Abstract
Upon temperature downshift below the lower threshold of balanced growth (~20°C), the Escherichia coli translational apparatus undergoes modifications allowing the selective translation of the transcripts of cold shock-induced genes, while bulk protein synthesis is drastically reduced. Here we were able to reproduce this translational bias in E. coli cell-free extracts prepared at various times during cold adaptation which were found to display different capacities to translate different types of mRNAs as a function of temperature. Several causes were found to contribute to the cold-shock translational bias: Cold-shock mRNAs contain cis-elements, making them intrinsically more prone to being translated in the cold, and they are selective targets for trans-acting factors present in increased amounts in the translational apparatus of cold-shocked cells. CspA was found to be among these trans-acting factors. In addition to inducing a higher level of CspA, cold shock was found to cause a strong (two- to threefold) stoichiometric imbalance of the ratio between initiation factors (IF1, IF2, IF3) and ribosomes without altering the stoichiometric ratio between the factors themselves. The most important sources of cold-shock translational bias is IF3, which strongly and selectively favors translation of cold-shock mRNAs in the cold. IF1 and the RNA chaperone CspA, which stimulate translation preferentially in the cold without mRNA selectivity, can also contribute to the translational bias. Finally, in contrast to a previous claim, translation of cold-shock cspA mRNA in the cold was found to be as sensitive as that of a non-cold-shock mRNA to both chloramphenicol and kanamycin inhibition.
Keywords: stress response, translational control, translational bias, initiation factors, RNA chaperone
INTRODUCTION
Following a sudden temperature downshift (e.g., from 37°C to 10°C), a mesophilic bacterium such as Escherichia coli undergoes a cold-adaptation period which lasts about 4 h at 10°C. During this time, a set of ~25 cold-shock proteins is selectively expressed, while bulk protein synthesis is drastically reduced. The situation reverses itself at the end of the acclimation phase, when the synthesis of the cold-shock proteins is reduced while bulk protein synthesis resumes (for review, see Gualerzi et al. 2003).
Although several mechanisms are likely responsible for turning on and off the expression of the cold-shock genes during the various phases of cold adaptation, the main regulatory events seem to occur at the posttranscriptional level, and entail both alterations of the mRNA half-lives and selective changes of the translational apparatus. These events ultimately determine an increased stability of cold-shock mRNAs and ensure their preferential utilization by the translational apparatus of cold-shocked cells (Brandi et al. 1996; Goldenberg et al. 1997).
Because a plausible explanation for how cold-shock mRNAs are preferentially translated during cold adaptation was lacking, in the present study we sought to characterize the properties acquired by the translational apparatus after cold shock. Our aim was to obtain a better understanding of the nature of the changes which determine this translational selectivity.
The present results indicate that more than one control element is involved in this preferential translation, and we identified at least some of these elements here: cis-elements located in the 5′ untranslated region (UTR) of cold-shock mRNAs, an increased level of some trans-acting elements such as the RNA-binding protein CspA and, most important, of the translational initiation factors IF3 and IF1. The increase of the latter, together with the known cold shock-induced increase of IF2, results in a transient imbalance in the initiation factors/ribosomes ratio never previously observed.
RESULTS
Characteristics of the mRNAs used
Our main purpose was to uncover the molecular bases of the translational bias which favors the expression in vivo of cold-shock mRNAs during cold acclimation (Goldenberg et al. 1997). Because the mechanisms underlying this event are very complex. For this reason we chose a reductionist approach, the first challenge of which was to reproduce this phenomenon in a cell-free system. Considering that the cold-shock transcripts affected by this translational bias represent only a small subset of the cellular mRNAs, an important strategic decision concerned the choice of the cold-shock and control mRNAs to be tested. As typical transcripts of cold-shock genes we chose cspA mRNA, and hns, encoding the single-stranded nucleic acid binding protein CspA (Goldstein et al. 1990) and the abundant nucleoid-associated protein H-NS (La Teana et al. 1991), respectively. HupA and cspD mRNAs represent two typical non-cold-shock mRNAs, the first encoding the α subunit of nucleoid-associated protein HU and the second encoding CspD, a protein localized in the nucleoid, unlike its paralog CspA, which is found at the cell poles (Giangrossi et al. 2001). Because the two non-cold-shock mRNAs (hupA and cspD) are translated at 37°C at a level similar to that of the cold-shock hns mRNA, yet to a much lower level than cspA mRNA and are translated very poorly at 15°C (Table 1 ▶), we also included in our comparisons three “cold-tolerant” mRNAs originating from the three promoters (P2, P3, and P4) of hupB, the gene encoding the β subunit of HU. The expression of this gene reaches its maximum in cells entering the stationary phase at 37°C (Claret and Rouviere-Yaniv 1997), and although its transcription and translation are stimulated by a cold stress, hupB cannot be regarded as a typical cold-shock gene, because the level of HUβ does not substantially increase after cold shock (Giangrossi et al. 2002). Thus, as indicated by their denomination, the three hupB transcripts are non-cold-shock mRNAs which can be translated to a fairly high level at both low and high temperature. Indeed, in the control system at 37°C, all of these mRNAs are translated better than hns mRNA and to almost the same level as cspA mRNA (Table 1 ▶).
TABLE 1.
Translation of various mRNAs by S30 extracts derived from control and cold-shocked cells after different times of cold exposure
| mRNA | Translation temperature | S30 (control) | S30 Cs (30 min) | S30 Cs (60 min) | S30 Cs (90 min) | S30 Cs (120 min) |
| wtcspA | 37°C | 13.65 ± 0.60 | 27.60 ± 0.60 | 26.20 ± 0.70 | 19.70 ± 0.53 | 24.40 ± 1.70 |
| 15°C | 4.95 ± 0.28 | 7.56 ± 0.70 | 8.44 ± 0.38 | 14.19 ± 1.50 | 13.70 ± 0.60 | |
| cspA (Δ1-180) | 37°C | 15.03 ± 0.70 | 31.90 ± 0.40 | 25.90 ± 1.10 | 23.37 ± 1.10 | 22.60 ± 0.86 |
| 15°C | 3.35 ± 0.34 | 4.10 ± 0.66 | 3.58 ± 0.65 | 6.54 ± 0.65 | 7.51 ± 0.06 | |
| hns | 37°C | 2.07 ± 0.07 | 7.56 ± 0.23 | 6.58 ± 0.60 | 4.32 ± 0.23 | 5.30 ± 0.73 |
| 15°C | 0.82 ± 0.15 | 0.87 ± 0.19 | 0.76 ± 0.10 | 1.61 ± 0.16 | 1.62 ± 0.13 | |
| cspD | 37°C | 1.16 ± 0.06 | 2.27 ± 0.22 | n.d. | 2.70 ± 0.60 | 2.24 ± 0.40 |
| 15°C | 0.23 ± 0.08 | 0.00a | 0.00a | 0.00a | 0.00a | |
| hupA | 37°C | 2.29 ± 0.18 | 1.80 ± 0.25 | 1.44 ± 0.12 | 1.70 ± 0.05 | 1.76 ± 0.36 |
| 15°C | 0.29 ± 0.07 | 0.22 ± 0.05 | n.d. | 0.27 ± 0.06 | 0.58 ± 0.04 | |
| P4hupB | 37°C | 16.2 ± 0.50 | 26.30 ± 0.51 | 24.40 ± 0.51 | 19.19 ± 0.16 | 21.50 ± 1.30 |
| 15°C | 3.35 ± 0.42 | 3.82 ± 0.37 | 3.51 ± 0.32 | 5.73 ± 0.48 | 5.83 ± 0.42 | |
| P3hupB | 37°C | 12.80 ± 0.28 | 21.17 ± 0.36 | 19.50 ± 0.32 | 17.46 ± 0.24 | 19.40 ± 0.05 |
| 15°C | 1.85 ± 0.46 | 2.60 ± 0.13 | 2.30 ± 0.24 | 4.10 ± 0.29 | 4.10 ± 0.40 | |
| P2hupB | 37°C | 13.4 ± 0.28 | 17.89 ± 1.40 | 15.70 ± 0.52 | 16.32 ± 0.42 | 18.9 ± 0.15 |
| 15°C | 2.52 ± 0.34 | 2.46 ± 0.16 | 2.98 ± 0.40 | 3.95 ± 0.46 | 4.40 ± 0.50 |
The experiment was carried out as described in the text and Materials and Methods. As indicated in the headings, the reaction mixtures contained different types of cell extracts programmed with different mRNAs and were incubated at 37° or 15°C. Samples were taken at different time intervals until saturation was reached. The data were analyzed by the Regression Wizard program (SigmaPlot) and could be fit in all cases to an exponential equation (see text). The correlation coefficients R, which indicate how well the regression model describes the data, were found to be in all cases very close to the maximum theoretical value of 1, ranging from 0.9418 (hns mRNA translated at 37°C by control S30) to 0.9996 (cspA mRNA translated at 37°C by S30 60 min cs). The standard error of the estimate, which measures the actual variability about the regression plane of the underlying population, ranged from ±0.40 to ±1.70.
The results presented in the table refer to the number of pmoles of product synthesized after 15 min incubation (± the standard error).
n.d. = not done.
aBarely detectable above background level.
In addition to the above-mentioned mRNAs, in some experiments we also used a shortened form of cspA mRNA (cspAΔ1-80) and two forms of infA* mRNA, one having the canonical initiation triplet AUG and the other the noncanonical AUU triplet. The mutated cspA mRNA carries a deletion of the first 80 (out of 159) nucleotides of its 5′ UTR thereby, completely eliminating the “cold-box” but not the “upstream-box sequence” which were reported to be important for transcription (Jiang et al. 1996) and translation (Yamanaka et al. 1999) in the cold, respectively. The infA*AUG mRNA is the transcript of a synthetic gene designed for high expression of E. coli initiation factor IF1 in vivo at 37°C; both codon usage and the sequence of the translation initiation region (TIR) of this mRNA are based on the E. coli consensus (Calogero et al. 1987).
Translational efficiency of cold-shock and non-cold-shock mRNA as a function of temperature and type of cell extract
To achieve a meaningful and rigorous comparison of the translational activity of the above-described mRNAs, our second task was that of dealing with the large number of variables which characterize the cold-shock response in vivo and bringing them under control in our cell-free systems. The most important of these variables are the intracellular milieu, which is continuously modified as a function of the time of cold shock, and the drastic temperature change (>20°C) which, among other effects, determines a large variation of the translational initiation/elongation rates (Farewell and Neidhardt 1998; see below).
With respect to the variable represented by the changing intracellular environment, to cover the most interesting period of cold adaptation, samples were routinely taken just before 30, 60, 90, and 120 min after shifting the cells to 10°C; during this time the cells reprogram their genetic expression, and cold-shock and non-cold-shock mRNAs are translated to the highest and lowest level, respectively (Goldenberg et al. 1997).
The effect of the temperature change was studied comparing the translational activities of the mRNA at 37°C and 15°C. In the first experiment we decided to record the entire time course of translation of each mRNA under each experimental condition, in order to determine the initial rates and saturation levels reached in each system. The amounts of protein synthesized “a” during each time course were fitted (see also Fig. 1 ▶ legend) to the exponential equation y = a(1 − 1/ebx) so that the rate of synthesis “b” could also be obtained from the fit. Our comparison revealed that the translational rates of both control and cold-shock extracts are reduced from 7.5- to 10-fold when the temperature is lowered from 37°C to 15°C. However, compared to the control extracts, the extracts from cold-shocked cells translate cold-shock and cold-tolerant mRNAs not only to a higher level but also at an increased rate, at any temperature tested (data not shown).
FIGURE 1.
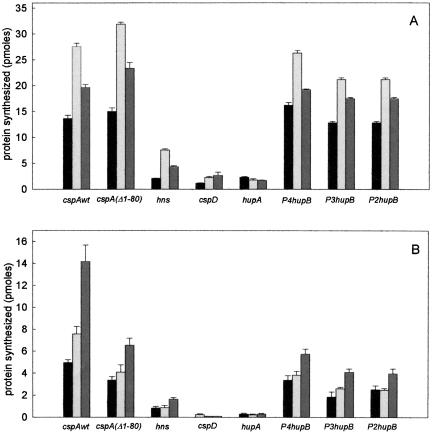
Translation of various mRNAs by S30 extracts derived from control and cold-shocked cells after different times of cold exposure. The histograms display the amount of proteins synthesized after 15-min incubation at either 37°C (A) or 15°C (B) by equivalent amounts (normalized for their ribosome content) of E. coli MRE600 S30 extracts prepared from cells harvested before (black) and after 30-min (light gray) and 90-min (dark gray) cold shock at 10°C; the extracts were programmed with 15 pmoles each of cold shock [cspAwt, cspA(Δ1-80), and hns], cold-tolerant (P2, hupB, P3 hupB, P4hupB) and non-cold-shock (cspD and hupA) mRNAs as indicated in the abscissa. The other conditions are described in Table 1 ▶ and in Materials and Methods.
Table 1 ▶ shows the amount of protein synthesized after 15-min incubation at either 37°C or 15°C by the S30 extracts prepared from control cells and from cells taken after 30, 60, 90, and 120 min in the presence of the various types of mRNAs. The histograms presented in Figure 1 ▶ are meant to give a general visual impression of the different translational activities, and they refer only to the experiments performed with control extract and with extracts of cells after 30- and 90-min cold shock.
It can be seen that the different templates are translated with different efficiency as a function of both incubation temperature and nature of the S30 extract used. Indeed, at 37°C the control S30 extract translates five mRNAs (wtcspA, cspAΔ1-80, and the three hupB transcripts) with high efficiency (≥10 pmole product after 15 min) and translates the other three (hupA, cspD, and hns mRNAs) with a much lower efficiency (≅ 1–2 pmole product). Furthermore, regardless of the time of cold shock, all cold-shock extracts translate with a higher efficiency than control extracts all mRNAs, with the exception of hupA mRNA.
The situation is quite different at low temperature (15°C), where the level of translation depends much more on the type of template and on the type of extract. In fact, although the amount of protein synthesized after 15 min is lower than at 37°C, due to the lower initiation/elongation rate, all cold-shock and cold-tolerant mRNAs are translated to fairly high levels, and the efficiency by which they are translated is higher with the cold-shock extracts compared to the controls and increases with the time of cold shock (Fig. 1B ▶; Table 1 ▶). Of the two non-cold-shock mRNAs, hupA is translated to very low yet clearly detectable levels, whereas cspD is translated to a detectable level only by the control extract (Fig. 1B ▶; Table 1 ▶). In fact, in the presence of the cold-shock extracts, translation of cspD mRNA after 15 min is reduced to a level barely above the background. However, if the translation is allowed to continue for 120 min, synthesis of CspD is clearly detectable even at 15°C with the extracts of cold-shocked cells (see below).
Finally, it should be noted that cspAΔ1-80 mRNA is translated with the same efficiency as wtcspA mRNA at 37°C but not at 15°C, where its activity is strongly reduced compared to that of wtcspA mRNA, especially in the presence of the cold-shock extracts; this indicates a partial loss of its capacity to respond to the cold shock-induced changes of the translational apparatus.
The above results clearly indicate that: (1) temperature has different effects on the translational rate of different mRNAs, (2) the cold-shock-modified translational apparatus better translates almost all templates at 37°C, whereas it (3) selectively favors the translation of cold-shock and cold-tolerant mRNAs at low temperature. Thus, our data demonstrate that, by and large, our cell-free systems can reproduce the cold-shock translational bias occurring in vivo.
The possibility that the different activities of the mRNAs could reflect their different half-lives was tested by measuring the stability of the mRNAs in the various cell-free extracts under translational conditions. As seen from Table 2 ▶, the difference in stability between the most and least translated mRNAs (wtcspA and cspD) is <20% in the control extracts at both 37°C and 15°C, whereas the different translational activities of wtcspA and cspA(Δ1-180) mRNAs cannot be correlated with stability differences, because they have very similar half-lives. The fact that the half-life of cspA mRNA at 15°C is significantly lower in the presence of S30 prepared from cold-shocked cells than that from control cells supports previous in vivo data (Goldenberg et al. 1996) which demonstrate the occurrence of structural/functional changes of the RNA-degrading machinery during the cold-acclimation phase which affect the stability of cspA mRNAs.
TABLE 2.
Half-lives of mRNAs during translation at 37°C and 15°C
| Translation at | |||
| mRNA | S30 from cells | 37°C | 15°C |
| cspA | control | 5.4 min | 80 min |
| cs | 4.4 min | 27 min | |
| cspA(Δ1–80) | control | 4.7 min | 80 min |
| cs | 3.4 min | 42 min | |
| cspD | control | 4.5 min | 83 min |
| cs | n.d. | 30 min | |
Experimental details are given in the text and in Materials and Methods.
n.d. = not done.
The results of these as well as of other experiments (Giangrossi et al. 2002; data not shown) clearly demonstrate that the mRNAs used in the present study display only minor stability differences which by no means could explain their several-fold differences in translational activity. Thus, other reasons must be found to explain the observed translational bias; the experiments described below were designed to investigate the nature of such reasons.
Both cis- and trans-acting elements contribute to the preferential translation of cold-shock mRNAs in the cold
Two types of tests were performed to determine whether the translational bias could be due to intrinsic structural properties (cis-elements) of the mRNAs, which would make them better or less suited for translation at low temperature and/or confer on them the capacity to respond differently to cold-shock factor(s) or to alterations of the cold-shock apparatus.
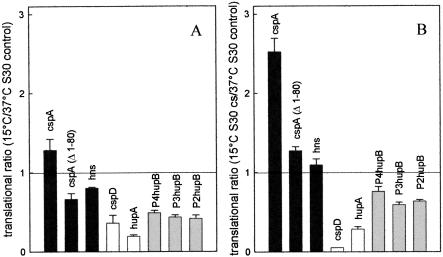
In the first test we compared the maximum (plateau) level of translation obtained at 15°C and 37°C with S30 extracts from control (non-cold-shocked) cells programmed with the various types of mRNAs (Fig. 2A ▶). In the second test, the maximum translational level of the same mRNAs obtained at 15°C with cold-shock extracts was compared to that obtained at 37°C with control extracts (Fig. 2B ▶). As seen in Figure 2A ▶, the 15°C/37°C translational ratio determined with the control S30 extract is highest with the cold-shock wtcspA (>1) and hns (≅0.8) mRNAs and lowest (≤0.35) with the non-cold-shock mRNAs (cspD and hupA), indicating that the two types of template have a high and scarce propensity to be translated at low temperature, respectively. In the same experiment, the three cold-tolerant hupB mRNAs yielded intermediate 15°C/37°C translational ratios (0.45–0.50).
FIGURE 2.
Different propensity of individual mRNAs to be translated in the cold is due to both cis- and trans-elements. The indicated cold-shock (black), cold-tolerant (gray), and non-cold-shock (white) mRNAs were used to program in vitro translational systems prepared with E. coli MRE600 S30 extracts from either control (37°C) or cold-shocked (120 min at 10°C) cells. The reactions were carried out to completion at either 37°C for 30 min or at 15°C for 120 min. The contribution of intrinsic cis-acting (A) and trans-acting (B) elements in determining the aptness of the individual mRNAs to be translated at low temperature were evaluated determining (A) the ratio between the maximum amount of product synthesized at 15°C and 37°C by the same control S30 extracts, and (B) the ratio between the maximum amount of product synthesized at 15°C by an S30 extract from cold-shocked cells and the amount synthesized at 37°C by a control S30 extract. Each ratio was calculated by dividing the average (±standard deviation) of three experimental points by the average (±sd) of three experimental points; the error bars represent the sd of the calculated ratios. The other conditions of translation were the same as described in Materials and Methods.
The results obtained in the second experiment (Fig. 2B ▶) demonstrate that the responsiveness of the mRNAs to the cold shock-modified translational apparatus is maximum with the cold-shock mRNAs, whose translational ratios increase to >2 (cspA) and >1 (hns), intermediate with the cold-tolerant hupB mRNAs (ratios = 0.6–0.7) and minimum (or even negative with cspD) with the non-cold-shock mRNAs (ratios <0.3). Finally, the 15°C/37°C translational ratio of cspA(Δ1-80) mRNA was lower compared to that of wt cspA mRNA in both experiments (Fig. 2A,B ▶), indicating that the deletion of the 5′ half of the 5′ UTR of cspA mRNA reduces the intrinsic capacity of this mRNA to be translated in the cold as well as, albeit to a lesser extent, its capacity to respond to the cold-shock modifications of the translational apparatus.
Thus, the results of these experiments clearly indicate that both an intrinsic propensity for translation in the cold due to cis-elements (Fig. 2A ▶) and a greater responsiveness to the modifications of the translational apparatus (Fig. 2B ▶) contribute to the selective translation of cold-shock and cold-tolerant mRNAs at low temperature. The latter data also suggest that the cold-shock and the cold-tolerant mRNAs could be the selective target of some trans-acting factor(s) present in the extracts of cold-shocked cells and capable of selectively stimulating their translation in the cold.
Finally, the above data concerning cspD mRNA confirmed that which was noticed above (Table 1 ▶), namely that the cold-shock extracts contain (or lack) some component capable of reducing (or stimulating) the expression of at least one non-cold-shock mRNA.
CspA stimulates mRNA translation at low temperature
To determine nature and localization of the trans-acting factor(s), the S30 extracts were fractionated into 70S ribosomes and S100 postribosomal supernatants, and translation of cspA mRNA at both low (15°C) and high (37°C) temperature was used as the most sensitive system to detect their presence.
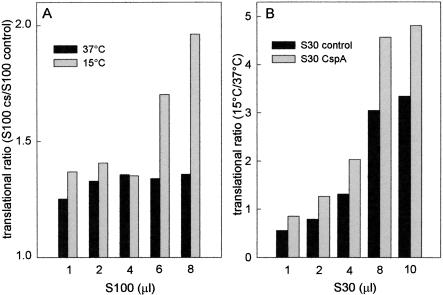
In the first series of experiments, cspA mRNA translation by control ribosomes was tested in the presence of increasing amounts of postribosomal supernatants (S100) from control or cold-shocked cells, and the results were plotted as the ratio between the maximum level of translation obtained with the same, normalized amounts of the two supernatants. As seen in Figure 3 ▶, the translational ratio S100cs/S100 control is >1 at both 37°C and 15°C, indicating that cspA mRNA is always translated with higher efficiency in the presence of the cold-shock postribosomal fraction; however, although at 37°C the cold-shock fraction is only 25%–30% more active than the control S100 (Fig. 3A ▶, black bars), translation at low temperature (gray bars) is almost two times more efficient with the cold-shock S100 when the level of the supernatant is increased above a certain point (i.e., >4 μL S100/reaction). These findings suggest that the postribosomal supernatant from cold shocked cells contains a trans-acting factor which, when its concentration is increased above a critical level, is capable of stimulating cspA mRNA translation, especially in the cold. These properties are similar to those of the cold-shock protein CspA, which produces a modest stimulation of cspA mRNA translation at 37°C (Brandi et al. 1996) and displays low affinity for its target, represented by a single-stranded nucleic acid (Jiang et al. 1997). For these reasons, the translational efficiencies of S30 extracts from control cells and from cells producing extra copies of CspA (S30CspA) were compared. In agreement with the previous findings, this experiment demonstrated that cspA mRNA translation becomes more efficient in the presence of S30CspA (Fig. 3B ▶). This effect is especially evident at low temperature (the results are plotted as the 15°C/37°C translational ratio), and when the cell extracts are added in amounts above a critical level (i.e., ≥4 μL S30).
FIGURE 3.
Presence of a trans-acting factor in the postribosomal supernatant (S100) of cold-shocked cells. (A) Translation of cspA mRNA was carried out to completion at 15°C (gray) and 37°C (black) in the presence of E. coli MRE600 70S ribosomes supplemented with the indicated amounts of postribosomal supernatants (S100) derived from either control or cold-shocked cells. These S100 fractions were prepared as described in Materials and Methods and exhaustively dialyzed against Buffer A and adjusted to the same protein concentration just before use. Translation was carried out as described in Materials and Methods, and the values reported in the ordinate are the ratios of the levels of protein synthesized with S100 from cold-shocked (60 min at 10°C) versus control cells. (B) Translation of cspA mRNA was carried out at 15°C and 37°C in the presence of increasing amounts of S30 extract from control cells (black) and from cells carrying extra copies of cspA (gray) as described in Materials and Methods. The other conditions are as described above. The values reported in the ordinate are the ratios between the maximum levels of protein synthesized at saturation by each S30 extract at 15°C and 37°C. No error bars are given for this experiment, as the translational ratios were obtained by dividing single experimental points obtained in a single experiment. However, essentially identical results were obtained when a similar experiment was repeated under somewhat different experimental conditions.
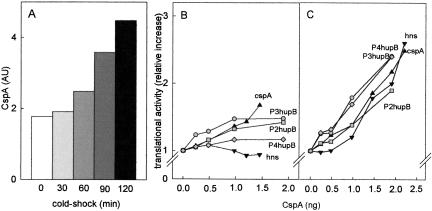
Due to the rather inefficient expression of the extra copies of cspA at 37°C, CspA does not increase more than two–threefold in the S30CspA (Brandi et al. 1996), so that its level in the S30CspA extracts is quantitatively similar to that found in the extracts of cells cold-shocked and harvested in early exponential growth (Fig. 4A ▶). A direct demonstration that CspA acts as a cold-shock translational enhancer is provided by the results of an experiment (Fig. 4 ▶) in which translation of other cold-shock and cold-tolerant mRNAs by both control 70S ribosomes and postribosomal supernatants was studied at high and low temperature in the presence of increasing amounts of purified CspA. The results obtained clearly indicate that CspA stimulates translation of all mRNAs examined without discriminating between them. However, this stimulation is small (from <10% to ~50%) at high (37°C) temperature (Fig. 4B ▶), whereas it is much more pronounced (> twofold) at low temperature (Fig. 4C ▶).
FIGURE 4.
Levels of CspA in extracts from cold-shock cells and stimulation of mRNA translation by purified CspA. The levels of CspA present in the S30 extracts from control and cold-shocked cells were determined as described (Brandi et al. 1999) by immunoblotting using polyclonal antibodies against CspA followed by densitometric quantification. The amounts of CspA normalized for the total amount of protein are expressed in (densitometric) arbitrary units and plotted as a function of the time of cold shock (A). The immunological quantification does not contain error bars, because in light of the excellent agreement of these data with the previously published results (La Teana et al. 1991; Brandi et al. 1999), this experiment was performed only once. In vitro translation of cspA mRNA (▴), hns mRNA (▾), P4hupB mRNA (♦), P3hupB mRNA (•), and P2hupB mRNA (▪) was carried out with 70S ribosomes and S100 supernatant from control cells in the presence of the indicated amounts of purified CspA. The other conditions are described in Materials and Methods, and the reaction mixtures were incubated for 30 min at 37°C (B) and for 90 min at 15°C (C). The results obtained are presented as the ratio of product synthesized in the presence of the indicated amounts of CspA versus the basal level product synthesized in the absence of CspA.
Taken together, these results identify CspA as a trans-acting factor, capable of improving translation at low temperature. The presence of higher levels of CspA can explain the increased translational activity displayed, mainly in the cold, by the extracts of cold-shocked cells, but cannot explain the mRNA selectivity which is characteristic of the cold-shock translational bias. These conclusions led us to search for additional trans-acting factors.
Cold-shock 70S ribosomes display some translational selectivity
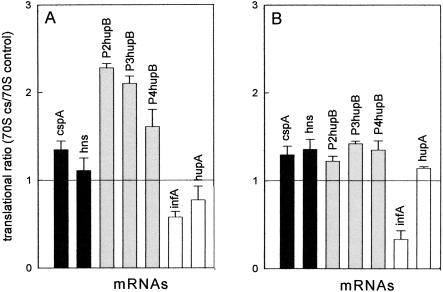
The translational activity of ribosomes purified from control and cold-shocked cells and programmed with various types of mRNAs were tested at 37°C and 15°C in the presence of control postribosomal supernatant (S100). As seen in Figure 5 ▶, both cold-shock and cold-tolerant mRNAs are translated somewhat better by the cold-shock than by the control ribosomes, the ratios between their maximum levels of translation being ≥1. This difference can be seen at both 37°C (Fig. 5A ▶) and 15°C (Fig. 5B ▶), but is more pronounced at high temperature with the cold-tolerant hupB mRNAs, which are translated ~two times better by the cold-stressed ribosomes. The control 70S, on the other hand, translates the non-cold-shock mRNAs, infA mRNA in particular, better than the cold-shock 70S (ratios ≤1). In agreement with a previous report (VanBogelen and Neidhardt 1990), these differences suggest that the cold stress may induce some structural modification of the 70S ribosomes, whose nature and significance in contributing to increasing the translational efficiency of cold-shock and cold-tolerant mRNAs remain to be established.
FIGURE 5.
Translational efficiency of different mRNAs by control and cold-shock ribosomes at different temperatures. The indicated cold-shock (black), cold-tolerant (gray), and non-cold-shock (white) mRNAs were used to program translational systems containing postribosomal supernatant (S100) from control cells and 70S ribosomes prepared from either control or cold-shocked cells. The results are expressed as the ratio between the average of three experimental points (derived from two or three separate experiments) representing the maximum amount of protein synthesized at saturation by the two types of ribosomes, namely after 30-min incubation at 37°C (A) and after 120 min at 15°C (B). Error bars, sd of the calculated ratios. Further details are given in Materials and Methods.
Cold shock induces a stoichiometric imbalance between initiation factors and ribosomes
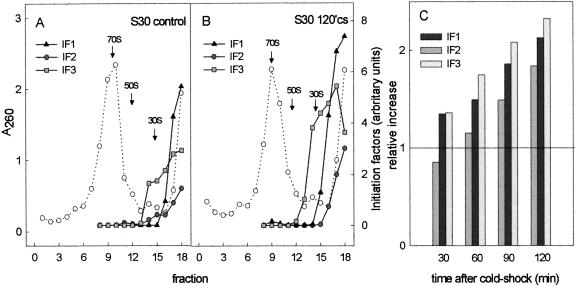
Translation initiation factor IF2 has been identified among the cold shock-induced proteins (Jones et al. 1987; Jones and Inouye 1994), but the functional significance of its induction is unclear. In light of this, we investigated the possibility that cold shock might also cause variations of the levels of IF1 and IF3. For this purpose, the S30 extracts from cells harvested immediately before (control) and 30, 60, 90, and 120 min after cold shock at 10°C were fractionated by sucrose density gradient centrifugation, and the gradient fractions were subjected to PAGE-SDS electrophoreses to resolve IF2 and IF1/IF3. The gels were then subjected to Western blotting, followed by immunodetection and quantification of the amount of the initiation factors present in each sample. The results presented in Figure 6 ▶ refer to samples taken before (Fig. 6A ▶) and after 2 h (Fig. 6B ▶) of cold shock. As seen in the figure, the levels of loosely ribosome-bound and free factors are substantially increased after the cold stress. Similar quantifications carried out on extracts prepared from cells taken after different times of cold shock showed that whereas the level of ribosomes remains more or less constant, the levels of the three initiation factors progressively increase as a function of the time of cold shock (Fig. 6C ▶). As a result, 2 h after the cold stress these levels, normalized to the amount of ribosomes, are approximately two times higher than in the pre-cold-shock controls. Separate experiments also indicated that at later times of cold acclimation, these levels may continue to increase up to ≅ threefold above the basal level (data not shown).
FIGURE 6.
The levels of the three initiation factors IF1, IF2, and IF3 increase after cold shock and determine an imbalance of the IF/ribosome stoichiometric ratio. S30 extracts prepared from control (A) and cold-shocked (120 min) (B) cells were fractionated by centrifugation on 10%–30% sucrose density gradients. The amount of each initiation factor present in the resulting gradient fractions (left to right from bottom to top of gradient) were determined following SDS-PAGE, Western blotting, and immunoquantification as described in Materials and Methods. An analysis similar to that shown in panels A and B was also carried out on S30 extracts derived from cells cold-shocked for 30, 60, and 90 min. (C) The total levels (i.e., the sum of the values of each gradient fraction) of IF1 (black), IF2 (dark gray), and IF3 (light gray), normalized with respect to the amount of ribosomes in each extract, as a function of the time of cold shock. The data in C do not contain error bars, as the quantification yielded only one value for each initiation factor at each indicated time of cold shock. However, similar experiments were carried out three times for IF1 and four times for IF2 and IF3, producing essentially the same results, namely, after 2-h cold shock at 10°C, the relative increases with respect to the pre-cold-shock levels were the same as those shown in C (±20% for IF3, ±25% for IF1 and IF2).
Inspection of the histogram in Figure 6C ▶ would indicate a lower increase and an overall lower level of IF2 compared to IF1 and IF3. It should be noted, however, that the reported quantification of IF2 and of its increase has taken into account only the longer form of the factor (IF2α), because a precise quantification of the shorter form IF2β was made difficult by the presence of an immunological cross-reaction with proteins having an electrophoretic mobility similar to that of IF2β. However, because the cold shock induces an increase of IF2β which is larger than that of IF2α, the total increase in the level of IF2 as a function of the time of cold shock is underestimated in Figure 6C ▶ by at least 30%.
Thus, taken together, these data demonstrate that the IF/ribosome ratio is not always maintained at a fixed level (i.e., ~0.15; Howe and Hershey 1983) but that cold shock causes a large imbalance of this ratio, which becomes ≥0.3 during the cold-acclimation phase. However, because the relative levels of the three initiation factors remain constant, our results indicate that their coordinate expression is not lost upon cold shock.
In light of these results, the following experiments were designed to investigate whether the increased levels of initiation factors could determine a cold-shock translational bias in favor of cold-shock mRNAs. For this purpose we analyzed the effect of increasing amounts of each initiation factor on the translational activity of control 70S ribosomes (high-salt washed and isolated from sucrose density gradient) programmed with different types of mRNAs. Considering the amount of these initiation factor-free ribosomes present in each reaction mixture, we estimate that an addition of 3 pmoles of each factor restores their normal in vivo levels, whereas adding 6–9 pmoles mimics the IF/ribosome ratio ensuing 2–4 h after cold shock.
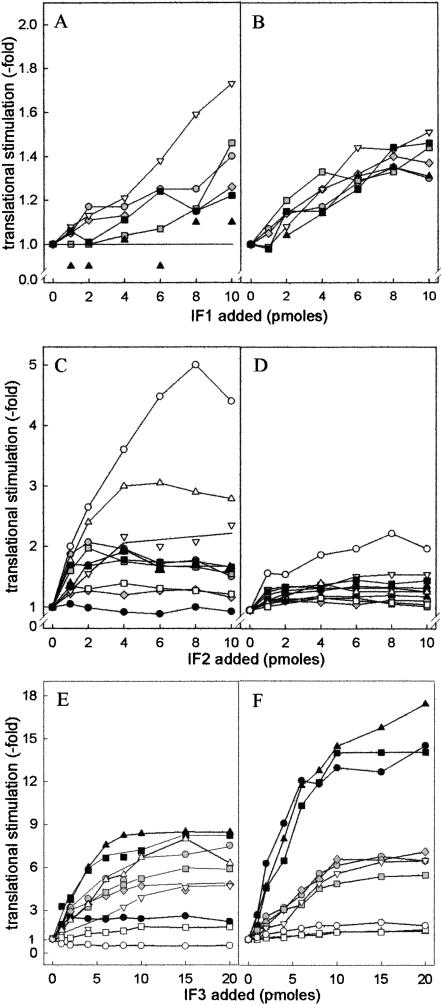
As seen from the results presented in Figure 7 ▶, increasing levels of IF1 produced only a modest translational stimulation. At 37°C, the stimulation seems to favor primarily the translation of the non-cold-shock hupA mRNA, whereas that of other mRNAs, cspA mRNA in particular, is not or is only marginally affected (Fig. 7A ▶). In the cold, IF1 stimulates (30%–40%) the translation of all mRNAs without preference (Fig. 7B ▶). Regarding IF2, at 37°C (Fig. 7C ▶) increasing amounts of this factor clearly stimulate translation of the mRNA beginning with the noncanonical AUU triplet (≥ fivefold) and of the other non-cold-shock cspD and hupA mRNAs (two–threefold), but affect either marginally or not at all the translation of cold-tolerant and cold-shock mRNAs. At low temperature, on the other hand, increasing amounts of IF2 do not produce any particular effect on the translation of any mRNA, with the notable exception of the infA*AUU mRNA, whose translational efficiency is almost doubled (Fig. 7D ▶). Finally, increasing amounts of IF3 affect to variable extents translation at 37°C, the maximum stimulation being observed with cspA mRNA (~eightfold), whereas the translation of other mRNAs is either not influenced (infA*AUG mRNA) or inhibited, as expected, in the case of infA*AUU mRNA (Fig. 7E ▶). However, in sharp contrast to the nonselective effects produced by IF3 at 37°C, the effects of this factor are clearly selective at 15°C (Fig. 7F ▶), the translation of the cold-shock mRNAs (cspA, cspAΔ1-80, and hns mRNA) being stimulated ≥ 15-fold, that of the “cold-tolerant” hupB mRNAs ~sixfold, and that of the non-cold-shock infA*, and cspD mRNAs ≤ twofold.
FIGURE 7.
Effect of individual initiation factors on the translation of cold-shock, cold-tolerant, and non-cold-shock mRNAs by 70S ribosomes at high and low temperature. Translation of cold-shock, cold-tolerant, and non-cold-shock mRNAs was performed under the conditions described in Materials and Methods with 70S ribosomes (20 pmoles) programmed with the indicated mRNAs (20 pmoles each) in the presence of S100 postribosomal fraction from control cells supplemented with 20 pmoles each of IF2 and IF3 (A,B), IF1 and IF3 (C,D), or IF1 and IF2 (E,F). In addition, each reaction mixture contained the amounts of IF1 (A,B), IF2 (B,C), or IF3 (D,E) indicated in the corresponding abscissa. Incubation was performed for 30 min at 37°C (A,C,E) or for 120 min at 15°C (B,D,F). For visualization of the translational stimulation produced by the increasing amounts of each factor, the results are presented as the ratio of the product obtained for each amount of IF1, IF2, and IF3 versus the basal level obtained in their absence. Cold-shock: cspAwt (▴), cspAΔ1-80 (▪), hns (•); cold-tolerant (gray symbols): P4hupB (♦), P3hupB (•), P2hupB (▪); non-cold-shock (white symbols): infA*AUU (○), infA*AUG (□), cspD (▵), hupA (▿). It should be noted that the translational stimulation displayed by each initiation factor is plotted using different scales in the ordinates.
In conclusion, our results indicate that although the increased level of IF2 is not responsible for the cold-shock translational bias, the increased levels of both IF3 and IF1 can substantially contribute to this phenomenon: IF3 by preferentially stimulating cold-shock mRNAs translation and IF1 by enhancing the effect of IF3 without influencing translational specificity. It is also noteworthy that the amounts of IF1 and IF3 eliciting the maximum translational stimulation in the cold correspond to the levels reached by these factors in vivo after cold shock.
Effect of antibiotics on cold-shock mRNA translation
In vivo translation of cold-shock transcripts such as cspA, cspB, and cspG mRNAs was reported to be insensitive at low temperature to both kanamycin and chloramphenicol, under conditions which normally completely block protein synthesis (Etchegaray and Inouye 1999). This surprising finding was attributed to the presence in these mRNAs of a downstream box (DB) which would permit bypassing the inhibition and was taken as compelling evidence that induction of the major cold-shock genes does not require the synthesis of new proteins. Alternatively, it was suggested that, due to its small size, the mRNAs transcribed from the csp genes represented a target too miniscule for these antibiotics (Etchegaray and Inouye 1999).
Kanamycin and chloramphenicol hit different targets on the ribosome (the 30S and 50S subunits, respectively) and inhibit different steps of translation (Gale et al. 1981). To be resistant to both of these antibiotics, an mRNA ought to be translated by a mechanism substantially different from the established one. To investigate whether this intriguing possibility could be true in the special case of translation at low temperature of a cold-shock mRNA by the cold shock-modified translational apparatus, we investigated the effects of kanamycin and chloramphenicol on the in vitro translation of a cold-shock (cspA) and a non-cold-shock (hupA) mRNA. As seen in Table 3 ▶, the IC50 determined for the two mRNAs are remarkably similar with both antibiotics and, if anything, the translation of cspA mRNA is somewhat more sensitive to the inhibition in the cold than at higher temperature. Furthermore, translation of cspA mRNA is somewhat more sensitive to kanamycin (IC50 ≥ threefold lower) than that of hupA mRNA. Thus, we conclude that translation at low temperature of the paradigm cold-shock transcript cspA mRNA is not unlike that of a non-cold-shock mRNA, at least as far as antibiotic sensitivity is concerned. Although a large number of hypotheses could be formulated, we are unaware of a satisfactory explanation for the conflicting results of Etchegaray and Inouye (1999).
TABLE 3.
Effects of antibiotics on the translation of cold-shock and control mRNAs
| mRNA | Temperature | Kanamycin IC50 (μM) | Chloramphenicol IC50 (μM) |
| cspA | 37°C | 4.8 | 3.8 |
| 15°C | 1.9 | 2.4 | |
| hupA | 37°C | 14.5 | 2.9 |
| 15°C | 19.0 | n.d. |
Experimental details are given in the text and in Materials and Methods.
n.d. = not done.
DISCUSSION
The experiments described here established that the translational bias which is normally observed in vivo after cold shock can be reproduced in vitro. We have shown in fact that the translational apparatus of cold-shocked cells translates at low temperature (15°C) cold-shock and, to a lesser extent, cold-tolerant mRNAs at a faster rate and to a higher level than non-cold-shock mRNAs, whose translation is sometimes inhibited. The preference in favor of cold-shock and cold-tolerant mRNAs is amplified within the first 2 h of cell exposure to the cold stress, reproducing the in vivo event. We also assessed the existence of cis-acting elements conferring to the mRNAs the capacity of being translated in the cold. Perhaps not surprisingly, this capacity was found to be maximal in the cold-shock mRNAs (cspA and hns), somewhat less in the cold-tolerant hupB transcripts, and lowest in the non-cold-shock mRNAs (hupA and cspD). These templates were also found to follow the same order in their responsiveness to trans-acting elements which determine their capacity to be preferentially translated in the cold by the cold-shocked translational apparatus.
As to the nature of the cis-elements, it is likely that these are constituted by elements of secondary/tertiary structure more than by sequence motifs. For this reason their identification may represent a difficult and elusive task, at least until the 3D structures of the 5′ UTRs of some of the most representative cold-shock mRNAs have been experimentally elucidated. Nevertheless, our results suggest that, at least in the case of cspA mRNA, they are in part located in the upstream half (nucleotides 1–80) of the 5′ UTR, as they seem to be partially lost in cspAΔ1-80 mRNA.
As to the nature of the trans-acting factors associated with the translational apparatus of cold-shocked cells, we found that cold shock causes a strong imbalance of the initiation factors/ribosomes ratio, and the increased level of IF3 was identified as being the most important trans-acting element capable of conferring translational selectivity in the cold in favor of cold-shock and, to a lesser extent, of cold-tolerant mRNAs. This effect stems from the exceptionally high (≥ 15-fold) IF3 dependence of the cold-shock mRNA translation in the cold. In comparison, translation of the “cold-tolerant” hupB mRNAs displays a lower IF3 dependence (~sixfold), and that of non-cold-shock mRNAs is almost completely insensitive to the presence of IF3. Regarding the mechanism responsible for this discrimination by IF3, we can speculate that the first-order isomerization of the “30S pre-ternary complex”, which leads to “30S initiation complex” formation and is kinetically controlled by IF3 (Gualerzi et al. 2001) is rate-limiting, especially at low temperature, for the translation of cold-shock mRNAs. Indeed, formation at low temperature of a translation initiation complex with cspA mRNA was found to be strongly stimulated by IF3, unlike the formation of an equivalent complex programmed with cspD mRNA, which was strongly inhibited by this factor (A.M. Giuliodori, C.O. Gualerzi, and C.L. Pon, unpubl.).
Concerning the role of the other two initiation factors, the increased level of IF1 could contribute to the cold-shock translational bias by potentiating the effect of IF3 with an ~0.4-fold stimulation, whereas a contribution of IF2 seems very unlikely in light of the fact that this factor was found to produce little or no stimulation of mRNA translation in the cold.
The unexpected finding of the cold-shock induction of all three initiation factors, whose levels increased by more than twice 2 h after cold shock and by three times after 4 h, deserves some additional comment. Because the levels of the initiation factors increase as a function of the time elapsed after cold shock both in absolute terms and with respect to the amount of ribosomes, this occurrence causes an imbalance of the IF/ribosome ratio and shatters the dogma stating that this ratio always remains constant (Howe and Hershey 1983). In turn, this finding raises an important question concerning the mechanism which generates this transient imbalance. The best explanation that we can offer is that immediately after cold shock, ribosome assembly slows down or stops completely, while translation of preexisting and/or de novo transcribed mRNAs encoding the initiation factors continues at a sustained rate. Preliminary results obtained in our laboratory fully support this hypothesis.
Our data also show that the relative stoichiometry of IF1, IF2, and IF3 is not altered following the cold shock, clearly demonstrating that the cold stress does not alter the coordinate expression of infA, infB, and infC at either the transcriptional or posttranscriptional level. The genes encoding the initiation factors are very differently organized in different parts of the chromosome. Thus, the genetic mechanism which ensures the expression of equivalent amounts of the three factors, even under conditions which induce extensive reprogramming of the genetic expression, remains a challenging dilemma to be resolved.
In addition to the initiation factors, we have identified the nucleic acid-binding protein CspA among the trans-acting factors present in higher amounts in the extracts of cold-shocked cells and capable of influencing translation. CspA was found to produce a very modest translational stimulation at 37°C and a larger (~2.5-fold) stimulation at low temperature. The most likely mechanism by which CspA could stimulate mRNA translation could be that of favoring unstructured mRNA conformations, especially in the cold, thereby facilitating their interaction with the ribosomes. This premise is in line with the single-stranded RNA binding properties of CspA and with its suggested role of RNA chaperone (Jiang et al. 1997; Brandi et al. 1999). Despite the absence of mRNA selectivity displayed by CspA in its stimulation, we suggest that this protein, like IF1, may also contribute to the translational bias by providing additional stimulation of the expression of those mRNAs which are efficiently translated in the cold.
Even though several elements capable of stimulating translation of cold-shock mRNAs in the cold were identified in this study, it seems likely that additional factors may exist which contribute to the translational bias. Future research should focus on pinning down the nature of the factors responsible for repressing translation of non-cold-shock mRNAs in the cold during the initial phases of cold acclimation, the elusive nature of the cis-elements present in cold-shock mRNAs, and the subtle structural differences which distinguish normal and cold-shock ribosomes. Indeed, in line with the suggestion that the ribosome may be the temperature sensor of the cell and direct the preferential translation of heat or cold-shock proteins, respectively (VanBogelen and Neidhardt 1990), it is clear that even after high-salt washing or after adding a full complement of initiation factors the cold-shock and control ribosomes translate different types of mRNAs with different and divergent efficiency. It is tempting to speculate that structural modifications of the ribosomes are capable of enhancing the effects of both cis- and trans-acting elements on the translational bias.
MATERIALS AND METHODS
Buffers
Buffer A: 10 mM Tris-HCl, pH 7.7, 60 mM NH4Cl, 10 mM Mg acetate, 1 mM DTT.
Buffer B: 10 mM Tris-HCl pH 7.7, 650 mM NH4Cl, 10 mM Mg acetate, 1 mM DTT.
Buffer C: 50 mM Tris-HCl, pH 8.3, 80 mM glycine, 0.04% SDS, 20% methanol.
Buffer D: 20 mM Tris-HCl, pH 7.5, 137 mM NaCl, 5% BSA, 0.1% Tween 20.
General preparations
Initiation factors IF1, IF2, and IF3 (Pawlik et al. 1981) and CspA (La Teana et al. 1991) were purified as described.
In vitro transcription and mRNA preparation
The templates used for the in vitro synthesis of the various mRNAs were plasmids derived from pTZ18R and pTZ19R (Pharmacia) propagated in and purified from Escherichia coli JM109. More specifically, these plasmids were: pTZ19hupA, pTZ19P4hupB, pTZ19P3hupB, pTZ19P2hupB (Giangrossi et al. 2001), pTZ18cspA(Δ1-80); (Brandi et al. 1996), pTZ18infA*AUG (Calogero et al. 1987), pTZ18infA*AUU (Petrelli et al. 2001), pSelect hns (Spurio et al. 1997), pTZ18cspAwt and pTZ18cspD, the latter two prepared as described below. Preparative transcription of the mRNAs with T7 RNA polymerase and mRNA purification were carried out essentially as described (Brandi et al. 1996).
Construction of pTZ18cspAwt and pTZ18cspD
pTZ18cspAwt was constructed replacing the truncated 5′ UTR of cspA present in pTZ18cspA(Δ1-80); (Brandi et al. 1996) with the entire 5′ UTR of cspA obtained by PCR amplification. CspD was amplified from chromosomal DNA using two primers (5′-CGG GATCCTAGAGTTGGCGAG and 5′-CCCAAGCTTCTCTGGCA CGAG), which created BamHI and HindIII recognition sites so as to allow the cloning of the fragment in the corresponding sites of pTZ18R. The forward primer annealed to the region containing the transcription initiation site, and the reverse primer annealed to a region located 77 bases downstream of the cspD termination codon.
Preparation of S30 fractions, ribosomes, and postribosomal supernatants (S100)
E. coli MRE600 cells were grown in LB supplemented with 0.5% glucose at 37°C to A620 = 1.2, and the cultures were divided into two portions, one harvested immediately and the other shifted to 10°C and harvested 30, 60, 90, and 120 min after the cold shock. Cell-free (S30) extracts, crude (unwashed) ribosomes, and postribosomal (S100) supernatants were prepared essentially as described (Brandi et al. 1996). The ribosome content of the cell-free extracts was determined by subjecting each S30 fraction to analytical sucrose density gradient (10%–30% in Buffer A) centrifugation in an SW60 rotor. For many experiments (as indicated in the appropriate figure legends) the ribosomes were purified by preparative sucrose density gradient centrifugations (15,000 rpm for 17 h at 4°C in the SW28 rotor) of the S30 extracts. After pooling of the appropriate gradient fractions, the 70S particles were pelleted by centrifugation for 15 h at 40,000 rpm at 4°C in a 50Ti rotor. Washed (initiation factor-free) ribosomes were obtained by resuspending and pelleting twice the ribosomes in a high-salt buffer (Buffer B). The 70S pellets were resuspended in Buffer A and stored in small aliquots at −80°C.
Preparation of an S30 fraction from cells carrying extra copies of cspA
E.coli K12ΔH1Δtrp cells transformed with pCspA1 or with pPLc2833 (Brandi et al. 1996) were grown at 30°C in LB containing ampicillin (60 μg/mL). To induce cspA expression, the cultures (at A620 = 0.75) were transferred to 42°C for 10 min and then to 37°C for 30 min. The cells were then harvested, washed in Buffer A, and lysed by sonication to prepare S30 cell-free extracts as described above.
In vitro translation
In vitro translation reactions were carried out in mixtures containing 30 mM Tris-HCl, pH 7.7, 10 mM Mg acetate, 75 mM NH4Cl, 2 mM DTT, 2 mM ATP, 0.4 mM GTP, 10 mM phosphoenolpyruvate, 0.025 μg of pyruvate kinase/μL reaction, 200 μM of each amino acid (minus Lys), 25 μM [14C] Lysine (309 mCi/mmol), 1 μg/μL reaction of E. coli MRE600 tRNA mixture, and 0.12 mM citrovorum (Serva). The reaction mixture also contained the in vitro transcribed mRNAs and either S30 fractions (normalized for their ribosome content) or 70S ribosomes and S100 postribosomal supernatant, in the amounts indicated in each experiment. After incubation for the indicated times and temperatures, samples (10-μL) were withdrawn from each reaction mixture and spotted on 3MM paper discs for determination of the hot-trichloroacetic acid (TCA)-insoluble radioactivity incorporated. Separate experiments indicated that the radioactivity incorporated measured by this method reflects the actual amount of electrophoretically resolved protein product.
Immunoquantification of the initiation factors
Aliquots of S30 extracts corresponding to 100 pmoles of 70S ribosomes were loaded onto sucrose density gradient (10%–30% in Buffer A) and centrifuged at 4°C for 2 h at 40,000 rpm in an SW60 rotor. Gradient fractions were then subjected to TCA precipitation, and the resulting pellets, resuspended in PAGE/SDS sample buffer, were subjected to PAGE/SDS at 7.5% and 15% polyacrylamide for the detection of IF2 and IF1/IF3, respectively. The proteins were electroblotted in Buffer C onto a cellulose nitrate membrane in a Trans-Blott cell (Biorad). To detect IF1, IF2, and IF3, the membranes were incubated with the corresponding polyclonal antibodies in Buffer D and revealed with Alkaline Phosphatase Conjugated anti-Rabbit IgG (Promega) using NBT/BCIP (Promega) as substrate. Quantification of Western blots was carried out by densitometric scanning using a Molecular Imager System GS-250 (Biorad).
Stability of mRNA under in vitro translational conditions
In vitro transcription was carried out in 50-μL reaction mixtures containing 40 mM Tris-HCl, pH 8.0, 15 mM MgCl2, 10 mM NaCl, 2 mM Spermidine, 10 mM DTT, 2.5 μg BSA, 20 U placental RNase inhibitor, 3.75 mM ATP, UTP, and CTP, 1.4 mM cold GTP, 50 μCi [α-32P] GTP (5000Ci/mmol), 25 U of purified T7 RNA polymerase, and 2 μg of pTZ18cspA wt, pTZ18cspA (Δ1-80), or pTZ18cspD as template. The reactions were incubated for 2 h at 37°C, and the transcripts were purified by LiCl precipitation followed by ammonium acetate-ethanol precipitation. Aliquots corresponding to 40 pmoles of each radiolabeled transcript were added to 90 μL of in vitro translation reaction mixtures containing the same components as those described in the Figure 1 ▶ legend except that [14C]-Lysine was replaced by cold lysine. After incubation for various times at either 37°C or 15°C, samples (5-μL) were withdrawn from each reaction mixture and immediately frozen in liquid N2.
The RNA was then extracted with phenol:chloroform, precipitated with sodium acetate (pH 5.2) and ethanol in the presence of 1 μg tRNA carrier, resuspended in formamide loading buffer, and analyzed by electrophoresis on 5% PAGE-urea gel. The amounts of cspA, cspA(Δ1-80), and cspD mRNA present in each sample were quantified with the Molecular Imager, and the in vitro half-lives were calculated.
Acknowledgments
This work was supported by MIUR grants PRIN 2001 (to C.O.G. and C.L.P.) and F.I.R.B. (to C.L.P.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5164904.
REFERENCES
- Brandi, A., Pietroni, P., Gualerzi, C.O., and Pon, C.L. 1996. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 19: 231–240. [DOI] [PubMed] [Google Scholar]
- Brandi, A., Spurio, R., Gualerzi, C.O., and Pon, C.L. 1999. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under nonstress conditions. EMBO J. 18: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero, R.A., Pon, C.L., and Gualerzi, C.O. 1987. Chemical synthesis and in vivo hyperexpression of a modular gene coding for Escherichia coli translational initiation factor. Mol. Gen. Genet. 208: 63–69. [DOI] [PubMed] [Google Scholar]
- Claret, L. and Rouviere-Yaniv, J. 1997. Variation in HU composition during growth of Escherichia coli: The heterodimer is required for long term survival. J. Mol. Biol. 273: 93–104. [DOI] [PubMed] [Google Scholar]
- Etchegaray, J.P. and Inouye, M. 1999. CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J. Bacteriol. 181: 1827–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farewell, A. and Neidhardt, F.C. 1998. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J. Bacteriol. 180: 4704–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, E.F., Cundliffe, E., Reynolds, P.E., Richmond, M.H., and Waring, M.J. 1981. Antibiotic inhibitors of ribosome function. In Molecular basis of antibiotic action, pp. 492–501. Wiley & Sons, London.
- Giangrossi, M., Exley, R.M., Le Hegarat, F., and Pon, C.L. 2001. Different in vivo localization of the Escherichia coli proteins CspD and CspA. FEMS Microbiol. Lett. 202: 171–176. [DOI] [PubMed] [Google Scholar]
- Giangrossi, M., Giuliodori, A.M., Gualerzi, C.O., and Pon, C.L. 2002. Selective expression of the β-subunit of nucleoid-associated protein HU during cold shock in Escherichia coli. Mol. Microbiol. 44: 205–216. [DOI] [PubMed] [Google Scholar]
- Goldenberg, D., Azar, I., Oppenheim, A.B., Brandi, A., Pon, C.L., and Gualerzi, C.O. 1997. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol. Gen. Genet. 256: 282–290. [DOI] [PubMed] [Google Scholar]
- Goldstein, J., Pollitt, N.S., and Inouye, M. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. 87: 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, C.O., Brandi, L., Caserta, E., Garofalo, C., Lammi, M., La Teana, A., Petrelli, D., Spurio, R., Tomsic, J., and Pon, C.L. 2001. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harbor Symp. Quant. Biol. 66: 363–376. [DOI] [PubMed] [Google Scholar]
- Gualerzi, C.O., Giuliodori, A.M., and Pon, C.L. 2003. Transcriptional and posttranscriptional control of cold-shock gene expression. J. Mol. Biol. 331: 527–539. [DOI] [PubMed] [Google Scholar]
- Howe, J.G. and Hershey, J.W. 1983. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J. Biol. Chem. 258: 1954–1959. [PubMed] [Google Scholar]
- Jiang, W., Fang, L., and Inouye, M. 1996. Complete growth inhibition of Escherichia coli by ribosome trapping with truncated cspA mRNA at low temperature. Genes Cells 1: 965–976. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Hou, Y., and Inouye, M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272: 196–202. [DOI] [PubMed] [Google Scholar]
- Jones, P.G. and Inouye, M. 1994. The cold-shock response-a hot topic. Mol. Microbiol. 11: 811–818. [DOI] [PubMed] [Google Scholar]
- Jones, P.G., VanBogelen, R.A., and Neidhardt, F.C. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169: 2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana, A., Brandi, A., Falconi, M., Spurio, R., Pon, C.L., and Gualerzi, C.O. 1991. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. 88: 10907–10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik, R.T., Littlechild, J., Pon, C., and Gualerzi, C. 1981. Purification and properties of Escherichia coli translational initiation factors. Biochem. Int. 2: 421–421. [Google Scholar]
- Petrelli, D., La Teana, A., Garofalo, C., Spurio, R., Pon, C.L., and Gualerzi, C.O. 2001. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J. 20: 4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurio, R., Falconi, M., Brandi, A., Pon, C.L., and Gualerzi, C.O. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 18: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen, R.A. and Neidhardt, F.C. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. 87: 5589–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka, K., Mitta, M., and Inouye, M. 1999. Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181: 6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]