics models as described in the text. The long arm on the A4 bulge side is shown as an RNA helix, and the various positions of the long helix on the K-turn side are shown as bars labeled with their corresponding spacer lengths. The relative rotational settings of these K-turn-side helices was fixed and the phase angle then varied while measuring end-to-end distances until the best fit between the calculated Lumpkin-Zimm migration factors (Equation 1) and the experimentally measured mobilities was obtained. The directions shown are those for the best fits in 2 mM EDTA (E) or 5 mM magnesium ions (F). For the data obtained in 5 mM magnesium ions (F), the anomalous points (14- and 16-bp spacers, ringed in D) are marked with asterisks; in these species the two long arms are closest to coplanarity, so the reflex kink angle of the K-turn could result in a clash of long arms.
In the presence of EDTA, the species migrate with the sinusoidal pattern of mobilities expected for two small bulges (Fig. 3A ▶). We can investigate the direction of bending using molecular graphics to construct a model of the series of species analyzed in these experiments. We start by placing two bends having the same direction (i.e., no phase shift) into an 88-bp duplex, and then construct the species with the same bend–bend spacing that we use experimentally. In this way we generate a series of models with a successive relative rotation of the outer arms, from which the end-to-end distance can be measured. This is then repeated for a number of new series, where phase shifts are introduced for each new set. The end-to-end distances (dee) are used to calculate Lumpkin-Zimm electrophoretic migration factors (fLZ; Lumpkin and Zimm 1982) according to:
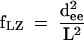 |
(1) |
where L is the contour length of the molecule. These are compared with the experimental data until the best fit is obtained (Fig. 3E ▶). A comparison between the experiment and the best fit is shown in Figure 3C ▶, giving a phase shift of < 15°, equivalent to ~0.5 bp. Thus the bend direction of the K-turn in the absence of magnesium ions is very similar to that of the An bulge, suggesting that under these conditions the element behaves as a simple three-purine bulge.
The pattern of mobilities in the presence of 5 mM magnesium ions deviated significantly from the simple sinusoidal dependence in EDTA (Fig. 3B ▶). The data can be explored using the same molecular graphics modeling that was applied to the data obtained in the absence of metal ions (Fig. 3F ▶). Using this approach we obtained the phase angle giving the best fit, indicating that we now require a phase shift of ~90° to get satisfactory agreement with the experimental data (Fig. 3D ▶). This corresponds to a difference in the direction of the bend of close to 75° in the presence and absence of magnesium ions (Fig. 3 ▶, cf. E and F). We noted however that using this fit, the data points corresponding to 14- and 16-bp spacing deviated significantly from the calculated fit. We constructed a further model using the direction for the A4 bulge determined in earlier related experiments (, in prep.), and the structure of the K-turn motif (Kt-7) found in the ribosome (Klein et al. 2001). This gave excellent agreement with the phase determined above, and also provided an explanation for the two anomalous data points. The bend angle of the K-turn is so severe that the 14 and 16 bp-spacing constructs result in the close approach of the outer arms, and therefore a distortion of the structure (or destabilization relative to the less-kinked structure) would be likely to occur in order to relieve the unfavorable interaction. There is no similar anomaly in the absence of magnesium ions because the bend angle is smaller, directing the long arm in the opposite direction and thus avoiding any steric clash. Other experiments reported in this paper indicate that even at 5 mM magnesium ion concentrations, the kinked RNA will be in equilibrium with a less bent conformation. Thus the phase angles measured by this technique will be smaller than would be expected for a fully-folded K-turn structure.
These data show that the K-turn has a defined direction in solution that is in good agreement with that found in the crystal structure of the 50S ribosomal subunit. The change in direction in the presence and absence of magnesium ions is a further demonstration of the importance of metal ion binding in determining the structure of the K-turn.
Ion-induced folding of the K-turn analyzed by fluorescence resonance energy transfer
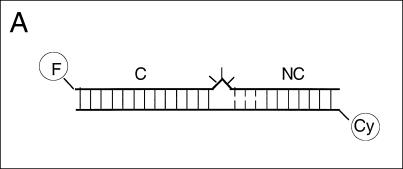
The electrophoretic results clearly indicate that the structure of the K-turn is strongly influenced by magnesium ions. This was further investigated using steady-state fluorescence resonance energy transfer (FRET). We constructed an RNA duplex of 20 bp, containing a Kt-7 sequence, with fluorescein (FRET donor) and Cy-3 (FRET acceptor) attached to the 5′-termini (Fig. 4A ▶). Efficiency of energy transfer (EFRET) depends on the inverse sixth power of the separation (R) between the two fluorophores, according to (Förster 1948):
FIGURE 4.
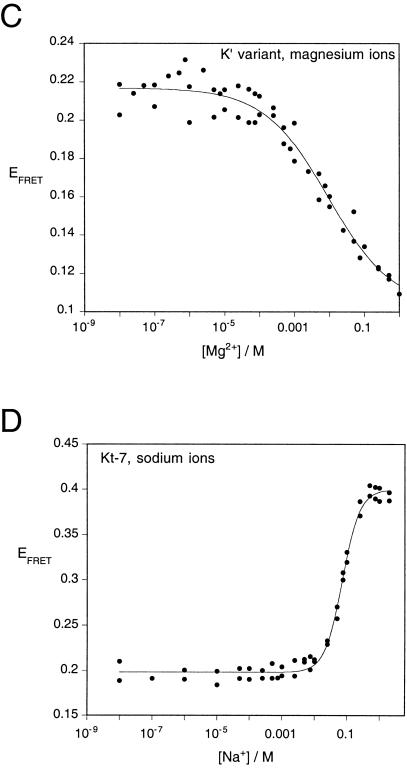
An ion-induced conformational transition in the K-turn structure followed by FRET. (A) The change in structure was followed as a change in FRET efficiency between fluorescein (F) and cyanine 3 (Cy) fluorophores attached to the 5′-termini of an RNA duplex containing a Kt-7 motif. The C arm was 13 bp and the NC arm 7 bp (not including the three G•A mismatches). An equivalent species in which the three mismatches were converted to Watson-Crick base pairs was also studied. FRET efficiency measured in the steady-state for these species as a function of ionic concentration, and in each case the change in EFRET was fitted using a two-state model of the transition (line). (B) EFRET measured for the Kt-7 structure as a function of magnesium ion concentration. (C) EFRET measured for the converted K′ structure as a function of magnesium ion concentration. In contrast to the K-turn, this species undergoes a decrease in EFRET with increased magnesium ion concentration. (D) EFRET measured for the Kt-7 structure as a function of sodium ion concentration.
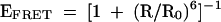 |
(2) |
where R0 is the Förster length, which is 56 Å for this donor-acceptor pair (Norman et al. 2000). FRET efficiency was measured in the steady-state, using the acceptor normalization method (Clegg 1992), as a function of the concentration of magnesium ions (Fig. 4B ▶). EFRET can be seen to increase significantly as the ion concentration is increased, showing that magnesium ions induce the formation of a more tightly kinked structure with a consequently shorter end-to-end distance. The data have been fitted to a simple two-state model in which the tightly kinked structure is stabilized relative to the more extended structure by the binding of x magnesium ions with an apparent association constant KMg. According to that model, the proportion (α) of tightly kinked RNA is given by:
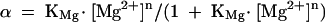 |
(3) |
where n is a Hill coefficient that provides a measure of the cooperativity of ion binding. The two-state model provides a good fit to the experimental data (R = 0.99), with a value of n = 0.84 ± 0.05. This indicates that the structure is stabilized by the noncooperative binding of magnesium ions. The Mg2+ ion concentration at which the transition is 50% complete ([Mg2+]1/2 = (1/KMg)1/n) is 1 mM.
We also performed the same FRET analysis on an equivalent Kt-7 sequence with converted G•A mismatches (Fig. 4C ▶). In the absence of magnesium ions, the two RNA species have closely similar FRET efficiencies. However, in contrast to the complete K-turn sequence, addition of magnesium ions to the converted sequence resulted in a small decrease in FRET efficiency, with a relatively high half-magnesium concentration of [Mg2+]1/2 = 12 mM.
Monovalent metal ions can induce an apparently very similar transition in the Kt-7 sequence. We performed an equivalent FRET analysis on the donor-acceptor labeled RNA as a function of sodium ion concentration (Fig. 4D ▶). Addition of sodium ions achieved the same end-point in FRET efficiency, indicating the formation of a similar global conformation. The data fitted the two-state model (R = 0.996), showing that the transition was induced by a more cooperative binding of sodium ions (n = 1.6 ± 0.1), with a half-sodium concentration of [Na+]1/2 = 72 mM.
Analysis of the solution structure and dynamics of the K-turn using time-resolved FRET
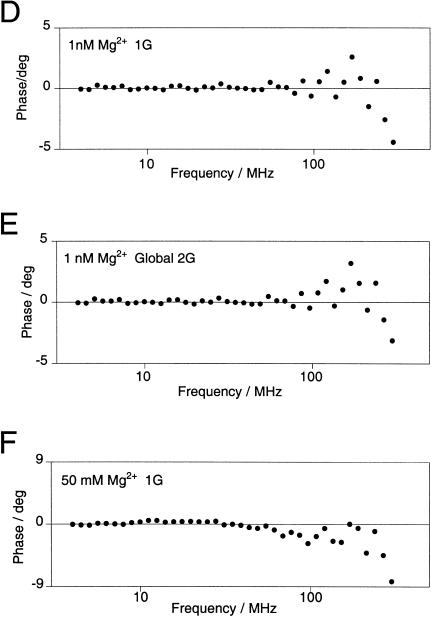
In principle, if the RNA has some dynamic character the donor-acceptor (D-A) distances could be better described by distance distributions with significant width. The two-state behavior of the K-turn RNA also suggests that there may be more than one conformation populated at equilibrium. Steady-state FRET data provide no information on possible distributions of D-A distances or multiple populations of distances, and we therefore employed time-resolved measurements of energy transfer to look for multiple distance distributions that could contribute to the observed average FRET. We made frequency-domain fluorescence lifetime measurements on the fluorophore-labeled K-turn RNA, analyzing the shortening of the excited state lifetime of the fluorescein donor arising from energy transfer to the Cy3 acceptor.
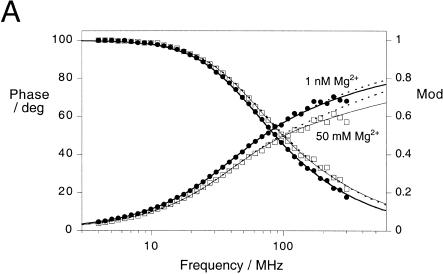
We measured the phase shift and demodulation of fluorescein emission as a function of the frequency of modulation of the excitation intensity at low (1 nM) and high (50 mM) concentrations of magnesium ions (Fig. 5A ▶). These data were fitted to a variety of models for the D-A distances. Inclusion of two distance populations described both data sets better than a single distance, especially for the high-salt conditions, with a better distribution of phase residuals and a halving of χ2 values. In the presence of 50 mM Mg2+, the second distance was similar to that expected for an extended conformation of the RNA. Neither the high nor the low magnesium ion concentration data provided enough information to determine two distances and half-widths, and the relative populations of each. Fitted together, however, it was possible to determine all six parameters. The best fits were obtained by globally fitting low- and high-magnesium data for two Gaussian distribution of interfluorophore distances with different centers. Although the two distance distributions were present under both conditions, the relative populations of the longer and shorter D-A distance varied with the magnesium ion concentration.
FIGURE 5.
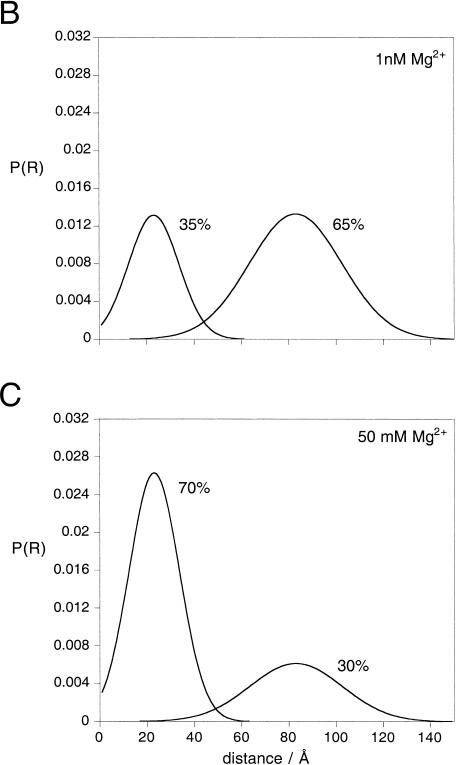
Analysis by time-resolved FRET shows that the Kt-7 RNA contains two populations of conformations. Donor-acceptor (D-A) distance distributions were analyzed using the same fluorescein-Cy3 end-labeled species analyzed by steady-state FRET (Fig. 4 ▶), using frequency-domain phase fluorometry. (A) Plots of phase shift and demodulation of the fluorescein emission as a function of the presence of 1 nM (•) and 50 mM (□) magnesium ions as a function of the modulation frequency. The best fits to the experimental data using one or two Gaussian distributions of D-A lengths are shown by the broken and solid lines, respectively. Detailed analysis of the distribution of residuals and Bootstrap analysis shows that the fits using two D-A length distributions are superior. Both sets of data were fitted globally to give the distributions shown in B and C, with mean D-A distances of 23 and 83 Å. (B) Two-Gaussian D-A length distributions [P(R) as a function of distance R] calculated from the fit to the data obtained in 1 nM magnesium ions. The major species (65%) corresponds to the longer donor-acceptor distance. The integrated areas under the curves total 100%. (C) Two-Gaussian D-A length distributions calculated from the fit to the data obtained in 50 mM magnesium ions. The major species (70%) now corresponds to the shorter D-A distance. (D,E) Comparison of the phase residuals when one (D) or two (E) Gaussian distributions of distance are applied to the data acquired at low Mg2+ concentration. The resulting χ2 values are 23.2 and 20.6, respectively. (F,G) Phase residuals when one (F) or two (G) Gaussian distributions of distance are applied to the data acquired at high Mg2+ concentration. The resulting χ2 values are 62.6 and 36.8, respectively. (H) Bootstrap error analysis of the probable distance and half-width for the shorter distance shows the scatter of values about the best fit from 800 cycles.
The results for the Kt-7 sequence in 50 mM magnesium ions are shown in Figure 5C ▶. The major species present in solution at 50 mM magnesium ions gives a relatively narrow distribution of D-A distances around a mean of 23 (21–32) Å with a half-width of 25 (16–39) Å. A minor species is present with a longer and wider distribution of D-A distances, around a mean of 83 (81–83) Å with a half-width of 46 (34–61) Å. Data obtained at lower magnesium ion concentrations globally fitted to the same two distributions, whereupon the major species was now the longer distance, that is, 65% (58%–89%) compared to 30% (26%–49%) in 50 mM magnesium ions (Fig. 5B ▶). The mean D-A distances suggest that the major species present in high magnesium ion concentration is the strongly kinked conformation, as observed in the crystal structure of the ribosome (see Discussion), whereas the minor species is more extended. The width of the Gaussian distribution of shorter lengths is similar to that obtained for a simple duplex species (Melcher et al. 2003), indicating that it arises largely from variation in the position of the fluorescein relative to the RNA, due to the flexible linker by which it is tethered. This suggests that the kinked form of the K-turn RNA does not have great intrinsic flexibility, that is, that the kink-turn is a rather tight, inflexible structure.
Effect of sequence changes in the G•A mismatches on the structure of the K-turn
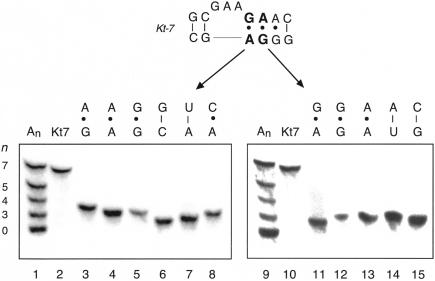
Inspection of the structure of Kt-7 and other K-turns in the ribosome indicates that the G•A mismatches of the NC stem should be very important in making the A-minor interaction that stabilizes the sharp kinking. We have shown that if these are collectively replaced by Watson-Crick base pairs, the resulting converted RNA migrates in polyacrylamide very similarly to an A3 bulge. We therefore explored in further detail the sequence requirement in this region of the K-turn, making a series of single-nucleotide substitutions within the two G•A mismatches adjacent to the bulge, and analyzing the kinking of the 120-bp RNA by gel electrophoresis in the presence of 5 mM magnesium ions as before (Fig. 6 ▶).
FIGURE 6.
The anomalous electrophoretic mobility of RNA containing the Kt-7 sequence is highly sensitive to the sequence of the G•A mismatches. A series of 120-bp duplexes containing a central Kt-7 (equivalent to that used in Fig. 2 ▶) and sequence variants were analyzed by polyacrylamide gel electrophoresis in the presence of 5 mM magnesium ions. Single-nucleotide substitutions were made at the G•A mismatch adjacent to the bulged GAA sequence (lanes 3–8) or the next A•G mismatch (lanes 11–15). Mobilities were compared with a mixture of species with An bulges of size n = 0 to 7 (lanes 1,9) and the natural Kt-7 sequence (lanes 2,10). The resulting base pair or mismatch created is shown above each lane. Note that all single-nucleotide changes investigated at these two positions led to loss of the anomalous mobility of the Kt-7 sequence.
It is immediately apparent that the two mismatches are highly intolerant of nucleotide substitution. All such changes that we made led to a loss of the highly retarded electrophoretic mobility. The largest effect arises from the replacement of the bulge-proximal G•A mismatch by the Watson-Crick G-C base pair, but we found no substitution at this position that did not affect the retardation in a major way. Even changes that resulted in other purine•purine mismatches (i.e., converting G•A to A•G, A•A, or G•G) resulted in the electrophoretic mobility expected for a simple bulge. The same was true for the next position out from the bulge, where all substitutions (including those giving the three alternative purine•purine mismatches at that position) gave RNA of very similar, faster, mobility. We conclude that the sequence requirements for presenting the adenine bases into the minor groove of the C stem are highly stringent, and the interaction is seriously disrupted by any change in sequence in this critical region.
DISCUSSION
It is clear that the Kt-7 K-turn motif adopts a tightly kinked conformation as an isolated motif in RNA in free solution, consistent with the structure observed in crystallized ribosomes (Klein et al. 2001). RNA containing this sequence exhibits highly retarded electrophoretic mobility, and reduced end-to-end distances. However, solution studies revealed that the K-turn structure is much more dynamic than might have been anticipated, and that the strongly kinked conformation is dependent on the binding of metal ions.
In the absence of added metal ions, a duplex RNA containing the Kt-7 sequence migrates in a manner very similar to that of a three-purine bulge. Under these conditions, both the bend angle and the direction of the bend are closely similar to a simple A3 bulge. This would suggest that the C and NC stems are intact in the structure, but that the A-minor interaction does not occur. The change in conformation between this less-bent structure and the full K-turn can be well fitted by a simple two-state transition, where the K-turn structure is stabilized by the noncooperative binding of magnesium ions with a [Mg2+]1/2 of 1 mM. Monovalent ions can also stabilize the K-turn conformation at high concentrations, suggesting that diffuse ion binding is sufficient to induce the transition. We also found that hexammine cobalt (III) is very efficient in inducing the formation of the K-turn structure (data not shown).
Time-resolved FRET studies show that the isolated K-turn sequence is polymorphic, with populations of the less kinked bulge-like structure predominant in the absence of metal ions, and the tightly kinked structure observed in the ribosome. The position of equilibrium is determined by the prevailing metal ion concentration, but even in relatively high magnesium ion concentrations we can detect a significant population of the more extended conformation. For this reason it is not possible to draw quantitative conclusions from electrophoretic mobilities or steady-state FRET efficiencies, because these properties will be a weighted average of the two forms. Indeed, we find that the mobility of the K-turn RNA becomes more retarded relative to the An bulge species with increased magnesium ion concentration, as the equilibrium becomes more biased to the highly kinked conformation.
For this reason we used the time-resolved FRET data to compare the solution structure of the K-turn RNA with that found in the crystal structure of the 50S ribosomal subunit. The species with shorter donor-acceptor length corresponds to the more tightly kinked form of the Kt-7 RNA. From these data we calculate an interfluorophore distance of 21–32 Å. To compare this with the structure observed in the crystal, we took the coordinates of the Kt-7 structure and superimposed arms of the correct length and the known positions (Norman et al. 2000) of the fluorescein and Cy3 fluorophores (Fig. 7 ▶). From this we measured an interfluorophore distance of 32 Å. Thus there is reasonable agreement between the donor-acceptor length predicted from the ribosomal K-turn structure, and that calculated for the tightly kinked species observed by time-resolved FRET. Moreover, the distribution of donor-acceptor lengths required to fit these data was relatively narrow; comparable to that obtained from a simple duplex. This indicates that the highly kinked form of the K-turn is constrained, unable to undergo any large-amplitude flexing motion while it is in this conformation. Additionally, our electrophoretic bend-phasing experiments indicate that the direction of the folded K-turn is in good agreement with that observed in the ribosomal crystals. Taken together these data show that the conformation of the tightly bent species is closely similar to that observed in the ribosome.
FIGURE 7.
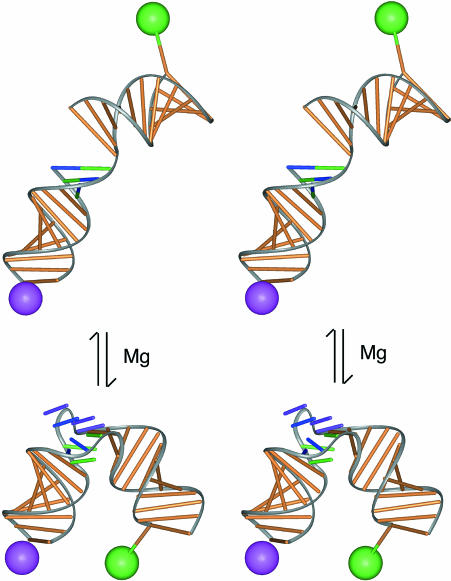
Models of the Kt-7 RNA in the tightly kinked and extended forms. The Kt-7 structure derived from the crystal structure (Klein et al. 2001) and the deduced form of the extended structure were fused to helical arms of the lengths used in the FRET experiments, and the fluorescein (represented as the green sphere) and Cy3 (magenta sphere) attached to their known positions (Norman et al. 2000). From this, interfluorophore distances of 32 Å and 85 Å were calculated for the kinked and extended forms, respectively. The local conformation of the GAA bulge in the extended structure is unknown, and is not represented in this model.
Steitz and coworkers (Klein et al. 2001) deduced a consensus sequence for the K-turn motif, shown in Figure 1 ▶. Examination of the different examples of K-turns in the ribosome suggests that the most invariant feature is the presence of the two A•G mismatches of the 3′ side of the (usually three) bulged nucleotides. The mismatches present the minor groove sides of their adenine bases into the groove of the C stem in order to make the A-minor interaction that generates the tight kinking of the K-turn. Simultaneous replacement of the three consecutive mismatches of Kt-7 by Watson-Crick base pairs reduced the retardation to that of a simple bulge. We found that the sequence requirements within the NC stem are extremely stringent, such that every single-nucleotide change investigated in either of the two A•G mismatches adjacent to the bulge resulted in almost complete loss of the anomalous electrophoretic retardation. Even A•A mismatches that might preserve an adenine base to make the interaction with the groove reduce the retardation to that of a simple bulge. Although the A•G mismatch immediately adjacent to the bulge is invariant in all of the known K-turn sequences, some changes can be found in the second position. For example, this position is A•A in Kt-58, and even an A-U base pair in Kt-23 in the 30 S subunit. Yet when either of these changes was examined in the Kt-7 context, it led to the loss of anomalous mobility.
The most surprising feature of the K-turn motif is the dynamic equilibrium with the less tightly kinked structure, and this has an important biological consequence. A significant population of the less-kinked species is present even at relatively high magnesium ion concentrations, and under physiological conditions the K-turn will not be fully folded into the form observed by crystallography of the ribosome. Moreover, as some of the naturally occurring K-turns contain sequence variations that we know destabilize the kinked form, it is likely that some or all of these structures will require stabilization. Most of the K-turn structures observed in the ribosome are bound by proteins (Klein et al. 2001), and this could help to stabilize the kinked conformation in situ. Tertiary interactions in the ribosome may also play a role in stabilizing the structure. All of this suggests that the K-turn motif is not of itself an organizing feature, in ribosomal biogenesis for example. Rather it seems that the tightly kinked structure will require stabilization as the ribosomal structure becomes organized. The same considerations will apply to nonribosomal K-turn structures (Mao et al. 1999; Vidovic et al. 2000). Further experiments are required to study the role of protein binding in the folding of K-turn RNA structures.
MATERIALS AND METHODS
Preparation of RNA
RNA was transcribed using T7 RNA polymerase (Milligan et al. 1987) from double-stranded DNA templates that were prepared by PCR reactions from synthetic DNA oligonucleotides. Where required, RNA was radioactively labeled by incorporation of [α-32P] ATP. RNA was purified by electrophoresis in 8% polyacrylamide gels containing 7 M urea. RNA was recovered by electroelution into 8M ammonium acetate solution. Eluted RNA was recovered by ethanol precipitation and dissolved in water.
The electrophoretic retardation of K-turn and related species was studied in 120-bp RNA duplexes (Figs. 2 ▶, 6 ▶). The bulge-strand sequence of the duplex containing the Kt-7 motif had the sequence (all sequences written 5′–3′):
GCGACACACGGACAAGCAAUGUGUGACGUAAUAUGGAGGGCGGACAAGCAAUGUGUGGGCGAAGAACCAUGUACUGUUCAACUUGACAGUCCUCGCAUGUACUGUUCAACUUAAGACCCACGC
where the K-turn motif is highlighted bold. The sequences of the converted K′ and An-containing duplexes can be derived from this.
The electrophoretic phasing experiments used to determine the relative direction of bending (Fig. 3 ▶) employed a series of 88-bp duplexes with an A4 bulge and a K-turn sequence symmetrically disposed and separated by a spacer of 8–22 bp (measured between the two bulges). The top strand sequence of the 8-bp spacer construct was:
GCGACACACGGACAAGCAAUGUGUGACGUAAUAUGGAGGGAAAACGUAUGGCGAAGAACCAUGCUCGCAUGUACUGUUCAACUUAAGACCCACGC
where the A4 and K-turn motif are highlighted bold.
Preparation of RNA for fluorescence experiments
Ribo-oligonucleotides were synthesized using phosphoramidite chemistry (Beaucage and Caruthers 1981). Fluorescein and Cy3 fluorophores were attached at 5′ termini as appropriate during synthesis. Labeled RNA was purified by gel electrophoresis and reversed-phase HPLC (Wilson et al. 2001). Equimolar quantities of the two strands were hybridized, and the resulting complexes purified by gel electrophoresis. Fluorescence experiments were performed in 90 mM Tris-borate, pH 8.3, and the indicated concentrations of salts pipetted directly into the cuvette. The sequences used in the FRET analyses were:
Bulge strand: Fluorescein-CCAGUCAGUGGCGAAGAACCAUGUCAGG
Nonbulge strand: Cy3-CCUGACAUGGGGAGCCACUGACUGG
For the variant with the complemented G•A mismatches, a new bulge strand was synthesized, and hybridized to the same nonbulge strand:
Fluorescein: CCAGUCAGUGGCGAAUCCCCAUGUCAGG
Gel electrophoresis
RNA was loaded onto 12% polyacrylamide gels (19:1::acrylamide:bis) and electrophoresed in 90 mM Tris-borate, pH 8.3, 2 mM EDTA or 5 mM Mg2+ (recirculated at > 1L/h) at room temperature for 24 or 72 h at 130 V. Gels were either exposed directly to phosphor plates at 4°C or dried onto Whatman 3MM paper and exposed to storage phosphor plates at room temperature.
Steady-state fluorescence
Fluorescence spectra were recorded at 4°C using an SLM-Aminco 8100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of EFRET were measured using the acceptor normalization method (Clegg 1992).
Time-resolved fluorescence
Time-resolved fluorescence lifetime measurements were performed at 4°C using a K2-Digital Phase Fluorimeter (ISS). Excitation at 488 nm was provided by a vertically polarized, argon ion laser (200 mW, Melles-Griot), intensity modulated at 39 frequencies between 4 and 300 MHz. The cross-correlation frequency was 400 Hz. Donor emission was measured using a 10-nm bandpass filter centered at 520 nm (520DF10, Omega Optical) to exclude scattered incident light and acceptor fluorescence and a polarizer set at 54.7° to remove instrumental artefacts. Measurements were referenced to fluorescein in 10 mM NaOH, with a lifetime of 4.05 nsec. A minimum of six and a maximum of 12 measurements were performed at each frequency, with data collection terminating if the phase error was below 0.1° and the modulation error was below 0.002.
Phase shift and modulation data were analyzed with the parameter estimation program CFS_LS (Johnson and Faunt 1992), according to the theory given in Melcher et al. (2003). The data were weighted using the standard error of each datum. Donor decay lifetimes were determined at 1 nM and 50 mM magnesium ion concentration for fluorescein attached to an RNA duplex with the same end sequence. This was fitted best by two lifetimes, which were then fixed for the appropriate magnesium ion concentration when fitting time-resolved data for the donor-acceptor labeled molecules. The effects of energy transfer on the donor lifetime were fitted by models where the donor-acceptor distances were described by either a single Gaussian or two Gaussian distributions, assuming an R0 of 56 Å (Norman et al. 2000). When fitting two Gaussian distributions, it was necessary to fix the relative amplitudes of each distribution in order to obtain estimates for the centers and half-widths. Best fits were obtained for a range of assumed relative amplitudes, and that with the lowest χ2 accepted. Data at both magnesium ion concentrations were therefore fitted simultaneously to two shared distances and half-widths with distinct relative proportions/amplitudes. Goodness of fit was evaluated by χ2, the distribution of residuals, and by a runs test. Confidence intervals (single standard deviation) were determined by the Bootstrap method (Efron and Tibshirani 1993).
Acknowledgments
We thank Tim Wilson for valuable discussion, Cancer Research-UK for financial support, and the EPSRC for a Ph.D. studentship (T.A.G.). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5176604.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Beaucage, S.L. and Caruthers, M.H. 1981. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22: 1859–1862. [Google Scholar]
- Bhattacharyya, A. and Lilley, D.M.J. 1989. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 17: 6821–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, A., Murchie, A.I.H., and Lilley, D.M.J. 1990. RNA bulges and the helical periodicity of double-stranded RNA. Nature 343: 484–487. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K.H., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996a. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K.H., Golden, B.L., Szewczak, A.A., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996b. RNA tertiary structure mediation by adenosine platforms. Science 273: 1696–1699. [DOI] [PubMed] [Google Scholar]
- Clegg, R.M. 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211: 353–388. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Moore, P.B., Freeborn, B., and Steitz, T.A. 1997. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell 91: 705–712. [DOI] [PubMed] [Google Scholar]
- Diekmann, S. and Wang, J.C. 1985. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J. Mol. Biol. 186: 1–11. [DOI] [PubMed] [Google Scholar]
- Efron, B. and Tibshirani, R.J. 1993. An introduction to the bootstrap. Chapman Hall, New York.
- Ferré-d’Amaré, A.R., Zhou, K., and Doudna, J.A. 1998. Crystal structure of a hepatitis δ virus ribozyme. Nature 395: 567–574. [DOI] [PubMed] [Google Scholar]
- Förster, T. 1948. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 2: 55–75. [Google Scholar]
- Gohlke, C., Murchie, A.I.H., Lilley, D.M.J., and Clegg, R.M. 1994. The kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. 91: 11660–11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, B.L., Gooding, A.R., Podell, E., and Cech, T.R. 1998. A preorganised active site in the crystal structure of the Tetrahymena ribozyme. Science 282: 259–264. [DOI] [PubMed] [Google Scholar]
- Hsieh, C.-H. and Griffith, J.D. 1989. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: Possible relevance to the folding of single-stranded nucleic acids. Proc. Natl. Acad. Sci. 86: 4833–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M.L. and Faunt, L.M. 1992. Parameter estimation by least-squares methods. Methods Enzymol. 210: 1–37. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: a new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov, A.S., Yang, X., Pan, T., and Mondragon, A. 2003. Crystal structure of the specificity domain of Ribonuclease P. Nature 421: 760–764. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B. and Westhof, E. 2003. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13: 300–308. [DOI] [PubMed] [Google Scholar]
- Luebke, K.J. and Tinoco, I. 1996. Sequence effects on RNA bulge-induced helix bending and a conserved five-nucleotide bulge from the group I introns. Biochemistry 35: 11677–11684. [DOI] [PubMed] [Google Scholar]
- Lumpkin, O.J. and Zimm, B.H. 1982. Mobility of DNA in gel electrophoresis. Biopolymers 21: 2315–2316. [DOI] [PubMed] [Google Scholar]
- Mao, H., White, S.A., and Williamson, J.R. 1999. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 6: 1139–1147. [DOI] [PubMed] [Google Scholar]
- Marini, J.C., Levene, S.D., Crothers, D.M., and Englund, P.T. 1982. Bent helical structure in kinetoplast DNA. Proc. Natl. Acad. Sci. 79: 7664–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, S.E., Wilson, T.J., and Lilley, D.M.J. 2003. The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA 9: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherall, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., and Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, D.G., Grainger, R.J., Uhrin, D., and Lilley, D.M.J. 2000. The location of Cyanine-3 on double-stranded DNA; importance for fluorescence resonance energy transfer studies. Biochemistry 39: 6317–6324. [DOI] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., and McKay, D.B. 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74. [DOI] [PubMed] [Google Scholar]
- Riordan, F.A., Bhattacharyya, A., McAteer, S., and Lilley, D.M.J. 1992. Kinking of RNA helices by bulged bases, and the structure of the human immunodeficiency virus transactivator response element. J. Mol. Biol. 226: 305–310. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B. and Ferré-D’Amaré, A.R. 2001. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410: 780–786. [DOI] [PubMed] [Google Scholar]
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., and Klug, A. 1995. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002. [DOI] [PubMed] [Google Scholar]
- Tang, R.S. and Draper, D.E. 1990. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry 29: 5232–5237. [DOI] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Luhrmann, R., and Ficner, R. 2000. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell 6: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Wilson, T.J., Zhao, Z.-Y., Maxwell, K., Kontogiannis, L., and Lilley, D.M.J. 2001. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40: 2291–2302. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Zacharias, M. and Hagerman, P.J. 1995. Bulge-induced bends in RNA: Quantification by transient electric birefringence. J. Mol. Biol. 247: 486–500. [DOI] [PubMed] [Google Scholar]