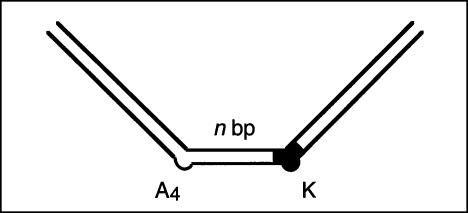
FIGURE 3.
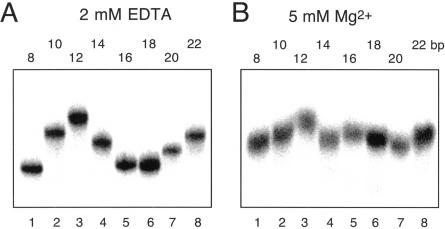
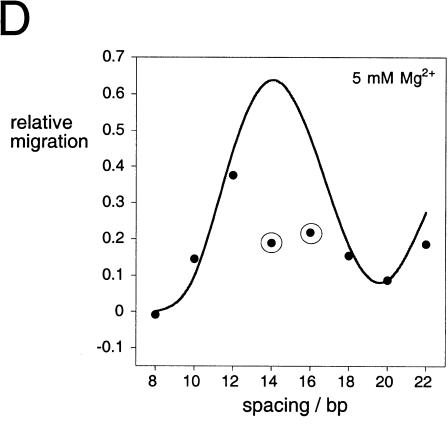
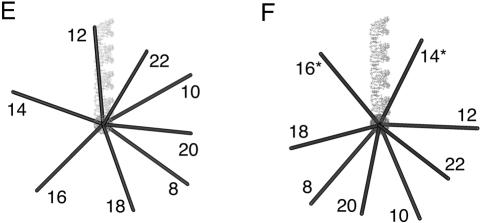
The direction of bending at the K-turn locus depends on the presence of metal ions. A series of 88-nt RNA duplexes containing an A4 bulge and the K-turn sequence symmetrically located about the center and separated by a spacer region of n bp (n = 8–22) were analyzed. (A,B) The radioactively [α′-32P]-labeled RNA was electrophoresed at 20°C in a 10% polyacrylamide gel in the presence of either 2 mM EDTA (A) or 5 mM magnesium ions (B). The RNA was visualized by phosphorimaging, and the migration of each species measured. (C,D) The experimentally determined relative inverse migrations in 2 mM EDTA or 5 mM magnesium ions are plotted as a function of spacing in C and D, respectively. The lines are smooth interpolations of the model derived migration factors that best fit the experimental data. (E,F) The data were analyzed using molecular graphics models as described in the text. The long arm on the A4 bulge side is shown as an RNA helix, and the various positions of the long helix on the K-turn side are shown as bars labeled with their corresponding spacer lengths. The relative rotational settings of these K-turn-side helices was fixed and the phase angle then varied while measuring end-to-end distances until the best fit between the calculated Lumpkin-Zimm migration factors (Equation 1) and the experimentally measured mobilities was obtained. The directions shown are those for the best fits in 2 mM EDTA (E) or 5 mM magnesium ions (F). For the data obtained in 5 mM magnesium ions (F), the anomalous points (14- and 16-bp spacers, ringed in D) are marked with asterisks; in these species the two long arms are closest to coplanarity, so the reflex kink angle of the K-turn could result in a clash of long arms.