Abstract
Retroviruses specifically package two copies of their RNA genome in each viral particle, along with some small cellular RNAs, including tRNAs and 7S L RNA. We show here that Rous sarcoma virus (RSV) also packages U6 snRNA at approximately one copy per virion. In addition, trace amounts of U1 and U2 snRNAs were detected in purified virus by Northern blotting. U6 snRNA comigrated with the RSV 70S genomic RNA dimer on sucrose gradients. We observed reverse transcription of U6 snRNA in an endogenous reaction in which RSV particles were the source of both reverse transcriptase and RNA substrates. This finding led us to examine mammalian genomic sequences for the presence of snRNA pseudogenes. A survey of the human, mouse, and rat genomes revealed a high number of spliceosomal snRNA pseudogenes. U6 pseudogenes were the most abundant, with approximately 200 copies in each genome. In the human genome, 67% of U6 snRNA pseudogenes, and a significant number of the other snRNA pseudogenes, were associated with LINE, SINE, or retroviral LTR repeat sequences. We propose that the packaging of snRNAs in retroviral particles leads to their reverse transcription in an infected cell and the integration of snRNA/viral recombinants into the host genome.
Keywords: U6 snRNA, retrovirus packaging, reverse transcription, snRNA pseudogenes
INTRODUCTION
Rous sarcoma virus (RSV) is a simple retrovirus that replicates in avian cells (Coffin 1996). It contains two identical copies of unspliced 9-kb genomic RNA inside its virion (Beemon et al. 1974; Kung et al. 1975). These RNA molecules are the templates for reverse transcription, the process by which RNA is copied into DNA by the packaged viral enzyme, reverse transcriptase (RT; for review, see Telesnitsky and Goff 1997). Reverse transcription occurs in the cytoplasm of a newly infected cell. The resulting double-stranded viral DNA is frequently a recombinant, due to an RT copy choice mechanism by which the enzyme jumps between templates (Hu and Temin 1990). The reverse-transcribed viral DNA, flanked by long terminal repeats (LTRs), is integrated into the host cell genome. If this occurs in a germ-line cell, the integrated provirus can be passed on vertically to subsequent generations. These retroviruses can ultimately become fixed in the population and are known as endogenous retroviruses. It is estimated that endogenous retroviruses account for about 8% of the human genome (Bock and Stoye 2000; Griffiths 2001; Lander et al. 2001).
In addition to the two viral genomic RNAs, retroviral virions contain nonviral RNAs, including ~ 50–100 copies of various tRNAs (Faras et al. 1973; Sawyer and Dahlberg 1973). Tryptophan tRNA base-pairs with the RNA genome at the primer binding site and is used as the primer for RSV reverse transcription (Faras et al. 1974; Sawyer et al. 1974). Viral particles have also been shown to contain cellular 7S L RNA (Chen et al. 1985) and some cellular mRNAs (Adkins and Hunter 1981). Many of these RNAs are reverse-transcribed and can recombine with the RSV genome (Dunn et al. 1992; Hajjar and Linial 1993). Furthermore, the SE21Q1b mutant virus, which lacks an RNA packaging signal (Shank and Linial 1980), can create processed pseudogenes from packaged cellular RNAs (Linial 1987).
Processed pseudogenes are typically created when RNA is copied into DNA and integrated into the genome via the enzymes coded for by a long interspersed nuclear element (LINE; Harrison and Gerstein 2002). LINEs are non-LTR retrotransposable elements that replicate via an RNA intermediate and jump throughout the human genome (Weiner 2002). LINEs encode a bifunctional polypeptide that contains both RT and DNA endonuclease activities, which catalyze the retrotransposition (Esnault et al. 2000). The L1 LINE can also transduce cellular sequences during retrotransposition (Pickeral et al. 2000). Short interspersed nuclear elements (SINEs) are thought to utilize LINE proteins in trans for their replication (Okada et al. 1997; Ogiwara et al. 1999, 2002). Reverse transcription of the 7S L RNA generates the Alu element, the most abundant SINE in human cells (Weiner 2002). These two classes of repetitive elements account for at least 34% of the human genome (Lander et al. 2001). Thus, reverse transcription and integration of retroelements can create multiple copies of cellular genes.
We report here that an average of one U6 snRNA is packaged in each RSV particle in association with the 70S viral genomic RNA. We also observed trace amounts of packaged U1 and U2 snRNAs by Northern blotting. Packaged U6 was reverse-transcribed by RSV RT in an endogenous reaction. We asked whether snRNAs might be integrated into the host genome, possibly in association with viral sequences. We observed that multiple copies of U1, U2, U4, U5, and U6 snRNA pseudogenes are present in the human, mouse, and rat genomes. Many of these spliceosomal snRNA genes are associated with LINEs, SINEs, or endogenous retroviral LTRs. The genomic association of snRNA sequences with retroviral sequences suggests that reverse-transcribed snRNAs may recombine with retroviral sequences in the viral particle and integrate together into the host cell genome.
RESULTS
U6 snRNA is selectively packaged into RSV virions
To further investigate the presence of small nonviral RNAs in RSV particles, we 3′ end-labeled total virion RNA with [32P]pCp. After fractionation of the end-labeled RNAs on an acrylamide gel, we observed multiple species between 100 and 200 nucleotides (data not shown). Because we had previously identified a negative regulator of splicing (NRS) element in RSV RNA that binds U1 and U11 snRNAs (Gontarek et al. 1993; Hibbert et al. 1999; McNally and McNally 1999), we asked whether any of the spliceosomal snRNAs were packaged in viral particles. To identify the packaged RNA, we purified wild-type RSV by pelleting and sucrose gradient sedimentation. RNA was extracted from virions and subjected to Northern analysis using probes complementary to spliceosomal snRNAs. As a negative control, media from uninfected cells was put through this same protocol.
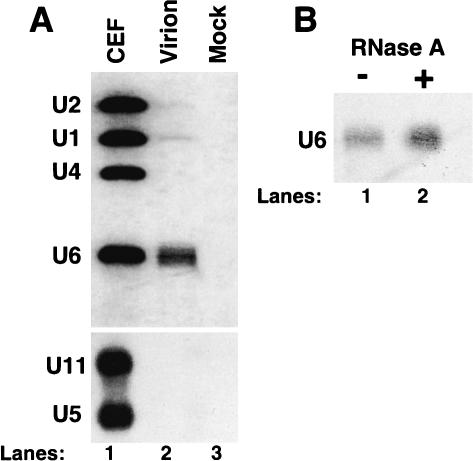
U6 snRNA was readily detected in viral particles by Northern analysis, whereas U1 and U2 snRNAs were barely detectable (Fig. 1A ▶). Comparison of the relative abundance of the different snRNAs in total chicken cellular RNA and in packaged virion RNA by Northern blotting (Fig 1A ▶, lanes 1,2) showed that U6 snRNA was selectively packaged in RSV particles relative to the other snRNAs. U4, U5, and U11 snRNAs were not detected in virions by Northern analysis (Fig. 1A ▶).
FIGURE 1.
U6 snRNA is packaged into RSV particles. (A) snRNAs were identified by Northern blotting with snRNA riboprobes after extraction from chicken embryo fibroblasts (CEFs; lane 1), from gradient-purified viral particles (lane 2), and from mock-infected particles (lane 3). U11 and U5 snRNAs were analyzed in a separate blot. (B) Purified RSV virions were incubated at 37°C for 60 min in the presence (lane 2) or absence (lane 1) of RNase A prior to RNA extraction and Northern analysis.
To confirm that the U6 snRNA was packaged inside the virions, rather than being associated on the outside, the viral particles were treated with RNase A prior to extraction of nucleic acids (Fig. 1B ▶, lane 2). As a control, virus was incubated in the absence of RNase A (Fig. 1B ▶, lane 1). This treatment showed that the virion-associated U6 snRNA was resistant to RNAse A digestion and therefore was packaged inside the virions.
To more accurately measure the relative amounts of packaged RSV genomic RNA and U6 snRNA in gradient-purified virus, we used an RNase protection assay with two probes assayed simultaneously. Quantitation of the viral and U6-protected probe bands revealed 1.09 ± 0.26 molecules of U6 snRNA per virion (data not shown), which had previously been shown to contain two identical molecules of genomic RNAs (Beemon et al. 1974; Kung et al. 1975).
U6 snRNA is associated with the viral genomic RNA inside virus particles
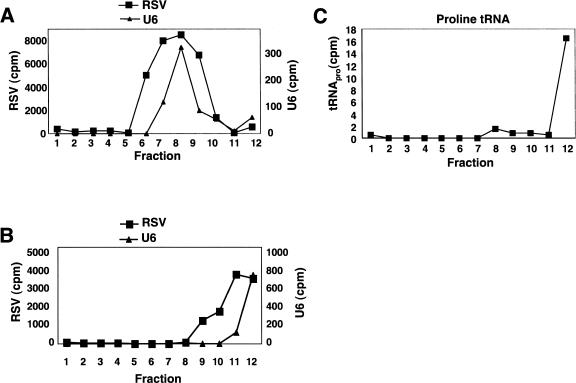
Because viral assembly is thought to occur at the plasma membrane (Swanstrom and Wills 1997) and U6 snRNA has no characterized cytoplasmic role (Will and Luhrmann 2001), it was surprising that U6 was preferentially packaged. As the other snRNAs, which pass through the cytoplasm, were much less abundant in virions, we asked what interactions led to U6 being packaged. To address this issue, we used velocity sedimentation analysis of virion RNA. Total RNA was extracted from viral particles and fractionated on a sucrose gradient without prior denaturation. The RNA content of individual gradient fractions was analyzed by RNase protection, using two antisense riboprobes specific for the viral and the U6 RNA. The sedimentation profiles of U6 snRNA and RSV genomic RNA were nearly identical (Fig. 2A ▶), suggesting that these RNAs were associated within the viral particle. When the virion RNAs were denatured by heating prior to gradient sedimentation, the viral RNA and U6 snRNA no longer comigrated, suggesting that they had been dissociated (Fig. 2B ▶). The U6 RNA was present at the top of the gradient (fractions 11 and 12), whereas the genomic RNA monomers were in fractions 9–12 (Fig. 2B ▶).
FIGURE 2.
RSV RNA and U6 snRNA comigrate during velocity sedimentation. (A) Native virion RNA was sedimented through a 10%–25% sucrose gradient. The RNA in each fraction was analyzed by an RNase protection assay with both viral and U6 specific probes. (B) The virion RNA was denatured by heating at 95°C for 5 min before layering onto the gradient. Sedimentation conditions and the RNase protection assay were performed as in A. (C) Native virion RNA was centrifuged as in A, and a Northern blot was performed to detect tRNApro.
As a control, we monitored the sedimentation profile of proline tRNA (which is similar in size to U6 snRNA) extracted from the same virion particles without denaturation. Although it is packaged, proline tRNA has no known interaction with RSV genomic RNA (Panet et al. 1978). Accordingly, the tRNA was observed in the top fraction of the gradient (Fig. 2C ▶). Our lab previously characterized a cis-acting NRS element within the gag gene of avian retroviruses that suppresses splicing to the src 3′ splice site and promotes polyadenylation (Arrigo and Beemon 1988; McNally et al. 1991; Hibbert et al. 1999; O’Sullivan et al. 2002; Polony et al. 2003). The NRS contains a 5′ splice site-like sequence that has 12 out of 13 nucleotides (nt) complementary to U6 snRNA (nt 33–45). Thus, we asked whether the packaged U6 snRNA base-paired with this NRS sequence. Viral substitution mutants were generated that disrupted eight of the 12 potential base pairs between the NRS and U6 snRNA without affecting the coding sequence of the overlapping Gag protein. When we performed velocity sedimentation on both wild-type and mutant virion RNAs as described in Figure 2 ▶, similar amounts of U6 snRNA were found comigrating with the viral 70S complex in both cases (data not shown). This indicates that the bulk of the packaged U6 snRNA does not interact with the complementary NRS region of RSV RNA.
U6 snRNA is reverse-transcribed in RSV virions
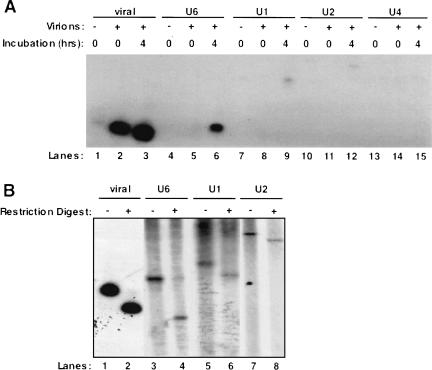
As cellular RNAs packaged in retroviral particles were previously observed to be reverse-transcribed into DNA (Dunn et al. 1992; Hajjar and Linial 1993), we next asked whether the endogenous RSV enzyme could reverse-transcribe the packaged U6 snRNA in detergent-permeabilized virus particles (Temin and Mizutani 1970). A fraction of the product of the endogenous RT assay was used as a template in a series of PCR reactions using primers for either the 5′ end of RSV (the initial “strong-stop” DNA product which is part of the LTR; Telesnitsky and Goff 1997) or for each of the spliceosomal snRNAs. As controls we collected media from uninfected chicken embryo fibroblasts (CEFs; mock) and harvested the nucleic acid from RSV without performing the endogenous RT assay (zero timepoint). It has been reported previously that much of the viral strong-stop DNA product in human immunodeficiency and murine leukemia viruses is generated within the extracellular virion particle (Trono 1992). In agreement with this, we see a large amount of reverse-transcribed RSV strong-stop DNA at the zero timepoint of the assay (Fig. 3A ▶, lane 2). However, the amount of strong-stop reverse transcription increased after a 4-h incubation (Fig. 3A ▶, cf. lanes 2 and 3). The mock treatment also contained a trace amount of viral DNA (Fig. 3A ▶, lane 1).
FIGURE 3.
Packaged snRNAs are reverse-transcribed by RSV in an endogenous RT reaction. (A) RSV virions were partially solubilized with detergent and incubated with dNTPs at 39°C for 4 h or placed immediately at −20.0°C. Media harvested from uninfected CEFs was used as the mock. Total nucleic acid was then harvested from the virions after the incubation and used as the template in a PCR reaction. Primer sets were used to detect the viral minus strand strong-stop, and DNA copies of U6, U1, U2, and U4 snRNAs. (B) The RSV, U6, U1, and U2 PCR products were restriction-digested to confirm their identities. RSV, U1, and U2 were digested with BstN1, and U6 was digested with MboII.
U6 snRNA was not reverse-transcribed in either the mock or the zero timepoint samples, but yielded a strong signal after a 4-h incubation (Fig. 3A ▶, lanes 4–6). U1 and U2 snRNAs had faint signals in both the mock and zero timepoint lanes but had slightly higher signals after 4 h, indicating that they were being reverse-transcribed (Fig. 3A ▶, lanes 7–12). U4 snRNA was not detected in any treatment (Fig. 3A ▶, lanes 13–15). To confirm the identities of the PCR products seen in Figure 3A ▶, restriction enzyme digestion was performed using restriction enzymes specific for each product as described in Materials and Methods (Fig. 3B ▶).
Multiple copies of the spliceosomal snRNA pseudogenes in the mouse, rat, and human genomes
Our finding that packaged snRNAs in RSV particles could be reverse-transcribed (Fig. 3 ▶) led us to wonder whether these DNAs were integrated into the host cell genome. Unfortunately, the chicken genome sequence is not yet complete. However, the finding that U6 snRNA is also packaged by Moloney MuLV (A. Onafuwa and A. Telesnitsky, pers. comm.) suggests that this may be a property shared with mammalian retroviruses. It has been reported that multiple copies of the major spliceosomal snRNA pseudogenes are present in mammalian genomes (Denison et al. 1981; Van Arsdell and Weiner 1984; Lund 1988; Bark and Pettersson 1989; Sibbald and Blencowe 1990). However, a complete determination of the number of these genes was not possible until the recent report of the human genome sequence (Lander et al. 2001). Buzdin and colleagues (2002) also observed numerous U6 pseudogenes in the human genome. We extended this analysis by searching for pseudogenes of all of the major spliceosomal snRNAs in the human, mouse, and rat genomes. Toward this end, we performed a BLAT search of the genomic sequences, using the Human Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway) with a minimum BLAT score of 50 as a cutoff point (Kent 2002; Kent et al. 2002).
By this criterion, we observed pseudogenes for all of the spliceosomal snRNAs in all three species examined (Table 1 ▶). Although U6 is only the third most abundant of the spliceosomal snRNAs in the cell (Yu et al. 1999), we observed the highest number of U6 pseudogenes in all three species analyzed. One hundred ninety copies of U6 snRNA pseudogenes were found in the human genome and 202 copies in both the mouse and rat genomes (Table 1 ▶). We also observed 86 U1 pseudogenes in the human genome, whereas mouse and rat have 60 and 58, respectively. In all three species, the U5 snRNA pseudogenes were the least abundant, varying from 11 to 20 copies. Similarly, U2 pseudogenes ranged from 17 to 41 copies. The number of U4 pseudogenes was more variable between the different species, ranging from 24 in mouse to 122 in rat. It will be interesting to analyze these pseudogenes in the chicken genome after it has been sequenced.
TABLE 1.
Spliceosomal snRNA pseudogenes in the human, mouse, and rat genomes
| Human | Mouse | Rat | |
| U1 | 86 | 60 | 58 |
| U2 | 41 | 17 | 35 |
| U4 | 61 | 24 | 122 |
| U5 | 20 | 11 | 11 |
| U6 | 190 | 202 | 202 |
Many human snRNA pseudogenes are associated with LINEs, SINEs, and LTRs
Next, we asked whether any of the spliceosomal snRNA pseudogenes in the human genome were associated with repeated elements, such as LINEs, SINEs, or retroviral LTRs. We propose that the reverse transcription of packaged snRNAs that we observed above (Fig. 3 ▶) may lead to recombination between retroviral and snRNA sequences through copy-choice reverse transcription of packaged RNAs. This could be followed by integration of snRNA sequences, along with the genome of the host virus, into the cellular genome. To identify such possible recombinants, we performed a BLAT alignment with the RepeatMasker program (A.F.A. Smit and P. Green, unpubl.) within the Human Genome Browser. RepeatMasker uses the RepBase database of repeats to identify repetitive genomic sequences (Jurka 2000).
We noticed that many of the human snRNA loci were associated with LINEs, SINEs, or retroviral LTRs using the RepeatMasker program. Although the majority of such snRNA pseudogenes were directly adjacent to one of these repeat elements, we included repeat elements within 50 bp of the snRNA sequence in our analysis. If two different repeats were within 50 bp of the snRNA sequence, it was scored as associated with the more conserved of the two elements. Finally, if more than two repeats existed near the snRNA it was scored as associated with none. The results of this analysis are summarized in Table 2 ▶. Eighty-four out of 190 U6 pseudogenes observed in the human genome were associated with LINE elements, and 62 were not associated with any known repeats. Buzdin et al. (2002) also observed that many of the U6 pseudogenes they identified were associated with LINEs. We found that 35 of the remaining U6 pseudogenes were associated with SINEs and nine were associated with retroviral LTRs. In contrast, the majority of the other snRNA pseudogenes (80% of U1, 66% of U2, 56% of U4, and 85% of U5) were not associated with any known repeat element. However, LINE associations similar to those seen with U6 were also found for a few of the U1, U2, and U4, but not the U5, pseudogenes. Association with SINE elements was seen for all of the snRNA pseudogenes. LTR associations were observed for all of the spliceosomal snRNAs except U5. About half of the U6 pseudogenes associated with LTRs were full-length or nearly full-length, and the remainder were truncated by about 20 nt at their 3′ end.
TABLE 2.
The association of human snRNA pseudogenes with LINE, SINE, or retroviral LTR sequences
| U1 | U2 | U4 | U5 | U6 | |
| LINE | 11 | 5 | 14 | 0 | 84 |
| SINE | 4 | 5 | 10 | 3 | 35 |
| LTR | 2 | 4 | 3 | 0 | 9 |
| None | 69 | 27 | 34 | 17 | 62 |
| Total | 86 | 41 | 61 | 20 | 190 |
Multiple genomic arrangements of LTRs and snRNAs in the human genome
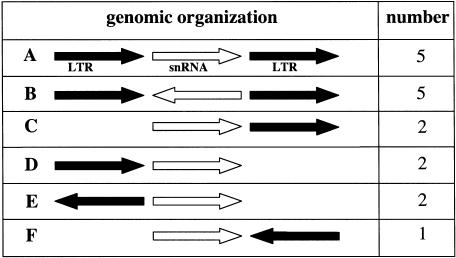
The LTRs identified by RepeatMasker in proximity to snRNA sequences in the human genome are homologous to LTRs of the endogenous retroviruses ERV1, ERVL, and MaLR (Table 3 ▶). Figure 4 ▶ shows a schematic representation categorizing the six types of snRNA/LTR genomic arrangements observed. We observed 10 cases in which two LTR sequences flank the snRNA pseudogene; these have been designated types A and B. In both types, the two LTR sequences are of the same family and type and in the same orientation. Five have the snRNA sequence in the same orientation as the LTRs (type A), and five have it in the opposite orientation (type B). We also observed seven examples of an snRNA sequence adjacent to a single LTR. In two of these cases the LTR is downstream of the snRNA (type C), and in two others the LTR is upstream (type D); types C and D have the snRNA and the LTR in the same orientation. There are also three cases in which the LTR and snRNA are in opposite orientations. In two cases the LTR and snRNA face away from one another (type E), and in one case the face towards one another (type F).
TABLE 3.
Human spliceosomal snRNA/LTR recombinant pseudogenesa
| snRNA | Position | LTR family | LTR name | Organizationb | BLAT score |
| U1 | chr12: 55985209-55985355 | ERV1 | MER39B | E | 109 |
| U2 | chr15: 93876266-93876452 | MaLR | MSTB | B | 187 |
| U2 | chr5: 157339330-157339493 | ERV1 | MER39 | B | 132 |
| U2 | chr11: 65165210-65165389 | MaLR | MLT1G1 | D | 127 |
| U2 | chr9: 12290276-12290451 | MaLR | MLT1J2 | E | 81 |
| U4 | chr15: 57083982-57084125 | MaLR | MLT1C | B | 102 |
| U4 | chr8: 101512746-101512865 | ERV1 | MER4D | A | 88 |
| U4 | chr8: 124136078-124136145 | ERVL | ERVL | C | 61 |
| U6 | chr10: 98003732-98003834 | MaLR | MLT1F | D | 94 |
| U6 | chr20: 42740082-42740187 | ERV1 | LTR35 | F | 92 |
| U6 | chr5: 86754877-86754977 | ERVL | ERVL-B4 | A | 87 |
| U6 | chr10: 79429135-79429238 | ERV1 | LOR1a | C | 87 |
| U6 | chr3: 132093278-132093382 | ERV1 | MER34B-int | A | 85 |
| U6 | chr2: 113751197-113751283 | MaLR | MLT1G1 | B | 73 |
| U6 | chr1: 11310931-11311006c | ERV1 | LTR48B | A | 70 |
| U6 | chr1: 42920646-42920721 | ERV1 | LTR48B | A | 68 |
| U6 | chr14: 102857349-102857415 | MaLR | MSTA | B | 55 |
FIGURE 4.
Six types of snRNA/retroviral LTR associations observed in the human genome. Each of the apparent snRNA/LTR recombinants observed using RepeatMasker and the Human Genome Browser is schematically represented. The black arrows represent the LTRs, and the white arrows represent the snRNAs. The arrows point in the 5′–3′ direction. The number of cases of each type of genomic organization is listed.
We have listed each of the snRNA loci associated with LTRs in Table 3 ▶. Four of the five type A recombinants involve LTRs of the ERV1 family. This family is homologous to the ERV1 human endogenous retrovirus that was originally found on chromosome 18 (O’Brien et al. 1983). Alignment of these flanking LTRs with the complete ERV1 LTR using Clustal W (http://www.cmbi.kun.nl/bioinf/tools/clustalw.shtml) demonstrated examples of both snRNA sequences interrupting a single LTR and snRNAs flanked by two LTRs (data not shown).
The other type A LTR is from the ERVL family. Four of the five type B recombinants are of the MaLR family (Fig. 4 ▶). This family is distantly related to human endogenous retroviruses, as it has no detectable homology to the RT gene (Smit 1993). The fifth type B is of the ERV1 family.
DISCUSSION
We have shown that approximately one molecule of U6 snRNA is packaged per RSV virion particle, along with trace amounts of U1 and U2 snRNAs. Moloney murine leukemia virus (MuLV) also packages U6 snRNA, but not U4 and U5 snRNAs, at similar stoichiometric levels (A. Onafuwa and A. Telesnitsky, pers. comm.), suggesting that this may be a property common to retroviruses. In an endogenous reaction involving purified RSV particles, U6, U1, and U2 snRNAs were reverse-transcribed. We propose that snRNAs may recombine with retroviral sequences and be integrated into the genome. Consistent with this, analysis of the human genome sequence revealed 17 examples of snRNA pseudogenes associated with endogenous retroviral LTRs.
The high levels of U6 snRNA found within RSV virions are surprising in light of the reports that U6 snRNA is not known to exist in the cytoplasm (Will and Luhrmann 2001) and that RSV virion particles bud from the plasma membrane (Pessin and Glaser 1980). However, the RSV Gag structural polyprotein was recently detected in the nucleus of RSV-infected cells (Scheifele et al. 2002). The transient nuclear localization of Gag opens the possibility that the initial interactions between RSV genomic RNA and Gag protein may occur in the nucleus. This could facilitate the association of U6 snRNA with unspliced viral RNA and its entry into viral particles. In the nucleus, U6 snRNA is base-paired with U4 snRNA to form an snRNP containing two snRNAs. During splicing, these RNA-RNA interactions are rearranged, causing the dissociation of U4 from U6 and from the spliceosome (Will and Luhrmann 2001). Because U6 is packaged at much higher levels than U4 snRNA in retroviruses, free U6 snRNA probably associates with the unspliced viral RNA. Alternatively, the U4/U6 di-snRNP may associate with viral RNA, followed by the subsequent dissociation of U4 snRNA.
It is unclear precisely what region of RSV RNA is associated with U6 snRNA. We initially thought that this interaction would involve the RSV NRS sequence due to the 12/13 base pair complementarity between the two sequences in this region. Furthermore, genetic suppression studies indicated that base pairing between U6 and the NRS was involved in splicing suppression (J. LeBlanc and K. Beemon, unpubl.). However, a mutant virus that reduced the putative interaction between the NRS and U6 had little effect on the levels of U6 snRNA found associated with the viral 70S complex. We identified several other regions within the RSV genome that are complementary to a region of U6 snRNA. It is possible that U6 snRNA associates with multiple sites on the RSV genome. Evidence that RSV virion RNA can be translated prior to packaging (Stoltzfus et al. 1983; J. LeBlanc and K. Beemon, unpubl.) suggests that U6 snRNA is not associated with the efficiently translated leader region or the gag gene.
We demonstrated that a wild-type retrovirus can reverse-transcribe packaged snRNA species of varying abundance. Similarly, a mutant form of RSV that is unable to package its own RNA can reverse-transcribe a packaged cellular RNA, 7S L RNA (Taylor and Cywinski 1984). We do not know how the snRNA reverse transcription reaction is primed nor whether the product is a DNA copy of the snRNA alone or a viral/snRNA recombinant. The latter could be generated by template switching by RT during reverse transcription, as proposed previously (Lum and Linial 1998). The reverse-transcribed molecules we identified by PCR annealed to primers near the ends of the various snRNAs, suggesting they were full-length or nearly full-length copies. Unfortunately, this assay does not allow us to determine whether these snRNAs have recombined with viral sequences. It is also possible that truncated cDNA copies of these snRNAs exist. We plan to clone and sequence reverse-transcribed species generated by the RSV endogenous reaction to address these issues.
Analysis of the human, mouse, and rat genomic sequences revealed hundreds of spliceosomal snRNA pseudogenes. Although U6 is not the most abundant cellular snRNA (Yu et al. 1999), it is the most abundant snRNA pseudogene we identified in all three genomes, with 190–202 copies. Buzdin and coworkers (2002) carried out a similar genomic analysis, restricted to full-length sequences, and identified 161 U6 snRNA pseudogenes in the human genome. Fifty-six of these integrants were adjacent to and in the same transcriptional orientation as a 3′ fragment of the L1 LINE, suggesting that they were created by 3′ transduction of U6 during L1 reverse transcription (Buzdin et al. 2002). It was recently shown that L1 reverse transcription can transduce many other sequences (Buzdin et al. 2003). The association of U1, U2, U4, U5, and U6 pseudogenes with LINEs and SINEs also appears to be mainly due to 3′ transduction (Buzdin et al. 2002, 2003; K. Giles, M. Caputi, K. Beemon, unpubl.). In contrast, 10 of 17 snRNA pseudogene loci associated with LTRs are flanked by LTR sequences on both sides. Four of these loci are flanked by LTR sequences of the ERV1 family, two by ERVL LTRs, and four by MALR LTRs (Table 3 ▶).
In contrast to the snRNA transduction by LINEs in which both sequences are in the same orientation (Buzdin et al. 2003), we observed snRNA pseudogenes in both the same and opposite orientations as the associated LTRs (Fig. 4 ▶). This could be the result of different recombination events between the retrovirus and the snRNA sequences. If RT jumped from the retroviral template to the snRNA while copying the antisense strand (the first strand synthesized), the snRNA would be in the same orientation as the virus. However, if template switching occurred during the synthesis of the sense strand, the snRNA would be in the opposite orientation. Similarly, snRNA sequences flanked by LTRs could be generated by jumps from the viral template to the snRNA and back to the viral sequences.
Table 3 ▶ shows that many snRNAs are not associated with any known repeat element. LTRs are the only characterized portions of endogenous retroviruses identified by RepeatMasker. There is no reason to believe that recombination between the virus and U snRNAs occurs solely in the LTR region. Thus, many of the loci not associated with repeat sequences may be inserted into an endogenous retroviral sequence somewhere other than the LTR. As more endogenous retrovirus sequences are identified, we may find the proposed recombination occurs more frequently than currently estimated.
MATERIALS AND METHODS
Purification of virion RNA
Chicken embryo fibroblasts (CEFs) were infected with the wild-type Prague C strain of RSV, and virus was harvested from the media daily. Virus particles were concentrated by pelleting in a Beckman SW28 Ti rotor for 2 h at 24,000 rpm at 4°C and resuspended in 1 mL of Standard Buffer (0.1 M NaCl, 0.01 M Tris-HCl, pH 7.4, and 1mM EDTA). Equivalent amounts of media from uninfected CEFs was put through the same protocol and used as a mock control. The virions were then split into three fractions; 500 μL were immediately loaded onto a 20%–55% continuous sucrose gradient made in Standard Buffer. The remaining 500 μL were split into two 250-μL fractions, and to one RNase A was added to a concentration of 10 μg/mL. Both fractions were incubated at 37°C for 1 h and then immediately loaded onto sucrose gradients as described above. All gradients were spun in a Beckman SW41 Ti rotor for 3 h at 34,000 rpm at 4°C. Purified virions were easily identified due to the visible formation of a band in the gradient. However, this was confirmed by carrying out an exogenous RT assay (Telesnitsky et al. 1995) on each fraction. RNA was extracted from viral particles with phenol/chloroform/isoamyl alcohol (24:24:1) and 1% sodium dodecyl sulfate (SDS) in Standard Buffer and precipitated with ethanol.
Northern blots
Northern blots were performed using the Trans Blot semi-dry apparatus (Bio-Rad) and Zeta-probe nylon membranes, according to the latter manufacturer’s formamide protocol. RNA was fractionated on a 6% acrylamide/8 M urea gel prior to transfer. Antisense riboprobes for the snRNAs were transcribed in vitro as described previously (Gontarek et al. 1993).
Velocity sedimentation of virion RNA
Virus from RSV-infected CEFs was harvested and pelleted as described above. Total virion RNA was extracted as above and loaded onto a 10%–25% sucrose gradient containing Standard Buffer and 0.1% SDS. The gradient was centrifuged for 1.5 h at 34,000 rpm in a Beckman SW41 Ti rotor at 20°C. The RNA in each fraction was precipitated and resuspended in 30 μL of hybridization buffer for RNase protection with the RSV splice donor probe as described in Arrigo and Beemon (1988) or a radiolabeled antisense U6 snRNA probe (Gontarek et al. 1993). All hybridizations were carried out at 55°C, and all RNase digestions were at 22°C for 1 h. The protected species were quantified using a Packard InstantImager. Proline tRNA was detected by Northern analysis of gradient fractions. As a probe, the following oligonucleotide, 5′-TCGTCCGGGATTTGAACCC-3′, was 5′ end-labeled with [γ32P]ATP using T4 polynucleotide kinase (New England Biolabs).
Endogenous reverse transcription reaction
We used a modification of the endogenous reverse transcription reaction of Temin and Mizutani (1970). Forty mL of RSV-infected CEF culture media were clarified and then pelleted as described above. The virions were resuspended in 100 μL of Standard Buffer and 100 μL of 2X reaction buffer (2 mM dATP, dTTP, dGTP, and dCTP; 40 mM Tris, pH 8.0, 2 mM EDTA, 40 mM KCl, 20 mM MgCl2, 10 mM DTT, 0.5% NP40). The reaction was carried out at 39°C for 4 h. Total nucleic acid was harvested from the virions as described above and resuspended in 50 μL of H2O. Ten μL of the reaction was used to amplify either the RSV minus strand strong-stop or the snRNAs. The primers for these reactions and the nucleotides they span are as follows:
RSV forward (24–48): 5′-GTGTGCACCTGGGTTGATGGCCGG-3′
RSV reverse (87–113): 5′-CGGGGTCACAAATGAAGCCTTC TGC-3′
U1 forward (8–34): CCTGGCAGGGGAGATACCATGATCACG-3′
U1 reverse (138–162): GGGGAAAGCGCGAACGCAGTCCCCC-3′
U2 forward (5–28): CTTCTTGGCCTTTTAGCTAAGATC-3′
U2 reverse (163–187): GGTGCACTGTTCCTGGAGGTACTGC-3′
U4 forward (2–23): GCTTTGCGCAGTGGCAGTATCG-3′
U4 reverse (117–144): CAGTCTCCGTAGAGACTGTCAAAAA TTG-3′
U6 forward (1–25): GTGCTCGCTTCGGCAGCACATATAC-3′
U6 reverse (79–99): GGAACGCTTCACGAATTTGCG-3′
The 25-cycle PCR reaction was carried out in 100 μL, using 1μM of both forward and reverse primers, 1.25 mM MgCl2, 2.5 mM dNTPs, 1μCi of [α32P] dGTP (Amersham), and 2.5 U of Taq (Invitrogen). The PCR products were restriction-digested as follows: RSV, U1, and U2 with BstN1, and U6 with MboII (all from New England Biolabs).
Identification of spliceosomal snRNA pseudogenes
The UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway) was used to carry out a BLAT search of the human, mouse, and rat genomes (April 2003 assembly date), using a BLAT score of 50 as a cut-off point (Kent 2002). The human consensus sequence for the spliceosomal snRNAs was used as a query for each of the three genomes. The LINE, SINE, or LTR sequences found tangential to each pseudogene were identified via RepeatMasker. This program searches the RepBase Update library of repetitive sequences from the Genetic Information Research Institute (Jurka 2000).
Acknowledgments
We thank the entire Beemon lab for thoughtful discussions and comments on the manuscript, and Alice Telesnitsky for the communication of results prior to publication. This work was supported by NIH grant RO1 CA48746 to KLB. MC was supported by NIH grant RO1 AI052820. K.E.G. was supported in part by NIH predoctoral training grant T32GM07231.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2150604.
REFERENCES
- Adkins, B. and Hunter, T. 1981. Identification of a packaged cellular mRNA in virions of Rous sarcoma virus. J. Virol. 39: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo, S. and Beemon, K. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell Biol. 8: 4858–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark, C. and Pettersson, U. 1989. Nucleotide sequence and organization of full length human U4 RNA pseudogenes. Gene 80: 385–389. [DOI] [PubMed] [Google Scholar]
- Beemon, K., Duesberg, P., and Vogt, P. 1974. Evidence for crossing-over between avian tumor viruses based on analysis of viral RNAs. Proc. Natl. Acad. Sci. 71: 4254–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, M. and Stoye, J.P. 2000. Endogenous retroviruses and the human germline. Curr. Opin. Genet. Dev. 10: 651–655. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., Ustyugova, S., Gogvadze, E., Vinogradova, T., Lebedev, Y., and Sverdlov, E. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics 80: 402–406. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., Gogvadze, E., Kovalskaya, E., Volchkov, P., Ustyugova, S., Illarionova, A., Fushan, A., Vinogradova, T., and Sverdlov, E. 2003. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31: 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.J., Cywinski, A., and Taylor, J.M. 1985. Reverse transcription of 7S L RNA by an avian retrovirus. J. Virol. 54: 278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin, J.M. 1996. Retroviridae: The viruses and their replication. In Virology (eds. B.N. Fields et al.), pp. 1767–1848. Lippincott-Raven Publishers, Philadelphia, PA.
- Denison, R.A., Van Arsdell, S.W., Bernstein, L.B., and Weiner, A.M. 1981. Abundant pseudogenes for small nuclear RNAs are dispersed in the human genome. Proc. Natl. Acad. Sci. 78: 810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, M.M., Olsen, J.C., and Swanstrom, R. 1992. Characterization of unintegrated retroviral DNA with long terminal repeat-associated cell-derived inserts. J. Virol. 66: 5735–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault, C., Maestre, J., and Heidmann, T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24: 363–367. [DOI] [PubMed] [Google Scholar]
- Faras, A.J., Garapin, A.C., Levinson, W.E., Bishop, J.M., and Goodman, H.M. 1973. Characterization of the low-molecular-weight RNAs associated with the 70S RNA of Rous sarcoma virus. J. Virol. 12: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faras, A.J., Dahlberg, J.E., Sawyer, R.C., Harada, F., Taylor, J.M., Levinson, W.E., Bishop, J.M., and Goodman, H.M. 1974. Transcription of DNA from the 70S RNA of Rous sarcoma virus. II. Structure of a 4S RNA primer. J. Virol. 13: 1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontarek, R.R., McNally, M.T., and Beemon, K. 1993. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes & Dev. 7: 1926–1936. [DOI] [PubMed] [Google Scholar]
- Griffiths, D.J. 2001. Endogenous retroviruses in the human genome sequence. Genome Biol. 2: REVIEWS1017–1017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar, A.M. and Linial, M.L. 1993. A model system for nonhomologous recombination between retroviral and cellular RNA. J. Virol. 67: 3845–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M. and Gerstein, M. 2002. Studying genomes through the aeons: Protein families, pseudogenes and proteome evolution. J. Mol. Biol. 318: 1155–1174. [DOI] [PubMed] [Google Scholar]
- Hibbert, C.S., Gontarek, R.R., and Beemon, K.L. 1999. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA 5: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W.S. and Temin, H.M. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. 87: 1556–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. 2000. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 16: 418–420. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, H.J., Bailey, J.M., Davidson, N., Vogt, P.K., Nicolson, M.O., and McAllister, R.M. 1975. Electron microscope studies of tumor virus RNA. Cold Spring Harbor Symp. Quant. Biol. 39 (Pt 2): 827–834. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Linial, M. 1987. Creation of a processed pseudogene by retroviral infection. Cell 49: 93–102. [DOI] [PubMed] [Google Scholar]
- Lum, R. and Linial, M.L. 1998. Retrotransposition of nonviral RNAs in an avian packaging cell line. J. Virol. 72: 4057–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E. 1988. Heterogeneity of human U1 snRNAs. Nucleic Acids Res. 16: 5813–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, L.M. and McNally, M.T. 1999. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J. Virol. 73: 2385–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, M.T., Gontarek, R.R., and Beemon, K. 1991. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology 185: 99–108. [DOI] [PubMed] [Google Scholar]
- O’Brien, S.J., Bonner, T.I., Cohen, M., O’Connell, C., and Nash, W.G. 1983. Mapping of an endogenous retroviral sequence to human chromosome 18. Nature 303: 74–77. [DOI] [PubMed] [Google Scholar]
- Ogiwara, I., Miya, M., Ohshima, K., and Okada, N. 1999. Retropositional parasitism of SINEs on LINEs: Identification of SINEs and LINEs in elasmobranchs. Mol. Biol. Evol. 16: 1238–1250. [DOI] [PubMed] [Google Scholar]
- Ogiwara, I., Miya, M., Ohshima, K., and Okada, N. 2002. V-SINEs: A new superfamily of vertebrate SINEs that are widespread in vertebrate genomes and retain a strongly conserved segment within each repetitive unit. Genome Res. 12: 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, N., Hamada, M., Ogiwara, I., and Ohshima, K. 1997. SINEs and LINEs share common 3′ sequences: A review. Gene 205: 229–243. [DOI] [PubMed] [Google Scholar]
- O’Sullivan, C.T., Polony, T.S., Paca, R.E., and Beemon, K.L. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses src mRNA splicing and promotes polyadenylation. Virology 302: 405–412. [DOI] [PubMed] [Google Scholar]
- Panet, A., Weil, G., and Friis, R.R. 1978. Binding of tryptophanyl-tRNA to the reverse transcriptase of replication-defective avian sarcoma viruses. J. Virol. 28: 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessin, J.E. and Glaser, M. 1980. Budding of Rous sarcoma virus and vesicular stomatitis virus from localized lipid regions in the plasma membrane of chicken embryo fibroblasts. J. Biol. Chem. 255: 9044–9050. [PubMed] [Google Scholar]
- Pickeral, O.K., Makalowski, W., Boguski, M.S., and Boeke, J.D. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polony, T.S., Bowers, S.J., Neiman, P.E., and Beemon, K.L. 2003. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J. Virol. 77: 9378–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, R.C. and Dahlberg, J.E. 1973. Small RNAs of Rous sarcoma virus: Characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 12: 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, R.C., Harada, F., and Dahlberg, J.E. 1974. Virion-associated RNA primer for Rous sarcoma virus DNA synthesis: Isolation from uninfected cells. J. Virol. 13: 1302–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele, L.Z., Garbitt, R.A., Rhoads, J.D., and Parent, L.J. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. 99: 3944–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank, P.R. and Linial, M. 1980. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: Characterization of a deletion in the provirus. J. Virol. 36: 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbald, P.R. and Blencowe, B.J. 1990. A completely conserved rat U6 snRNA pseudogene coding sequence is sandwiched between a cytochrome c retropseudogene and a LINE-like sequence. Nucleic Acids Res. 18: 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F. 1993. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 21: 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, C.M., Dimock, K., Horikami, S., and Ficht, T.A. 1983. Stabilities of avian sarcoma virus RNAs: Comparison of subgenomic and genomic species with cellular mRNAs. J. Gen. Virol. 64 (Pt 10): 2191–2202. [DOI] [PubMed] [Google Scholar]
- Swanstrom, R. and Wills, J.W. 1997. Synthesis, assembly, and processing of viral proteins. In Retroviruses (eds. J.M. Coffin et al.), pp. 263–334. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Taylor, J.M. and Cywinski, A. 1984. A defective retrovirus particle (SE21Q1b) packages and reverse transcribes cellular RNA, utilizing tRNA-like primers. J. Virol. 51: 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky, A., Blain, S., and Goff, S.P. 1995. Assays for retroviral reverse transcriptase. Methods Enzymol. 262: 347–362. [DOI] [PubMed] [Google Scholar]
- Telesnitsky, A. and Goff, S.P. 1997. Reverse Transcriptase and the generation of retroviral DNA. In Retroviruses (ed. J.M. Coffin et al.), pp. 121–160. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Temin, H.M. and Mizutani, S. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226: 1211–1213. [DOI] [PubMed] [Google Scholar]
- Trono, D. 1992. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J. Virol. 66: 4893–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell, S.W. and Weiner, A.M. 1984. Pseudogenes for human U2 small nuclear RNA do not have a fixed site of 3′ truncation. Nucleic Acids Res. 12: 1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, A.M. 2002. SINEs and LINEs: The art of biting the hand that feeds you. Curr. Opin. Cell. Biol. 14: 343–350. [DOI] [PubMed] [Google Scholar]
- Will, C.L. and Luhrmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Yu, Y.T., Scharl, E.C., Smith, C.M., and Steitz, J.A. 1999. The growing world of small nuclear ribonucleoproteins. In The RNA world (eds. R.F. Gesteland et al.), pp. 487–524. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.