Abstract
Pre-mRNA splicing is executed by the spliceosome, a complex of small nuclear RNAs (snRNAs) and numerous proteins. One such protein, 15.5K/Snu13p, is associated with the spliceosomal U4/U6.U5 tri-snRNP and box C/D small nucleolar ribonucleoprotein particles (snoRNPs), which act during preribosomal RNA (rRNA) processing. As such, it is the first splicing factor to be identified in two functionally distinct particles. 15.5K binds to an internal helix–bulge–helix (K-turn) structure in the U4 snRNA and two such structures in the U3 snoRNA. Previous work has concentrated on the structural basis of the interaction of 15.5K with the RNAs and has been carried out in vitro. Here we present a functional analysis of Snu13p in vivo, using a galactose inducible SNU13 strain to investigate the basis of three lethal mutations in Saccharomyces cerevisiae. Two are point mutations that map to the RNA-binding domain, and the third is a C-terminal deletion. These mutations result in accumulation of unspliced pre-mRNA, confirming a role for Snu13p in pre-mRNA splicing. In addition, these mutants also display rRNA processing defects that are variable in nature. Analysis of one mutant in the RNA-binding domain reveals a reduction in the levels of the U4 snRNA, U6 snRNA, and box C/D snoRNAs, but not H/ACA snoRNAs, supporting a role for Snu13p in accumulation and/or maintenance of specific RNAs. The mutations in the RNA-binding domain exhibit differential binding to the U4 snRNA and U3 snoRNA in vitro, suggesting that there are differences in the mode of interaction of Snu13p with these two RNAs.
Keywords: U4 snRNA, U3 snoRNA, Snu13p, pre-mRNA splicing, pre-rRNA processing
INTRODUCTION
Introns are removed from premessenger RNA (pre-mRNA) in a two-step transesterification reaction catalyzed in the nucleus by the spliceosome (for review, see Burge et al. 1999). The spliceosome is composed of four small nuclear ribonucleoprotein particles (U1, U2, U4/U6, and U5 snRNPs) and additional non-snRNP proteins. Each snRNP contains a small nuclear RNA (U1, U2, U4, U5, or U6 snRNA) and proteins that can be defined as specific to one snRNP or common to all four (for review, see Kambach et al. 1999).
In a newly proposed model for spliceosome assembly, the snRNPs form a U1.U2.U4/U6.U5 penta-snRNP containing the snRNAs and numerous proteins before interacting with the pre-mRNA (Stevens et al. 2002). The previously identified base-pairing interactions between the snRNAs and the pre-mRNA then occur within the penta-snRNP rather than in discrete stages of spliceosome assembly (for review, see Brow 2002). These include the interaction of the U1 snRNA with the 5′ splice site (Zhuang and Weiner 1986; Séraphin et al. 1988) and the U2 snRNA with the branchpoint sequence (Parker et al. 1987). After these initial interactions, a complex series of RNA rearrangements occurs (Nilsen 1998). During these rearrangements, the U6 snRNA displaces U1 at the 5′ splice site. Base-pairing between the U4 and U6 snRNAs is disrupted to allow the interaction of the U2 and U6 snRNAs during the formation of the spliceosome catalytic site (Madhani and Guthrie 1992). Evidence suggests that the U6 snRNA is responsible for catalysis of the splicing reaction (Yean et al. 2000; Valadkhan and Manley 2001). The U4 snRNA is thought to act as a chaperone by delivering U6 to the spliceosome while sequestering its catalytic residues.
In addition to changes in RNA–RNA interactions, spliceosomal rearrangements also involve changes in protein–RNA and protein–protein interactions. The protein components of the spliceosome, particularly members of the DEXD/H-box helicase family, are involved in driving many of these rearrangements (de la Cruz et al. 1999; Linder 2000). Many spliceosomal proteins are involved in stabilizing interactions between the snRNAs and the pre-mRNA, with some implicated in splicing catalysis (Staley and Guthrie 1998; Will and Lührmann 2001). Several novel protein splicing factors were identified by using mass spectrometric techniques after purification and fractionation of the yeast U4/U6.U5 tri-snRNP particle (Gottschalk et al. 1999; Stevens and Abelson 1999). One protein, Snu13p, the product of an essential gene, is highly conserved across eukaryotes and contains an RNA-binding motif shared by the yeast ribosomal protein L32, human ribosomal protein L7a, and yeast protein Nhp2p (Koonin et al. 1994; Nottrott et al. 1999; Stevens et al. 2001).
The human homolog of Snu13p, 15.5K, exhibits 71% identity and binds specifically to the conserved 5′ stem loop of U4 snRNA (Nottrott et al. 1999). A U4 fragment, corresponding to the 15.5K binding site, inhibited the first step of in vitro splicing reactions and prevented spliceosome assembly after complex B formation, suggesting a late role for 15.5K during spliceosome assembly (Nottrott et al. 1999). In a crystal structure of 15.5K in complex with a fragment of the U4 snRNA, the RNA binds to a pocket within the protein, resulting in a sharp bend in the RNA (Vidovic et al. 2000).
Snu13p has recently been found to copurify with components of the U3 small nucleolar RNP (snoRNP), a member of the box C/D class (Watkins et al. 2000; Dragon et al. 2002; Galardi et al. 2002). The U3 snoRNP acts as an RNA chaperone during 18S ribosomal RNA (rRNA) synthesis and is essential for cleavage of the pre-rRNA precursor at sites A0, A1, and A2 (Hughes and Ares 1991; Beltrame and Tollervey 1992; Beltrame et al. 1994; Beltrame and Tollerrey 1995). Snu13p also co-immunoprecipitates with other members of the box C/D family of snoRNAs (Watkins et al. 2000), the majority of which function as guides during 2′-O methylation of ribosomal RNA precursors (for review, see Tollervey and Kiss 1997; Venema and Tollervey 1999). This has led to the conclusion that Snu13p, with Nop1p, Nop56p, and Nop58p, is a core component of the box C/D snoRNPs, required for their correct accumulation and localization (Verheggen et al. 2001).
Electrophoretic mobility shift assays have demonstrated that 15.5K binds specifically to both the box C′/D and box B/C motifs in U3 snoRNA and generally to the C/D motif across the whole family of box C/D snoRNAs (Watkins et al. 2000). Interestingly, the secondary structure formed by the box C/D motif is similar to that formed by the 5′ stem loop in U4 snRNA and also that formed by the B/C motif in U3. These motifs can form a two-stranded helix I–bulge–helix II motif with a characteristic kink in the phosphodiester backbone, which has been termed a kink-turn (K-turn; Klein et al. 2001). Mutagenesis suggests it is the helix–bulge structure, rather than sequence of base-pairing regions, that is crucial for 15.5K binding (Nottrott et al. 1999; Watkins et al. 2002). Assembly of 15.5K with a radiolabeled fragment of the human U3 snoRNA, containing only the B/C box, was observed directly in HeLa cell nuclear extracts (Granneman et al. 2002). Nucleotide analog interference mapping (NAIM) of a box C/D snoRNP reconstituted in Xenopus oocytes suggests that 15.5K interacts only with the box C/D motif in vivo and not with the box C′/D′ (Weinstein Szewczak et al. 2002). An asymmetric association of the box C/D snoRNA core proteins with the C′/D′ and C/D motifs is also supported in Xenopus by differential cross-linking of Nop58, Nop56, and fibrillarin to the C/D and C′/D′ motifs, although this cross-linking did not detect 15.5K (Cahill et al. 2002). However, recent analysis of the interaction of Snu13p with both the C′/D and B/C motifs in yeast U3 snoRNA in vitro demonstrated that both are bound independently, although affinity for the B/C motif is higher (Marmier-Gourrier et al. 2003).
As Snu13p is the first yeast protein to be identified as a component of two functionally distinct RNP particles, this raises interesting questions about its function. In addition to the structural similarities, there are functional similarities between U4 snRNA and U3 snoRNA, with both acting as RNA chaperones through base-pairing with the U6 snRNA or the pre-rRNA, respectively. Moreover, there are similarities between the protein constituents of the U4 snRNP and the U3 snoRNP. The central NOP domain conserved between U3 snoRNP proteins Nop56 and Nop58 is also found within the U4/U6 snRNP protein Prp31p/61K (Gautier et al. 1997; Makarova et al. 2002). Given these similarities, it is conceivable that Snu13p has related roles in the two processing pathways.
Apparently contradictory data have been obtained with regard to a potential role for Snu13p in pre-mRNA splicing. The identification of Snu13p as a spliceosomal tri-snRNP component supports a role for the protein in the splicing process (Gottschalk et al. 1999; Stevens and Abelson 1999). The observation that a U4 snRNA fragment containing the 15.5K binding site will inhibit in vitro splicing reactions, likewise supports a role for 15.5K in pre-mRNA splicing (Nottrott et al. 1999). In contrast, genetic depletion studies determined that there was no accumulation of unspliced pre-mRNA associated with depletion of Snu13p, suggesting that the protein is not essential for efficient splicing (Watkins et al. 2000). Instead, Snu13p depletion was found to result initially in a reduction in levels of the box C/D snoRNAs, followed by a general reduction in the levels of all cellular RNAs (Watkins et al. 2000). More recently, genetic depletion of Snu13p resulted in an accumulation of unspliced U3 snoRNA precursor, again implicating Snu13p as a splicing factor (Stevens et al. 2001).
One possible role that has been proposed for Snu13p is as a nucleation factor for the binding of other proteins to RNA. Immunoprecipitation and GST pull-down assays have demonstrated that binding of the 15.5K protein to U4 is a prerequisite for binding of 61K (Prp31p in yeast) and the 20/60/90K complex to the U4/U6 duplex (Nottrott et al. 2002). Based on UV cross-linking data and the cocrystal structure, it has been suggested that 15.5K binding to U4 is necessary to produce an RNA conformation required for 61K binding (Vidovic et al. 2000; Nottrott et al. 2002). Binding of 15.5K is also necessary for binding of the U3-specific protein hU3–55K to the B/C box of the U3 snoRNA in nuclear extracts (Granneman et al. 2002). Likewise, sequestering 15.5K prevents the association of the box C/D snoRNP core proteins Nop56, Nop58, and fibrillarin, and also Tip48 and Tip49 with the U14 snoRNA (Watkins et al. 2002).
There are many questions remaining about the role of Snu13p. Inconsistencies remain in the genetic depletion data, and a likely role for Snu13p in splicing must be confirmed. In particular, the data obtained to date have largely been from experiments completed in vitro, and a functional analysis of regions of the protein has not yet been carried out in vivo. In this work, mutations were made in Snu13p to investigate the effects of changes in the protein on its function in yeast. Three lethal mutations were identified: two were substitutions (E59A and R84A/V81L) and one a C-terminal deletion of 16 amino acids (Δ111-126). The behavior of these mutants was investigated in a galactose-inducible SNU13 strain in which all three mutations were shown to prevent growth under repressed conditions. Each mutation resulted in a decrease in levels of mature U3 snoRNA accompanied by a build up of unspliced U3, indicative of a splicing defect. Analysis of the splicing of several intron-containing yeast genes suggests that mutation E59A is associated not only with a defect in splicing of pre-U3 snoRNA but also with a global splicing defect. All three lethal mutants were also found to have varying affects on the processing of pre-rRNA. The effect of the E59A mutation on the levels of various RNAs was investigated by primer extension, and a dramatic decrease in the levels of the spliceosomal U6 snRNA and box C/D snoRNAs was apparent. This was accompanied by a similar, but less pronounced, reduction in levels of the U4 snRNA. Levels of H/ACA snoRNAs appeared to remain constant. Taken together, these results suggest that Snu13p is involved in the accumulation and/or maintenance of specific RNAs. Electrophoretic mobility shift assays indicated that the substitution mutants (E59A and R84A/V81L) exhibited differential binding in vitro to the U4 snRNA and a region of U3 snoRNA containing the B/C and C′/D boxes. These data suggest that there are differences in the way in which Snu13p interacts with the U4 snRNA and U3 snoRNA.
RESULTS
Functional analysis of Snu13p
Although the crystal structure of the 15.5K protein in complex with a fragment of the U4 snRNA has been solved (Vidovic et al. 2000), the structure of Snu13p has not yet been determined, nor has a comprehensive functional analysis of the protein been carried out. To determine which amino acid residues are functionally important in vivo, charged-to-alanine scanning mutagenesis was undertaken throughout the SNU13 open reading frame. Charged residues were chosen initially to provide a series of mutations spanning the entire open reading frame.
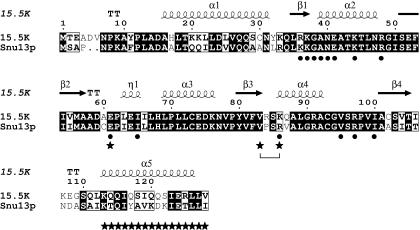
A total of 26 single-alanine substitutions were produced, and their effects on cell viability were analyzed by plasmid shuffle in a haploid SNU13 knockout strain (YSNU13KO). In addition, two double mutants came up fortuitously during the mutagenesis procedure, and their effects on cell viability were also studied. Two substitutions were found to have a detrimental effect on the growth of the knockout strain after plasmid shuffle at 30°C (Table 1 ▶). These mutants were E59A and the double substitution R84A/V81L. The remaining substitution mutants had little or no effect on growth at 30°C. Mutations V81L and R84A were tested as single substitutions but had no effect individually. Based on the crystal structure and sequence alignment of the two proteins (Fig. 1 ▶), equivalent residues in 15.5K to the Snu13p residues E59 and R84 play a key role in protein–RNA interactions (Vidovic et al. 2000). These mutagenesis data confirm that the homologous residues (E59 and R84) in Snu13p may have a similar role in yeast. Viable mutants were subsequently tested for temperature and cold sensitivity by incubating at 37°C or 16°C after plasmid shuffle. None of the mutations had any effect on growth at either temperature.
TABLE 1.
Viability of Snu13p mutants
| Mutation | Growth at 30°C |
| K7A | + |
| D13A | + |
| D22A | + |
| R31A | + |
| K34A | + |
| K35A | + |
| E39A | + |
| K42A | + |
| R46A | + |
| E50A | + |
| D57A | + |
| E59A | − |
| E62A | + |
| H66A | + |
| E72A | + |
| D73A | + |
| K74/C71S | + |
| K74A | + |
| R84A/V81L | − |
| V81L | + |
| R84A | + |
| R89A | + |
| R95A | + |
| D106A | + |
| K111A | + |
| K118A | + |
| D119A | + |
| K120A | + |
| E122A | + |
| Δ1–11 | + |
| Δ1–30 | + |
| Δ111–126 | − |
+ indicates growth; −, no growth. Plates were scored after 2 d at 30°C.
FIGURE 1.
Amino acid sequence alignment of S. cerevisiae Snu13p with the human homolog 15.5K. The 15.5K residues involved in U4 snRNA interaction are indicated by solid circles below the sequences (Vidovic et al. 2000). Positions of Snu13p lethal mutations E59A, R84A/V81L, and Δ111-126 are indicated by stars. Sequence alignment was completed by using CLUSTALW (Thompson et al. 1994). Schematic representation of secondary structure was generated by using ESPript 2.0 (Gouet et al. 1999) based on the 15.5K crystal structure (Protein Data Bank identifier 1E7K; Vidovic et al. 2000).
Because the majority of point mutations had no effect on growth, three deletion mutants were constructed in Snu13p, covering larger regions of the protein. These deletions removed amino acids at the N or C terminus, in sections that were some distance from the equivalent RNA-binding site in the 15.5K protein and in regions that were surface-exposed (Vidovic et al. 2000). This approach was taken in an attempt to identify residues that were important during functions other than RNA binding. Deletion of the N-terminal 11 or 30 amino acids had no visible effect on growth after plasmid shuffle at 30°C (Table 1 ▶). However, deletion of the C-terminal 16 amino acids (Δ111-126) was lethal at 30°C. Taken together, these results indicate that although Snu13p is the product of an essential gene, it is surprisingly tolerant to mutation of its charged residues and to larger deletions.
Analysis of lethal mutations in a galactose-inducible SNU13 strain
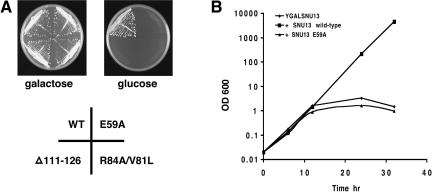
To investigate the function of Snu13p, mutants E59A and R84A/V81L were analyzed in a galactose-inducible SNU13 strain (YGALSNU13). In this strain, the endogenous SNU13 gene was placed under the control of the GAL1 promoter. Expression of SNU13 is, therefore, induced during growth in media containing galactose and repressed in glucose. Such a strain allows the study of lethal SNU13 mutations during the delay that occurs between repression of wild-type SNU13 transcription and cell death. The strain YGALSNU13, transformed with a plasmid copy of wild-type SNU13, was viable when grown on media containing galactose, in which expression of chromosomal SNU13 was induced, or glucose, in which expression was repressed (Fig. 2A ▶). Transformants containing plasmid copies of SNU13 mutants E59A, R84A/V81L, and Δ111-126 were viable on galactose but nonviable on glucose in which chromosomal SNU13 was repressed (Fig. 2A ▶). This confirmed the effects of all three mutations on cell viability as found by plasmid shuffle.
FIGURE 2.
Growth of SNU13 mutants. (A) Single colonies of strain YGALSNU13 transformed with plasmid-based SNU13 mutants E59A, R84A/V81L, Δ111-126, or wild-type SNU13 were streaked on glucose or galactose-containing medium. Plates were photographed after 2-d incubation at 30°C. (B) Strain YGALSNU13 was grown in liquid culture in the presence and absence of plasmid-based wild-type SNU13 or mutant E59A. Cells were grown overnight initially in galactose-containing medium, washed, and then transferred to medium containing glucose. Optical density at 600 nm was monitored at the indicated time points.
Growth of YGALSNU13 in the presence of plasmid copies of each of the lethal SNU13 mutations or wild-type SNU13 was also monitored in liquid culture after transfer to glucose-containing medium. In all cases, cells divided normally for the first 12 h (Fig. 2B ▶; data not shown). After this time point, cells containing wild-type SNU13 continued to divide normally. However, in the presence of SNU13 mutation E59A, growth was impaired at approximately the 12-h time point. After this point, the growth curve obtained for this mutant was almost identical to that obtained with strain YGALSNU13 alone. Analysis of the growth of lethal mutants R84A/V81L and Δ111-126 in liquid culture revealed growth curves almost identical to that for mutant E59A (data not shown).
Mutations in Snu13p cause pre-mRNA splicing and pre-rRNA processing defects
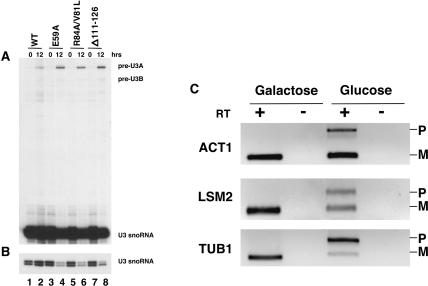
Strain YGALSNU13 was next used to investigate the ability of the various SNU13 mutants to support splicing of pre-mRNA in vivo. Total RNA was harvested from YGALSNU13 in the presence of plasmid copies of wild-type SNU13, or mutants E59A, R84A/V81L and Δ111-126, after transfer to glucose-containing medium in order to monitor splicing of the U3 snoRNA. Genes SNR17A (U3A) and SNR17B (U3B) encode the U3 snoRNA and have introns of different sizes. The levels of spliced and unspliced U3 snoRNA were analyzed by using primer extension from a primer complementary to exon 2 (Table 2 ▶). In the case of the wild-type SNU13, unspliced U3 snoRNA was barely detectable at the time points tested (Fig. 3A ▶). However, in the case of mutants E59A, R84A/V81L, and Δ111-126, an accumulation of unspliced RNA from U3A and U3B was detectable 12 h after the switch to glucose-containing medium, indicating that splicing was inhibited (Fig. 3A ▶). In contrast to results obtained from similar analysis of mutations in other splicing factors, these SNU13 mutations also have a detrimental effect on cellular levels of the mature U3 snoRNA compared with the wild type (Fig. 3B ▶). This may be due to the observed splicing defect, to a decrease in stability of the U3 snoRNA, or to a combination of the two.
TABLE 2.
Oligonucleotides used in this work
| Use | Name | Sequence 5′–3′ |
| SNU13 into pRS416 | Snu13F | GGGGTACCAGCTTCTGAATCACTAAAGC |
| Snu13B | CGGGATCCAATAAATCTCCTACGAACGA | |
| K7A | 13-1 | TCTGCCCCAAACCCAGCTGCTTTCCCATTAGCC |
| D13A | 13-2 | GCTTTCCCATTAGCCGCTGCTGCATTAACCCAA |
| D22A | 13-3 | CAGGCTGCTAACTTGGCACAATTGAAGAAAGGT |
| R31A | 13-4 | AACTTGAGACAATTGGCTAAAGGTGCCAACGAA |
| K34A | 13-5 | TTGAGACAATTGAACGCAGGTGCCAACGAAGCT |
| K35A | 13-6 | AAGAAAGGTGCCAACGCAGCTACCAAAACTTTG |
| E39A | 13-7 | GCCAACGAAGCTACCGCAACTTTGAACCGTGGT |
| R46A | 13-8 | ACCAAAACTTTGAACGCTGGTATCTCCGAATTC |
| E50A | 13-9 | AACCGTGGTATCTCCGCATTCATCATTATGGCT |
| D57A | 13-10 | ATCATTATGGCTGCTGCCTGTGAACCTATTGAA |
| E59A | 13-11 | ATGGCTGCTGACTGTGCACCTATTGAAATCCTA |
| E62A | 13-12 | GACTGTGAACCTATTGCAATCCTATTGCATTTA |
| H66A | 13-13 | ATTGAAATCCTATTGGCTTTACCATTACTATGT |
| E72A | 13-14 | TTACCATTACTATGTGCAGACAAGAATGTTCCA |
| D73A | 13-15 | CCATTACTATGTGAAGCCAAGAATGTTCCATAT |
| K74A | 13-16 | TTACTATGTGAAGACAAGAATGTTCCATATGTC |
| R84A | 13-17 | GTCTTTGTTCCATCTAGAGTTGCTTTAGGTAGA |
| R89A | 13-18 | AGAGTTGCTTTAGGTGCAGCTTGTGGTGTTTCT |
| R95A | 13-19 | GCTTGTGGTGTTTCTGCACCAGTTATTGCTGCT |
| D106A | 13-20 | TCTATTACCACCAACGCTGCTTCTGCTATCAAG |
| K111A | 13-21 | CAAATCTACGCTGTCGCTGACAAGGATTGAACT |
| K118A | 13-22 | CAAATCTACGCTGTCGCTGACAAGATTGAAACT |
| D119A | 13-23 | ATCTACGCTGTCAAGGCCAAGATTGAAACTTTA |
| K120A | 13-24 | TACGCTGTCAAGGACGCTATTGAAACTTTATTA |
| E122A | 13-25 | GTCAAGGACAAGATTGCAACTTTATTAATTTAA |
| D22A | 13-26 | ACCCAACAAATCTTGGCTGTCGTTCAACAGGCT |
| V81L | 13-27 | CCATATGTCTTTCTTCCATCTAGAGTT |
| Δ1-11 | 13-Ndel1 | ACTATATTAAAAATGGATGCTGCATTAACC |
| Δ1-30 | 13-Ndel2 | ACTATATTAAAAATGCAATTGAAGAAAGGT |
| Δ111-126 | 13-Cdel | ATGCTTCTGCTATCTAGACCCAAATCTACG |
| SNU13 into pET21d | Snu13pET21F | GCGCTAGCTCTGCCCCAAACCCAAAG |
| Snu13pET21R | CAGCCGCTCGAGAATTAATAAAGTTTCAAT | |
| SNR14 into pRS416 | U4GF | CGGGATCCTATAGAGGGACAAACATACGC |
| U4GB | GGGGTACCTCTCTGCTGTTTTAGTTTACCG | |
| SNR17 into pBluescript IIKS+ | T7U3F | TAATACGACTCACTATAGTCGACGTACTTCATAGGAT |
| T7U3R | ACTTGTCAGACTGCCATTTG | |
| U3snoRNA truncation | U3del234 | ACCTTTGTACCCCAGAGTGAGAAACGCGATGATCTT |
| ATGGGTACAAATGG | ||
| GAL1::SNU13 strain | 13GALF | GTCCTACTTTCAAATCTTAATTTAAATCATCATTCGA |
| GCCAGCTTGAATTCGAGCTCGTTTAAAC | ||
| 13GALB | TAATGCAGCATCGGCTAATGGGAAAGCCTTTGGGTT | |
| TGGGGCAGACATTTTGAGATCCGGGTTTT | ||
| RT, U3 snoRNA | U3snoRNA-RT | CCAAGTTGGATTCAGTGGCTC |
| RT, U2 snRNA | U2RT2 | GGGTGCCAAAAAATGTGT |
| RT, U6 snRNA | U6B | AAAACGAAATAAATCTCTTTGTAAAACGG |
| RT, U4 snRNA | U4S2 | GACGGTCTGGTTTATAATTAAATTTC |
| RT, U14 snoRNA | U14snoRNA-RT | CACTCAGACATCCTAGGAAGGTCTC |
| RT, U18 snoRNA | U18RT | CGTCAGATACTGTGATAGTC |
| RT, snR4 snoRNA | snr4RT | CAGTTATTAATAGTTAAAGC |
| RT, snR3 snoRNA | R3snoRNA-RT | CCCGGAAATTGATCTCTCTCCCAC |
| RT, snR30 snoRNA | R30snoRNA-RT | GAGACTCTGCCTCTCGAGCG |
| RT, snR31 snoRNA | snr31 | TCATGACCAACTGTATCAGG |
| RT, 18S rRNA 5′ ETS | 5ETS | CATGGCTTAATCTTTGAGAC |
| RT, 18S rRNA ITS1 | ITS1 | GGCCAGCAATTTCAAGTTA |
| U3 snoRNA PCR | T7U3F | TAATACGACTCACTATAGTCGACGTACTTCATAGGAT |
| T7U3R | ACTTGTCAGACTGCCATTTG | |
| U4 snRNA PCR | U4F | TAATACGACTCACTATAGATCCTTATGCACGGGAAA |
| TACG | ||
| U4B | AAAAGGTATTCCAAAAATTCCC |
FIGURE 3.
Mutations in SNU13 inhibit pre-mRNA splicing. (A) Primer extension analysis of pre-U3 snoRNA splicing. Strain YGALSNU13 was grown in the presence of plasmid-based wild-type SNU13 or mutants E59A, R84A/V81L, or Δ111-126. Total RNA was harvested before (0) and 12 h after transfer to glucose-containing medium. Splicing of U3A and U3B was analyzed by using an end-labeled primer complementary to exon 2 and RNA extracted from the wild-type SNU13 (lanes 1,2), mutant E59A (lanes 3,4), R84A/V81L (lanes 5,6), or Δ111-126 (lanes 7,8). Pre-U3A, pre-U3B, and mature U3 snoRNA are indicated to the right of the panel. (B) Shorter exposure of the mature U3 snoRNA primer extension products from A. (C) RT-PCR analysis of the effects of SNU13 mutation E59A on splicing of intron-containing genes. Inverted images of ethidium bromide–stained agarose gels. Strain YGALSNU13 was grown in the presence of plasmid-based wild-type SNU13 in galactose-containing medium or in the presence of SNU13 mutant E59A in glucose-containing medium, as indicated. Poly(A)+ RNA was prepared after 12-h incubation and analyzed by RT-PCR using primers complementary to the exons of each gene. The gene analyzed in each case is indicated to the left of the panel. Bands representing unspliced pre-mRNA (P) and spliced mRNA (M) are indicated to the right of each panel. + and − denote reactions carried out in the presence or absence, respectively, of reverse transcriptase.
To investigate whether the observed splicing defect was global or limited to the U3 genes, the effect of SNU13 mutations on splicing of alternative genes was analyzed. RT-PCR from poly(A)+ mRNA was used as a means of studying levels of other spliced genes. Primers were designed that were complementary to exon sequences, allowing the amplification of spliced and unspliced RNA. RNA samples were taken from YGALSNU13 grown in glucose in the presence of a plasmid containing SNU13 mutant E59A and compared with RNA taken from the same strain grown in galactose in the presence of a plasmid containing wild-type SNU13. In the presence of the wild-type SNU13, only spliced RNA was detected for all the transcripts analyzed. In contrast, in the case of mutant E59A, an accumulation of unspliced RNA for genes ACT1, LSM2, and TUB1 was visible (Fig. 3C ▶). This indicates that the splicing defect observed in this mutant is not limited to the U3 snoRNA but includes other pre-mRNAs. This suggests that this SNU13 mutation results in a general splicing defect.
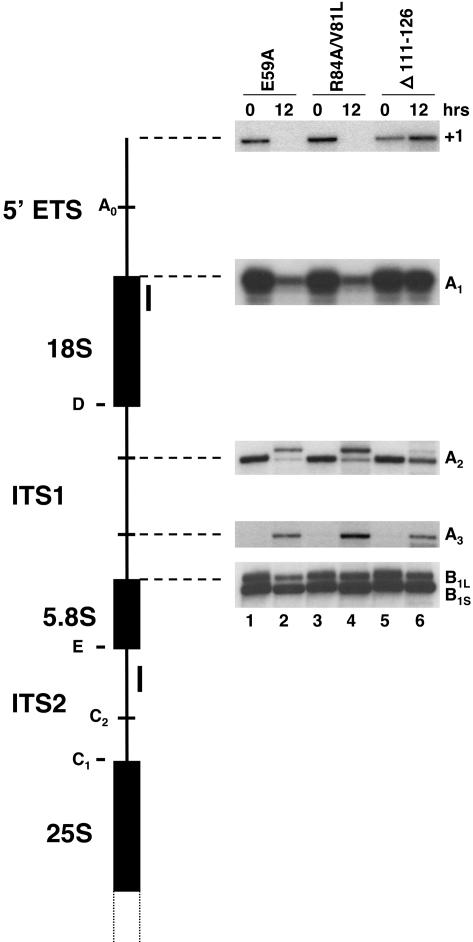
The U3 snoRNA is required for processing of the 35S pre-rRNA at sites A0, A1 and A2 (Hughes and Ares 1991; Beltrame and Tollervey 1992; Beltrame et al. 1994; Beltrame and Tollervey 1995). The decrease in the levels of mature U3 snoRNA found with the three lethal SNU13 mutations could prevent pre-rRNA processing. To assess pre-rRNA processing, primer extension analysis was carried out with total RNA harvested from YGALSNU13 in the presence of plasmid copies of mutants E59A, R84A/V81L, and Δ111-126 before, or 12 h after, transfer to glucose-containing medium (Fig. 4 ▶). Primer extension through the 5′ ETS from a primer complementary to the 5′ end of 18S rRNA showed a decrease in the stops at position +1 and A1 for mutants E59A and R84A/V81L. In contrast, the level of stops at position +1 is increased and the stop at A1 is unchanged or slightly decreased for mutant Δ111-126. Primer extension through ITS1 from a primer complementary to the 5′ region of ITS2 showed a decrease in the stop at A2 for all three mutants, with the decrease being more pronounced for mutants E59A and R84A/V81L. In addition, a new primer extension stop appears 5′ of A2 with mutants E59A and R84A/V81L, which is less apparent with mutant Δ111-126. The primer extension stop at A3 increases with all three mutants, whereas stops at B1L and B1S remained unchanged. These results suggest that mutations in SNU13 cause defects in pre-rRNA processing.
FIGURE 4.
Mutations in SNU13 disrupt pre-rRNA processing. Strain YGALSNU13 was grown in the presence of plasmidbased SNU13 mutants E59A (lanes 1,2), R84A/V81L (lanes 3,4), or Δ111-126 (lanes 5,6). Total RNA was prepared 0 and 12 h after transfer to glucose-containing medium and was subjected to primer extension by using end-labeled primer 5ETS or ITS1. Primer stops at the 5′ end of the 5′ ETS (+1) and at sites A1, A2, A3, B1L, and B1S are indicated.
Mutation of SNU13 causes a specific decrease in levels of the U4 snRNA, U6 snRNA, and box C/D snoRNAs, but not box H/ACA snoRNAs
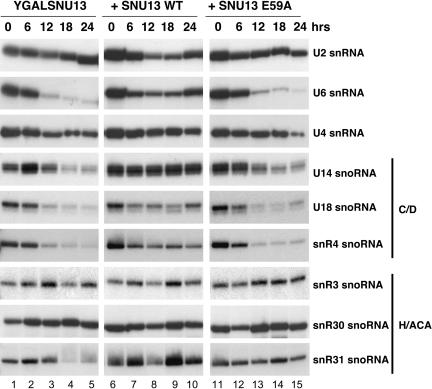
To further analyze the effects of the lethal mutations in SNU13 and to establish the basis of their effect on pre-mRNA splicing, the levels of various cellular RNAs were analyzed by primer extension (Fig. 5 ▶). Strain YGALSNU13 was grown in the absence of plasmid-based SNU13 and in the presence of plasmid-based SNU13 wild-type or SNU13 mutant E59A. RNA was isolated at 0-, 6-, 12-, 18-, and 24-h intervals after transfer to glucose. Primer extension reactions contained an equal quantity of total RNA at each time point; therefore, any differences observed reflected changes relative to the total RNA population.
FIGURE 5.
SNU13 mutation E59A results in reduced levels of specific cellular RNAs. Total RNA was prepared 0, 6, 12, 18, and 24 h after transfer to glucose-containing medium and was subjected to primer extension analysis by using an end-labeled primer complementary to specific cellular RNAs. Strain YGALSNU13 was grown alone (lanes 1–5) or with plasmid-based wild-type SNU13 (lanes 6–10) or SNU13 mutant E59A (lanes 11–15). The RNA analyzed in each case is indicated to the right of the panels.
The U4, U6, and U2 snRNAs were first chosen for analysis as SNU13 may impact either directly or indirectly on their levels. U4 snRNA is known to interact with Snu13p, whereas U6 snRNA interacts first with the U4 snRNA and then with the U2 snRNA during assembly of the spliceosome and formation of the catalytic site. The most dramatic result was found for U6 snRNA. Levels of U6 were found to decrease for YGALSNU13 after 12 h of growth in glucose, in which expression of chromosomal SNU13 is repressed (Fig. 5 ▶). Levels of U6 snRNA displayed a similar decrease in the presence of plasmid-based SNU13 mutant E59A, yet remained high at the end of the time course in the presence of wild type (Fig. 5 ▶). To a lesser extent, levels of U4 snRNA were also found to decrease, at approximately the same time, for both strain YGALSNU13 and for YGALSNU13 in the presence of SNU13 mutant E59A (Fig. 5 ▶). Again, levels of the U4 snRNA remained high at the end of the time course in the presence of wild-type plasmid-based SNU13. Levels of the U2 snRNA (Fig. 5 ▶) and U5 snRNA (data not shown) remained high at the end of the time course in all three SNU13 strains analyzed. The U14, U18, and snR4 snoRNAs were chosen for analysis as representatives of the box C/D snoRNAs, and the snR3, snR30 and snR31 as representatives of the box H/ACA class. Levels of the U14, U18, and snR4 snoRNAs decreased in strain YGALSNU13 and in the presence of plasmid-based mutant E59A after 12-h incubation in glucose-containing media (Fig. 5 ▶). In contrast, levels of the snR3 and snR30 snoRNAs remained high at the end of the time course for all strains (Fig. 5 ▶). The levels of snR31 remained high at the end of the time course for YGALSNU13 in the presence of SNU13 mutant E59A but showed a smear of primer extension stops in YGALSNU13. Taken together, these results suggest that the effect of Snu13p mutation is to decrease the stability or synthesis of specific cellular RNAs.
Differential interaction of SNU13 mutants with the U4 snRNA and U3 snoRNA
15.5K is known to interact with both the U4 snRNA and the U3 snoRNA (Nottrott et al. 1999; Watkins et al. 2000). The ability of recombinant Snu13p mutants E59A and R84A/V81L to bind to RNAs in vitro was investigated by using electrophoretic mobility shift assays. It was hypothesized that the phenotypes associated with mutations E59A and R84A/V81L may be due to the inability of the mutant proteins to bind to U4 snRNA and/or U3 snoRNA. This hypothesis was based on the involvement of the equivalent 15.5K protein residues E61 (E59) and K86 (R84; Fig. 1 ▶) in U4 snRNA binding in the cocrystal structure (Vidovic et al. 2000).
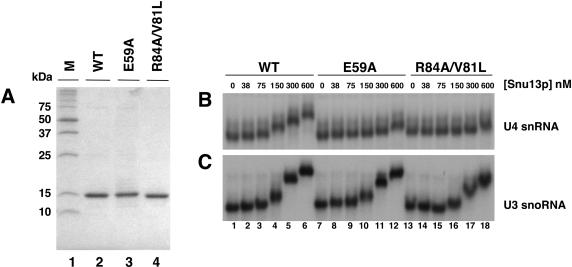
Recombinant His-tagged Snu13p was purified from wild type, mutant E59A, and mutant R84A/V81L constructs for use in electrophoretic mobility shift assays. Analysis of equal concentrations of mutant and wild-type protein preparations by SDS-PAGE demonstrated that the three were comparable in purity and quantity (Fig. 6A ▶). When in vitro transcribed wild-type U4 snRNA and recombinant Snu13p were used in electrophoretic mobility shift assays, binding of wild-type Snu13p resulted in a shift in mobility of the RNA at 75 nM Snu13p (Fig. 6B ▶). After this point, increasing Snu13p concentrations resulted in increased mobility shift up to 600 nM protein. A shift in mobility for Snu13p (E59A) and Snu13p (R84A/V81L) equivalent to that seen for 75 nM wild-type protein was only evident at 600 nM mutant protein. This indicates that interaction with the U4 snRNA is severely impaired for both mutants.
FIGURE 6.
Snu13p mutations E59A and R84A/V81L interact with the U3 snoRNA but not the U4 snRNA in vitro. (A) SDS-PAGE showing purified, His-tagged, Snu13p wild-type (lane 2), mutant E59A (lane 3), and mutant R84A/V81L (lane 4). Sizes of molecular weight markers (lane 1) are indicated to the left of the panel in kilodaltons. (B) Electrophoretic mobility shift assay (EMSA) using end-labeled in vitro transcribed U4 snRNA with increasing concentrations of recombinant wild-type Snu13p (lanes 1–6), Snu13p mutant E59A (lanes 7–12), and mutant R84A/V81L (lanes 13–18). (C) EMSA using end-labeled in vitro–transcribed U3 snoRNA containing both the B/C and C′/D boxes but lacking stem-loops 2, 3, and 4 with increasing concentrations of recombinant wild-type Snu13p (lanes 1–6), Snu13p mutant E59A (lanes 7–12), and mutant R84A/V81L (lanes 13–18). Recombinant protein concentrations (nM) are indicated above the panels. The specific activities of the U4 snRNA and U3 snoRNA are the same in B and C.
A region of the U3 snoRNA containing both the B/C and C′/D boxes but lacking stem-loops 2, 3, and 4, was in vitro transcribed and tested for binding to recombinant wild-type Snu13p, Snu13p (E59A), and Snu13p (R84A/V81L; Fig. 6C ▶). The yeast U3 snoRNA lacking stem loops 2, 3, and 4 is functional in vivo (Mereau et al. 1997; Samarsky and Fournier 1998; Marmier-Gourrier et al. 2003). Wild-type Snu13p was found to interact with U3 as expected, with concentrations comparable to those required for interaction with U4. As the concentration of Snu13p was increased, however, complexes with distinct mobilities were observed. These complexes have been previously observed for Snu13p and 15.5K and most likely represent the differences between the binding of one or two Snu13p molecules to the B/C and C′/D boxes in U3 (Watkins et al. 2000; Marmier-Gourrier et al. 2003). In the case of Snu13p (E59A), the binding observed was almost identical to that of wild type (Fig. 6C ▶). The mobility shifts observed for Snu13p (R84A/V81L) were only fractionally less than those of wild type at the same concentration. In contrast to results obtained with the U4 snRNA, both Snu13p mutants were able to bind to the U3 snoRNA as well as wild type. This differential binding exhibited by the Snu13p mutants to the U4 snRNA and U3 snoRNA indicates that there may be differences in the way the two RNAs are bound by Snu13p.
DISCUSSION
Snu13p is a component of both the U4/U6.U5 tri-snRNP and the U3 snoRNP (Gottschalk et al. 1999; Stevens and Abelson 1999; Watkins et al. 2000). The unprecedented discovery of this splicing factor as a core component of the box C/D snoRNPs raised questions about the role of the protein during pre-mRNA splicing and pre-rRNA processing. It is of particular interest to establish whether it is providing a common or related function in the two distinct processes. Previous work focused on the structural basis of the interaction of 15.5K with the U4 snRNA and U3 snoRNA with respect to the RNA structure and sequence (Nottrott et al. 1999; Vidovic et al. 2000; Watkins et al. 2000). Here we investigated the function of Snu13p in vivo by using mutagenesis to determine the role of individual amino acids or regions of the protein. Mutagenesis revealed that E59, R84/V81, and the C-terminal 16 amino acids are essential for Snu13p function and cell viability. Use of an inducible SNU13 strain has allowed investigation of the effects of these lethal SNU13 mutations on splicing of specific transcripts, rRNA processing, and the levels of specific RNAs. First, we have shown that the three lethal mutations E59A, R84A/V81L, and Δ111-126 resulted in defective splicing of the U3 snoRNA precursor. Mutation E59A also resulted in defective splicing of the intron-containing Saccharomyces cerevisiae genes ACT1, TUB1, and LSM2. Second, we found that the three lethal mutations E59A, R84A/V81L, and Δ111-126 all disrupted pre-rRNA processing. Next, we demonstrated that mutation E59A resulted in a decrease in the levels of the U4 snRNA, U6 snRNA, U3 snoRNA, and other box C/D snoRNAs, but not the H/ACA snoRNAs. Last, we demonstrated a decreased ability of Snu13p mutants E59A and R84A/V81L to bind to U4 snRNA but not the U3 snoRNA in vitro.
Three, of a total of 32 mutants, were found to have a negative effect on cell viability at 30°C, including just one of the 26 single-alanine substitutions. This surprisingly low proportion may reflect the functional importance of residues other than these charged amino acids or simply indicate that changes that are more substantial than single base substitutions are required to affect cell viability. Functional importance for substituted residues E59, R84, and V81 is implied by their evolutionary conservation (Nottrott et al. 1999). Snu13p residue glutamate 59 is absolutely conserved across species, as is valine 81, whereas arginine 84 is replaced by a lysine in all but S. cerevisiae (Nottrott et al. 1999). In the crystal structure of 15.5K bound to a U4 snRNA fragment, the equivalent 15.5K residues E61 (E59 in yeast) and K86 (R84) form part of a pocket in the protein that is the site of interaction with U4 snRNA residue U31 (summarized in Fig. 1 ▶). Both the amino group of K86 and the main-chain amide of E61 are involved in hydrogen bonding with the O4 atom of U31 (Vidovic et al. 2000). Given the high degree of sequence identity between the two proteins, a similar role can be predicted for Snu13p residues R84 and E59 in hydrogen bonding to the U4 snRNA. Although the equivalent residue to V81 (V83) is not directly in contact with the RNA in the cocrystal structure, an indirect role, perhaps in positioning R84, can be hypothesized from the mutagenesis data.
Although this is the first mutational analysis of Snu13p, there has been a study of a region of the mammalian SECIS-binding protein 2 (SBP2), which exhibits high sequence similarity to the RNA-binding domains of Snu13p and 15.5K (Allmang et al. 2002). In this analysis, a series of alanine mutants was constructed based on the residues that interact with the U4 snRNA in the 15.5K cocrystal structure and tested for RNA binding in vitro. The most dramatic results were obtained for residues E699 (equivalent to E59 in Snu13p) and R731 (equivalent to R84), which decreased binding to 13.5% and 0% of wild type, respectively. Our in vitro RNA-binding data of the two mutants (E59A and R84A/V81L) with the U4 snRNA support these findings.
Snu13p was initially identified as a tri-snRNP component and was therefore implicated in pre-mRNA splicing. Consistently, in cell extracts, splicing can be inhibited by adding an RNA containing the 15.5K binding site to sequester 15.5K (Nottrott et al. 1999). However, previous reports on the role of Snu13p in pre-mRNA splicing according to genetic depletion data have been contradictory. Initial depletion studies did not demonstrate a build up of unspliced pre-mRNA, implying that Snu13p was not necessary for efficient splicing (Watkins et al. 2000). However, more recent studies have shown that depletion of Snu13p is associated with defective splicing of U3 snoRNA precursors (Stevens et al. 2001). By using the galactose inducible strain YGALSNU13, we have demonstrated that lethal SNU13 mutations E59A, R84A/V81L, and Δ111-126 resulted in defective splicing of pre-U3 snoRNA and confirmed that a similar defect is associated with Snu13p depletion. We have also shown that the level of the mature U3 snoRNA is decreased with both depletion and mutation of Snu13p. It is likely that the observed defect in splicing of pre-U3 snoRNA affects the accumulation of mature U3 snoRNA. However, given the observed effects of mutations in SNU13 on the stability of the U4 and U6 snRNAs, it is not possible to rule out a similar effect on the stability of the mature U3 snoRNA. The observed reduction in levels of the U3 snoRNA may be attributed to either of these factors or a combination of the two. RT-PCR has demonstrated that splicing of genes ACT1, TUB1, and LSM2 is also adversely affected by SNU13 mutation E59A. This confirms a role for Snu13p in pre-mRNA splicing and suggests that Snu13p may be involved in splicing of all pre-mRNAs.
As SNU13 interacts with the U3 snoRNA, it was possible that the lethal mutations identified may also perturb some aspect of pre-rRNA processing. The U3 snoRNA is responsible for cleavages in the pre-rRNA at site A0, A1, and A2 (Hughes and Ares 1991; Beltrame and Tollervey 1992; Beltrame et al. 1994; Beltrame and Tollervey 1995). Analysis of pre-rRNA processing for the three SNU13 mutants revealed that they all displayed varying defects when the expression of the wild-type SNU13 was turned off. Specifically, mutation of the RNA-binding domain of SNU13 appears to influence pre-rRNA processing differently than does mutation in the C terminus. The C-terminal deletion displayed an accumulation of the 35S pre-rRNA as indicated by an increase in primer extension stops at +1, whereas mutants E59A and R84A/V81L did not. Levels of the mature 18S rRNA also varied between the C-terminal deletion and the E59A and R84A/V81L mutants. Furthermore, cleavage at site A2 was only slightly inhibited in the C-terminal deletion but was drastically reduced in mutants E59A and R84A/V81L with a concomitant appearance of a new primer extension stop. Although these differences may reflect interesting functional variations between the two mutant types, further detailed analysis of rRNA processing is required to determine if this is indeed the case.
Previous analysis of RNA levels using an inducible strain showed an initial decrease in levels of the box C/D snoRNAs on Snu13p depletion, followed by a decrease in levels of all cellular RNAs by 24 h (Watkins et al. 2000). Our data also indicate an initial decrease in levels of box C/D snoRNAs, as well as an initial decrease in levels of the U6 snRNA. However, our data reveal that by 24 h after Snu13p depletion, only the U4 snRNA levels drop, whereas the levels of the U2 snRNA, U5 snRNA, and H/ACA snoRNAs remain constant. Mutation E59A displayed a similar profile of RNA stability/instability as our depletion of Snu13p. Our data suggest that Snu13p is required to maintain the steady-state levels of a specific set of cellular RNAs (U4 snRNA, U6 snRNA, and box C/D snoRNAs) but not the majority of cellular RNAs, as previously proposed (Watkins et al. 2000).
Consistently, both mutations E59A and R84A/V81L resulted in decreased ability to bind to U4 snRNA in our electrophoretic mobility shift assay. However, different results were obtained for binding of these mutants to the U3 snoRNA. Mutants E59A and R84A/V81L appeared to bind as efficiently to U3, containing both the B/C and C′/D boxes but lacking stem-loops 2, 3, and 4, as the wild-type Snu13p. These results suggest that there are differences in the way that Snu13p interacts with U4 snRNA and U3 snoRNA. These differences could be due to the structures of the two RNAs before Snu13p binding. The helix–loop–helix that Snu13p binds in the U4 snRNA has been found to be, in part, preformed in structure to allow Snu13p binding (Mougin et al. 2002). In contrast, although the C′/D motif in the U3 snoRNA can form before Snu13p binding, the B/C motif does not (Marmier-Gourrier et al. 2003). Even though the B/C motif does not have a preformed structure before Snu13p binding, it still has higher affinity for Snu13p than the box C′/D motif (Marmier-Gourrier et al. 2003). In addition, mutations in the B/C motif affect growth in vivo more strongly than the corresponding mutations in the C′/D motif (Marmier-Gourrier et al. 2003). The mutations we have introduced in Snu13p may prevent its binding to a preformed helix–loop–helix such as that found in U4 and the C′/D motif of U3. Therefore, the ability of our Snu13p mutants to bind to the U3 snoRNA could be due to their initial binding to the unstructured box B/C, which may then allow binding to the box C′/D. Further analysis will determine whether the Snu13p mutants can bind an RNA containing only the U3 snoRNA box C′/D.
The Snu13p RNA-binding sites and the lethal mutations discussed involve only a small surface area of the protein. Further work must be carried out to elucidate the role of regions of the protein distant from this RNA-binding site. More specifically, the identity of the protein factors that are interacting with Snu13p to provide accurate function during pre-mRNA splicing and pre-rRNA processing must be determined. Given the similarities between the protein components of the U3 snoRNP and the U4/U6 snRNP, it is of particular interest to determine whether Snu13p is involved in similar protein–protein interactions in the two particles. Our binding data suggest that there are differences in the mode of interaction of Snu13p with the U4 snRNA and the U3 snoRNA. A cocrystal structure of Snu13p bound to a U3 snoRNA fragment similar to that obtained for 15.5K and the U4 snRNA will facilitate the detailed analysis of this binding interaction.
MATERIALS AND METHODS
Plasmid construction and mutagenesis
The SNU13 gene was cloned by PCR amplification with Vent DNA polymerase (New England Biolabs) of yeast genomic DNA (Promega) by using primers Snu13F and Snu13B complementary to the 5′ and 3′ untranslated regions of the gene and containing restriction sites for BamHI and KpnI. The PCR product was digested with BamHI and KpnI, then ligated into pRS416 (Sikorski and Hieter 1989) digested with the same enzymes to produce plasmid pRS416-Snu13, or digested with BamHI then ligated into pRS413 (Sikorski and Hieter 1989) digested with BamHI and HincII to produce plasmid pRS413-Snu13. The identity of the inserts was confirmed by sequencing. Mutagenesis of pRS413-Snu13 was carried out according to the method of Kunkel (1985), using primers 13–1 through 13–27. Plasmids containing mutated SNU13 were named according to the mutation introduced (e.g., pRS413-Snu13-K7A, pRS413-Snu13-D13A). pET-Snu13 was created by PCR amplifying the SNU13 coding region from plasmid pRS413-Snu13 by using primers Snu13pET21F and Snu13pET21R (containing an XhoI restriction site) and Pfu polymerase (Stratagene). Products were digested with XhoI, ligated into vector pBluescriptIIKS+ (Stratagene) cut with EcoRV and XhoI to give pBS-Snu13. After sequencing, mutagenesis was repeated, using pBS-Snu13 as a template and primers 13–11 and 13–17 to generate SNU13 mutants E59A and R84A/V81L. Resulting plasmids were sequenced to confirm that mutagenesis was successful, and digested with NheI and XhoI for ligation into the NheI and XhoI sites of pET21d (Novagen) to give pET-Snu13-E59A and pET-Snu13-R84A/V81L. The SNR14 gene encoding the U4 snRNA was cloned by PCR amplification with Vent DNA polymerase of yeast genomic DNA by using primers U4GF and U4GB complementary to the 5′ and 3′ untranslated regions of the gene and containing restriction sites for BamHI and KpnI. The PCR product was digested with BamHI and KpnI and then ligated into pRS416 digested with the same enzymes to produce plasmid pRS416-U4. The identity of the insert was confirmed by sequencing. The coding region of the SNR17 gene encoding the U3 snoRNA was amplified by RT-PCR from total RNA extracted from strain YGALSNU13 (see below) grown in galactose in the presence of pRS413-Snu13; 0.2 nmole primer T7U3R was hybridized to 0.7 μg total RNA by heating for 10 min to 70°C and placing on ice. First-strand cDNA was produced in a 35 μL reaction containing RT buffer (25 mM Tris-HCl at pH 8.3, 50 mM KCl, 2 mM DTT, and 5 mM MgCl2), 0.6 mM each dNTP, 40 U RNasin (Promega), and 10.5 U Super RT (HT Biotech) for 1 h at 42°C. Two microliters of this cDNA preparation was used in a PCR reaction by using primers T7U3R and T7U3F and Pfu polymerase. RT-PCR products were purified on a 1% agarose gel and inserted into the EcoRV site of pBluescriptIIKS+ to give pBS-U3 and then were sequenced. A truncation of the U3 snoRNA lacking stem-loops 2, 3, and 4 was produced by oligo-directed mutagenesis of pBS-U3, using primer U3del234, to give pBS-U3del234.
Yeast manipulation
For analysis of SNU13 mutations, the diploid strain Y20267 (BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YEL026w::kanMX4/YEL026w) was obtained from EUROSCARF and was transformed with pRS416-Snu13 by using the lithium acetate transformation procedure (Gietz et al. 1992). Viable G418-resistant and Ura+ transformants were sporulated, and tetrads were dissected. The haploid progeny were analyzed to identify the strain YSNU13KO (Mat a; his3Δ1; leu2Δ0; lys2Δ0; MET15; ura3Δ0; YEL026w::kanMX4; pRS416-Snu13). This strain was transformed with mutant pRS413-Snu13 plasmids, and cells were tested for viability by plasmid shuffle on 5-fluoro-orotic acid. A yeast strain with the SNU13 gene under control of the GAL1 promoter was constructed by PCR of plasmid pFA6a-kanMX6-PGAL1 (Longtine et al. 1998) with primers 13GALF and 13GALB. The resulting PCR product was transformed into the yeast strain BY4743 (Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; LYS2/lys2Δ0; met15Δ0/MET15; ura3Δ0/ura3Δ0), and G418-resistant colonies were selected. PCR of genomic DNA from G418-resistant colonies was used to confirm the correct integration of the GAL1 promoter, and these strains were sporulated and tetrads dissected. The haploid progeny were analyzed to identify the strain YGALSNU13 (Mat a; his3Δ1; leu2Δ0; met15Δ0; LYS2; ura3Δ0; kanMX6:PGAL1-SNU13). This strain was transformed with plasmids pRS413-Snu13, pRS413-Snu13-E59A, pRS413-Snu13-R84A/V81L, and pRS413-Snu13-ΔC. Growth curves in liquid culture were performed by initially growing yeast in YP-Gal overnight. Cells were then collected by centrifugation, washed with water, and resuspended in YPD or YP-Gal to an OD600 of 0.02. OD measurements were taken over a period of 36 h to monitor growth.
Primer extensions and RT-PCR
Total RNA was extracted by using the hot phenol method (Kohrer and Domdey 1991) from strain YGALSNU13 alone or in the presence of plasmid-based SNU13 mutants E59A, R84A/V81L, Δ111-126 or wild-type SNU13 after incubation in YPD for 0, 6, 12, 18, or 24 h. Total RNA was then hybridized to a 32P end-labeled oligonucleotide primer (3.5 μg with primers U3snoRNA-RT, U2RT2, U6B, U4S2, U14snoRNA-RT, U18RT, snr4RT, R3snoRNA-RT, R30snoRNA-RT, and snr31; or 9.5 μg with primers 5ETS, ITS1) by heating for 10 min to 55°C in RT buffer (50 mM Tris-HCl at pH 8.3, 100 mM KCl, 4 mM DTT, 10 mM MgCl2). Primer extension was carried out by adding 0.5 mM each dNTP, 0.1 U/μL RNasin, and 0.2 U/μL Super RT and incubating for 30 min at 41°C. The extension reaction was stopped by heating for 1 min to 100°C and then was incubated with 10 μg RNase A for 15 min at 37°C before 200 μL stop solution (0.3 M sodium acetate at pH 5.3, 1 mM EDTA, 0.1% SDS, 25 μg/mL Escherichia coli tRNA) was added. Reactions were extracted with phenol/chloroform/iso-amyl alcohol (25:24:1) and precipitated with ethanol. After resuspension in formamide gel-loading buffer, primer extension products were run on a 6% or 8% polyacrylamide gel containing 6 M urea. Dried gels were exposed to X-ray film overnight at −80°C in the presence of an intensifying screen. Poly(A)+ mRNA was prepared from 200 μg total RNA by using the GenElute mRNA miniprep kit (Sigma) for YGALSNU13 grown for 12 h at 30°C in the presence of plasmid based SNU13 mutant E59A or wild-type SNU13 in YPD or YPGal, respectively. RT-PCR was carried out as previously described (O’Keefe 2002).
Expression and purification of recombinant Snu13p
E. coli strain BL21(DE3) (Novagen) was transformed with pET-Snu13, pET-Snu13-E59A, or pET-Snu13-R84A/V81L and grown at 37°C to an OD550 of 0.6. IPTG (iso-propylthiogalactoside) was added to a concentration of 0.4 mM to induce protein expression, and ethanol was added to a concentration of 2%. After incubation for 4 h at 30°C, cells were pelleted and placed overnight at −80°C. Pellets were resuspended in 15 mL ice-cold lysis buffer (50 mM NaH2PO4 at pH 8, 300 mM NaCl, 10 mM imidazole) and incubated for 30 min at 4°C in the presence of 1 mg/mL lysozyme (Sigma). Cells were then subjected to sonication by using a tapered microtip with four 20-sec bursts at an amplitude of 30%. Insoluble material was pelleted by centrifugation at 35,000g for 15 min, and the cleared lysate was loaded directly onto a 1 mL Ni2+-NTA affinity column (Qiagen). Bound protein was washed and eluted in 50 mM NaH2PO4 and 300 mM NaCl buffer (pH 8) containing either 20 mM (wash) or 250 mM (elution) imidazole. Recombinant Snu13p eluted across the first two 1 mL fractions and was dialyzed against buffer containing 50 mM NaH2PO4 (pH 8), 300 mM NaCl, 1 mM DTT, and 20% glycerol overnight at 4°C to remove imidazole. Protein was visualized on a 15% SDS-PAGE gel, and protein concentrations were determined by using the BioRad dye binding reagent with bovine serum albumin as the standard.
RNA transcription and electrophoretic mobility shift assay
A truncated U3 snoRNA containing the B/C and C′/D boxes, but lacking stem-loops 2, 3, and 4, was transcribed from PCR products amplified from plasmid pBS-U3del234 by using primers T7U3F and T7U3R. U4 snRNA was transcribed from PCR products amplified from plasmid pRS416-U4 by using primers U4F and U4B. UpG-primed transcription reactions contained 2 mM UpG; l U/μL RNasin; 1 mM each of ATP, CTP, and UTP; 0.4 mM GTP; and 1 U/μL T7 RNA Polymerase (Promega) in transcription buffer (40 mM Tris-Cl at pH 8, 2 mM spermidine, 10 mM DTT, and 20 mM MgCl2). Transcribed RNAs were treated with 20 U DNase (Promega) for 15 min at 37°C, phenol extracted, and purified on a 6% denaturing acrylamide gel. For electrophoretic mobility shift assays, recombinant Snu13p was incubated with 25,000 dpm 32P end-labeled U3 snoRNA or U4 snRNA for 30 to 45 min on ice. Resulting protein–RNA complexes were resolved on native 6% acrylamide gels. Dried gels were exposed to X-ray film overnight at −80°C in the presence of an intensifying screen.
Acknowledgments
We thank Oliver Sinclair for constructing some of the Snu13 mutants and Lily Novak Frazer for critical reading of the manuscript. This work was supported by a grant and studentship (H.C.D.) from the Biotechnology and Biological Sciences Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5970404.
REFERENCES
- Allmang, C., Carbon, P., and Krol, A. 2002. The SBP2 and 15.5K/Snu13p proteins share the same RNA binding domain: Identification of SBP2 amino acids important to SECIS RNA binding. RNA 8: 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame, M. and Tollervey, D. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14: 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame, M., Henry, Y., and Tollervey, D. 1994. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 22: 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosome. In The RNA world, 2nd ed. (eds. R.F. Gesteland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Cahill, N.M., Friend, K., Speckmann, W., Li, Z.-H., Terns, R.M., Terns, M.P., and Steitz, J.A. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21: 3816–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz, J., Kressler, D., and Linder, P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24: 192–198. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi, S., Fatica, A., Bachi, A., Scaloni, A., Presutti, C., and Bozzoni, I. 2002. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 22: 6663–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, T., Berges, T., Tollervey, D., and Hurt, E. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17: 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk, A., Neubauer, G., Banroques, J., Mann, M., Lührmann, R., and Fabrizio, P. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18: 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. 1999. ESPript: Multiple sequence alignments in Postscript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Granneman, S., Pruijn, G.J.M., Horstman, W., van Venrooij, W.J., Lührmann, R., and Watkins, N.J. 2002. The hU3–55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in vitro. J. Biol. Chem. 277: 48490–48500. [DOI] [PubMed] [Google Scholar]
- Hughes, J.M.X. and Ares, M. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10: 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach, C., Walke, S., and Nagai, K. 1999. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9: 222–230. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer, K. and Domdey, H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., Bork, P., and Sander, C. 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22: 2166–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T.A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. 82: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, P. 2000. Quick guide: DEAD-box proteins. Curr. Biol. 10: R887. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D. and Guthrie, C. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817. [DOI] [PubMed] [Google Scholar]
- Makarova, O.V., Makarov, E.M., Liu, S., Vornlocher, H.-P., and Lührmann, R. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6.U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21: 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmier-Gourrier, N., Clery, A., Senty-Segault, V., Charpentier, B., Schlotter, F., Leclerc, F., Fournier, R., and Branlant, C. 2003. A structural, phylogenetic, and functional study of 15.5kD/Snu13 protein binding on U3 small nucleolar RNA. RNA 9: 821–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereau, A., Fournier, R., Gregoire, A., Mougin, A., Fabrizio, P., Lührmann, R., and Branlant, C. 1997. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: Protein–RNA contacts and base-pair interaction with the pre-ribosomal RNA. J. Mol. Biol. 273: 552–571. [DOI] [PubMed] [Google Scholar]
- Mougin, A., Gottschalk, A., Fabrizio, P., Lührmann, R., and Branlant, C. 2002. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell’s and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J. Mol. Biol. 317: 631–649. [DOI] [PubMed] [Google Scholar]
- Nilsen, T.W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function (eds. R.W. Simons and M. Grunberg-Manago), pp. 279–307. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nottrott, S., Hartmuth, K., Fabrizio, P., Urlaub, H., Vidovic, I., Ficner, R., and Lührmann, R. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18: 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott, S., Urlaub, H., and Lührmann, R. 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: A biochemical role for U4/U6 proteins. EMBO J. 21: 5527–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe, R.T. 2002. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res. 30: 5476–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., Siliciano, P.G., and Guthrie, C. 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239. [DOI] [PubMed] [Google Scholar]
- Samarsky, D.A. and Fournier, R. 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 18: 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin, B., Kretzner, L., and Rosbash, M. 1988. A U1 snRNA: Pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 7: 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Stevens, S.W. and Abelson, J. 1999. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. 96: 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Barta, I., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6.U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Ryan, D.E., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9: 31–44. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. and Kiss, T. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9: 337–342. [DOI] [PubMed] [Google Scholar]
- Valadkhan, S. and Manley, J.L. 2001. Splicing-related catalysis by protein-free snRNAs. Nature 413: 701–707. [DOI] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Verheggen, C., Mouaikel, J., Thiry, M., Blanchard, J.-M., Tollervey, D., Bordonné, R., Lafontaine, D.L.J., and Bertrand, E. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 20: 5480–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Lührmann, R., and Ficner, R. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Ségault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C., and Lührmann, R. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Dickmanns, A., and Lührmann, R. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22: 8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein Szewczak, L.B., DeGregorio, S.J., Strobel, S.A., and Steitz, J. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9: 1095–1107. [DOI] [PubMed] [Google Scholar]
- Will, C.L. and Lührmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Yean, S.L., Wuenschell, G., Termini, J., and Lin, R.J. 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, Y. and Weiner, A.M. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835. [DOI] [PubMed] [Google Scholar]