Abstract
Positive-strand RNA viruses replicate their RNA genome within a ribonucleoprotein (RNP) complex that is associated with cellular membranes. We used a two-step method of purification to isolate hepatitis C virus (HCV) RNP complexes from human hepatoma cell line Huh7, which stably expresses HCV subgenomic replicons. The procedure involved hybridization of replicon-expressing cellular lysates with oligonucleotides tagged with biotin and digoxigenin at their respective termini complementary to subgenomic replicon RNA followed by avidin-agarose enrichment of the mixture and subsequent immunoprecipitation of biotin-eluted material with anti-digoxigenin antibody. The immunoprecipitates were immunoblotted with antisera against HCV nonstructural (NS) proteins. The analysis revealed the association of all the HCV NS proteins (NS3, NS4a, NS4b, NS5a, and NS5b) that are encoded by the subgenomic replicon RNA. The HCV RNP complex migrated in a native polyacrylamide gel with an approximate molecular mass of 450 kD. The association of these viral proteins in the RNP complex reinforces the widely acknowledged notion that RNA viruses accomplish replication within a membranous RNP complex.
Keywords: HCV, HCV replicon, ribonucleoprotein complex, biotin, digoxigenin
INTRODUCTION
About 3% of the world population is chronically infected with hepatitis C virus (HCV). More than 20% of HCV-infected patients develop cirrhosis, and 1%–5% of those develop liver cancer (Di Bishceglie 1997). HCV is an enveloped positive-strand RNA virus and belongs to the genus Hepacivirus in the family Flaviviridae (Murphy et al. 1995). The 9.6-kb RNA genome consists of a single open reading frame (ORF) flanked by 5′ and 3′ noncoding regions (NCRs). The 5′ NCR contains an internal ribosome entry site (IRES), which regulates cap-independent translation of the long open reading frame (ORF) of ~3011 amino acids (Tsukiyama et al. 1992; Wang et al. 1993; Rijnbrand and Lemon 1999). Although the overall sequence of HCV RNA displays significant diversity within the coding region among the various isolates, the 5′ and 3′ NCRs are relatively conserved (Bartenschlager and Lohmann 2000; Reed and Rice 2000). Among the several host proteins that have been shown to bind the 5′ and 3′ HCV-NCR, interactions with PTB and La autoantigen have been studied in considerable detail (Ali and Siddiqui 1995; Ali and Siddiqui 1997; Ito and Lai 1997; Spangberg et al. 1999).
HCV proteins are generated from a polyprotein precursor that is co- and posttranslationally processed by cellular and viral proteases to yield the mature structural and nonstructural proteins (Hijikata et al. 1993). The structural proteins include the core protein, which forms the viral capsid, and the envelope glycoproteins E1 and E2. These are followed by a series of nonstructural proteins, p7, NS2, NS3, NS4a, NS4b, NS5a, and NS5b (Bartenschlager and Lohmann 2000; Reed and Rice 2000). A new protein termed F is thought to be produced by −2/+1 ribosomal frameshift during translation (Walewski et al. 2001; Xu et al. 2001). NS3 protein contains an N-terminal protease and C-terminal helicase activity (Bartenschlager and Lohmann 2000; Reed and Rice 2000). NS5b encodes an RNA-dependent RNA polymerase (RdRp) that is responsible for viral RNA synthesis. NS5a has drawn considerable interest largely because of its possible contribution to interferon resistance (Gale et al. 1997; Reyes 2002). To date, a wide variety of host cellular proteins have been shown to interact with NS5a (Reyes 2002). It has also been implicated in a variety of intracellular events, none of which shows any functional relevance to RNA synthesis. The most convincing arguments for its indispensable role in replication is its inclusion in the subgenomic replicon and the frequent appearance of cell culture-adaptive mutations in the NS5a-coding sequences, which are believed to contribute to efficient RNA replication (Lohmann et al. 1999; Blight et al. 2000). However, the exact mechanism of how NS5a plays this functional role in replication remains to be investigated. HCV RNA is translated on the rough ER and replicates within the RNP complexes in the ER membrane (Dubuisson et al. 2002). A recent study has described the association of RNA replication with lipid rafts (Shi et al. 2003). All known positive-strand RNA viruses replicate their RNA on intracellular membranes in association with vesicles, Golgi, or other membrane structures (Lai 1998; Ahlquist 2002). Although it is known that membrane association is important for viral RNA synthesis, the composition, organization, and function of the ribonucleoprotein complexes have not been fully characterized. The HCV viral proteins required for RNA replication are genetically defined by the minimal HCV coding sequences included in the subgenomic replicons (Lohmann et al. 1999). These include NS3, NS4a, NS4b, NS5a, and NS5b, and have been recently shown to interact with each other (Dimitrova et al. 2003). The distribution of adaptive mutations in the replicon, which contributes to efficient RNA replication, reinforces the functional significance of the NS proteins (Lohmann et al. 1999; Blight et al. 2000). In this study, we describe a two-step affinity-coupled immunoprecipitation purification scheme for the isolation of the HCV ribonucleoprotein (RNP) complex. We demonstrate the association of all the HCV NS proteins whose coding sequences are included in the subgenomic replicon with the RNP complex that is likely engaged in RNA replication. The stoichiometry of viral proteins binding to RNA was not readily apparent owing to different sensitivities of antiserum used in the Western blot assays.
RESULTS
General strategy for the isolation of HCV RNP complexes
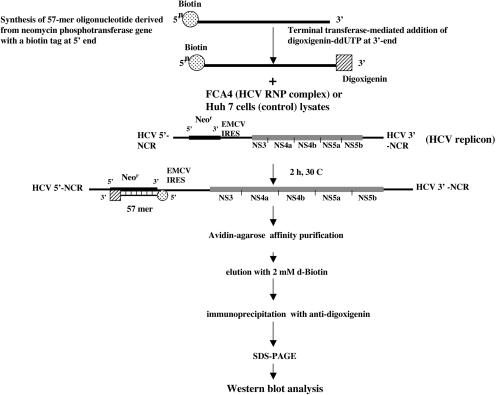
The general scheme for RNP isolation used in this study is illustrated in Figure 1 ▶. The HCV subgenomic replicon contains the neomycin-resistance gene under the translational control of the HCV IRES. An oligonucleotide complementary to the neomycin-resistance gene was synthesized with a biotin tag at the 5′ end. Subsequently, digoxigenin was added to the 3′ end of the same oligonucleotide using the terminal transferase enzyme. Similarly, we also synthesized an anti-sense neomycin oligonucleotide to isolate the RNP complex containing minus-strand HCV RNA. An unrelated biotin- and digoxigenin-tagged oligonucleotide bearing no homology to subgenomic replicon RNA was also included in these assays to serve as a negative control. Neomycin oligonucleotides (57-mer) with biotin and digoxigenin tags were added to Huh7 or FCA4 lysates (see Materials and Methods) prepared according to Ali et al. (2002). After incubating the lysates for 2 h with neomycin oligonucleotides, avidin-agarose beads were added to the mixture. The mixture was then incubated overnight at 4°C and washed several times. The material bound to the avidin beads was eluted with biotin, and the eluate was immunoprecipitated with the anti-digoxigenin monoclonal antibody. The immunoprecipitates were eluted and subjected to SDS-PAGE and immunoblotted with the respective anti-HCV NS proteins sera.
FIGURE 1.
Two-step purification scheme for the isolation of the HCV RNP complexes. The organization of the HCV subgenomic replicon is presented. Replicon RNA serves as a substrate for the hybridization of a 57-nt (5′) biotin- and (3′) digoxigenin-tagged oligonucleotide representing neomycin-resistance gene sequences in the replicon. Both sense and anti-sense neomycin oligonucleotides complementary to either plus or minus replicon RNA strands were synthesized. A flowchart illustrates the avidin-affinity and anti-digoxigenin immunoprecipitation procedures used in the purification scheme.
HCV RNP complex contains HCV NS proteins
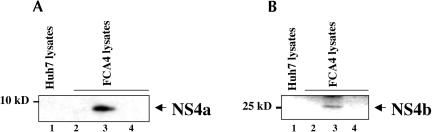
The development of the HCV subgenomic replicon system, pioneered by Bartenschlager and colleagues (Lohmann et al. 1999), represents a major breakthrough in the HCV field. It opened avenues of investigations relating to the HCV gene expression, replication, and impact of viral gene expression on infected hepatocytes. We chose to use the subgenomic replicon system to investigate the HCV RNP complexes. The FCA4 cell line developed by Guo et al. (2001) represents a human hepatoma cell line, which stably replicates the HCV subgenomic replicon RNA and is maintained in G418-containing medium as it has a neomycin-resistant gene sequence. The cellular lysates containing HCV RNP complexes were prepared by the scheme outlined in Figure 1 ▶, and subjected to a series of Western blot analyses using several anti-HCV sera. In the first analysis, we used antisera against NS4a and NS4b to determine the association of these proteins in the RNP complexes. The Western blot analysis carried out with anti-NS4a and NS4b sera clearly shows the association of these two HCV NS proteins in the RNP complexes that were purified from FCA4 lysates using the neomycin oligonucleotide complementary to the plus-strand replicon RNA (Fig. 2A,B ▶, lanes 3). However, when Huh7 cellular lysates were treated with neomycin oligonucleotides, neither protein was observed (Fig. 2A,B ▶, lanes 1). In two separate control experiments, a similar absence of NS4a and NS4b was noted (Fig. 2A,B ▶, lanes 2,4). The first control involves the incubation of FCA4 lysates with a nonspecific (unrelated) oligonucleotide tagged with biotin and digoxigenin at respective termini similar to the neomycin oligonucleotides (Fig. 2A,B ▶, lanes 4). The second control is the direct immunoprecipitation of FCA4 lysates with the anti-digoxigenin antibody, omitting the first step of hybridization with biotin- and digoxigenin-tagged oligonucleotides (Fig. 2A,B ▶, lanes 2). NS4a is a 6-kD polypeptide that has been shown to function as a cofactor for NS3 protease activity (Ishido et al. 1998). The function of NS4b is presently unknown. The functional relevance of both proteins in RNA replication remains to be investigated.
FIGURE 2.
HCV NS4a/b proteins are associated with the HCV RNP complex. Western blot analysis of the RNP complexes was carried out after two-step purification using antisera against NS4a and NS4b (see Materials and Methods). (A,B, lanes 1) Lysates prepared from Huh 7 cells hybridized with oligonucleotides specific to the neomycin gene that is complementary to the plus replicon RNA strand; (lanes 2) FCA4 lysates directly immunoprecipitated with anti-digoxigenin monoclonal antibody, omitting the first step of hybridization with biotin- and digoxigenin-tagged oligonucleotides; (lanes 3) FCA4 lysates hybridized to oligonucleotides specific to the neomycin gene that is complementary to the plus replicon RNA strand; (lanes 4) FCA4 lysates hybridized to nonspecific (unrelated) oligonucleotides.
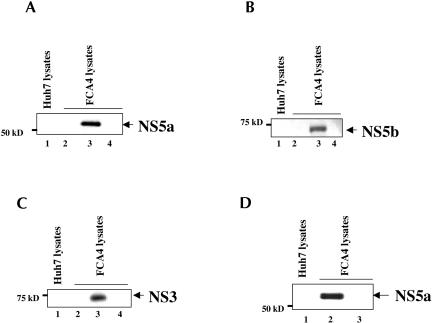
Next, we continued a second series of immunoblot assays, which involved the use of antisera against NS5a, NS5b, and NS3 proteins to determine the association of these HCV NS proteins in the HCV RNP complexes. The results shown in Figure 3A–C ▶ demonstrate the presence of NS5a, NS5b, and NS3 proteins in the RNP complex (lanes 3). Several control experiments, similar to those described in Figure 2 ▶, were included in this analysis, and none of these revealed the presence of respective HCV NS proteins (Fig. 3A–C ▶, lanes 1,2,4). These include treatment of Huh7 lysates with neomycin oligonucleotides (Fig. 3A–C ▶, lanes 1), FCA4 lysates with nonspecific (unrelated) oligonucleotides (lanes 4), and direct immunoprecipitation with anti-digoxigenin antibody (lanes 2). These results demonstrate the association of HCV NS proteins with the HCV RNP complexes. Because we used a neomycin oligonucleotide complementary to plus-strand replicon RNA in the purification scheme (Fig. 1 ▶), the HCV RNP complexes so purified contained the plus-strand HCV RNA in the RNP complexes.
FIGURE 3.
HCV NS5a, NS5b, and NS3 proteins are associated with the HCV RNP complex. Western blot analysis carried out with Huh7 or FCA4 lysates purified by the two-step purification scheme outlined in Figure 1 ▶, with (A) anti-NS5a, (B) anti-NS5b, and (C) anti-NS3 antibodies. (Lanes 1) Huh7 lysates subjected to hybridization with the neomycin oligonucleotide complementary to the plus-strand RNA replicon; (lanes 3) FCA4 lysates hybridized with the same neomycin oligonucleotide and subjected to the two-step purification scheme; (lanes 2) FCA4 lysates directly immunoprecipitated with anti-digoxigenin antibody; (lanes 4) FCA4 lysates hybridized to a nonspecific (unrelated) oligonucleotide of similar length and subjected to the two-step purification scheme. (D) The HCV RNP complexes contain minus-strand RNA in the HCV RNP complexes. Equal amounts of FCA4 and Huh7 lysates were incubated with the biotin- and digoxigenin-tagged neomycin-resistant oligonucleotide that is complementary to the minus-strand RNA replicon and purified as described in Figure 1 ▶, followed by Western blot analysis with anti-NS5a antibody. (Lanes 1,2) Huh7 and FCA4 lysates hybridized with complementary oligonucleotides specific to the minus-strand RNA neomycin gene; (lane 3) FCA4 lysates hybridized with the nonspecific (unrelated) oligonucleotides.
To demonstrate the presence of minus-strand replicon RNA in the complex, we used a neomycin oligonucleotide complementary to the minus HCV RNA strand during affinity purification of the RNP complexes (Fig. 1 ▶). FCA4 cellular lysates were incubated with the neomycin oligonucleotides complementary to the minus-strand replicon RNA, and Western blot analysis was carried out with anti-NS5a antibody (Fig. 3D ▶). NS5a was found to be associated with RNP complexes containing minus-strand HCV replicon RNA (lane 2). We used NS5a as representative of the HCV NS proteins. Huh7 cellular lysates treated with anti-sense neomycin oligonucleotides (lane 1) or FCA4 lysates incubated with an unrelated oligonucleotide, purified by avidin-affinity and immunoprecipitated with the anti-digoxigenin antibody and subsequently immunoblotted with anti-NS5a antibody, did not reveal the association of NS5a (lane 3). These results indicate that RNP complexes formed in replicon-expressing Huh7 cells contain either and or plus/minus strand viral RNAs.
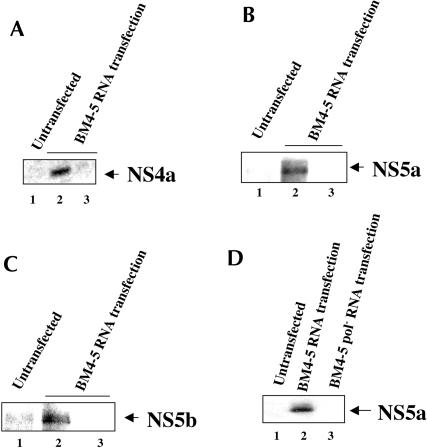
To demonstrate that the formation of RNP complex is not specific to the FCA4 stable cell line, we performed transient transfections of Huh7 cells with an in vitro synthesized subgenomic replicon BM4–5 RNA (Guo et al. 2001). Huh7 lysates prepared from BM4–5 RNA transiently transfected cells were subjected to the two-step purification scheme outlined in Figure 1 ▶. The results of this analysis clearly show the association of three tested HCV NS proteins (NS4a, NS5a, NS5b; Fig. 4A–C ▶) in the Huh7 cellular lysates transiently transfected with BM4–5 replicon RNA and hy-bridized with neomycin oligonucleotides complementary to plus-strand replicon RNA (Fig. 4A–C ▶, lanes 2). The same cellular lysates when hybridized with unrelated oligonucleotides did not show the association of any of the HCV NS proteins (Fig. 4A–C ▶, lanes 3). Similarly, the untransfected Huh7 cells (lanes 1) or BM4–5 RNA pol− transfected Huh7 cellular lysates subjected to hybridization with neomycin oligonucleotides failed to reveal HCV NS proteins in the Western blot analysis (Fig. 4D ▶, lane 3). These studies together demonstrate the successful isolation of HCV RNP complex using this novel two-step procedure and document the presence of all the HCV proteins encoded by the subgenomic replicon in the HCV RNP complexes.
FIGURE 4.
Association of the HCV NS proteins in the RNP complexes purified from Huh7 cells transiently transfected with BM4–5 HCV replicon RNA. Huh 7 cells were transiently transfected with BM4–5 replicon RNA as described in Materials and Methods. The cellular lysates were purified according to the scheme outlined in Figure 1 ▶. The Western blot analysis was carried out with anti-NS4a (A), anti-NS5a (B, D), and anti-NS5b (C) antibodies. Untransfected (lanes 1) or BM4–5 replicon RNA transiently transfected Huh7 cellular lysates were either hybridized with the neomycin-resistant oligonucleotides complementary to the plus-strand replicon (lanes 2) or hybridized with a nonspecific oligonucleotide (lanes 3). (D, lane 1) Untransfected; (lanes 2,3) BM4–5 replicon RNA and BM4–5 replicon pol− RNAs were transiently transfected in Huh7 cells. Cellular lysates were hybridized with the neomycin-resistant oligonucleotides complementary to the plus-strand replicon.
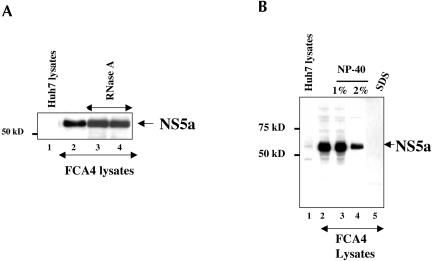
Addition of increasing amounts of RNase A to the RNP complexes purified from FCA4 lysates hybridized with plus-strand-specific neomycin oligonucleotides left the RNP complex intact as evidenced by the appearance of NS5a protein. This result indicates that the viral replicon RNA is protected against the nuclease attack owing to its association with the RNP complex (Fig. 5A ▶, lanes 3,4). The NS5a protein band in lane 2 represents a positive control in which FCA4 lysates were not treated with RNase A, incubated with the neomycin oligonucleotides, and purified as described above (Fig. 1 ▶). Our results are consistent with previous reports in which viral RNA was found to be protected from nucleases as RNP complexes are membrane-associated (Ahlquist 2002; Schwartz et al. 2002).
FIGURE 5.
Effect of RNase A and detergent treatments on the HCV RNP complexes. (A) Mixture of FCA4 replication lysates and labeled neomycin oligonucleotides were either untreated (lane 2) or treated with 1 μg/mL RNase A (lane 3) or 3 μg/mL RNase A (lane 4). (Lane 1) The Huh7 lysates hybridized with labeled neomycin oligonucleotides. (B) Effect of NP-40 and SDS on HCV RNP complexes. Equal concentrations of Huh7 and FCA4 lysates were incubated with biotin- and digoxigenin-tagged neomycin oligonucleotides, and RNP complexes were purified as described in Figure 1 ▶, followed by Western blot analysis with anti-NS5a antibody. (Lanes 1,2) Untreated Huh7 and FCA4 lysates; (lanes 3,4) FCA4 lysates treated with 1% or 2% NP-40, respectively; (lane 5) FCA4 lysates treated with 0.5% SDS.
The HCV RNP complexes have been recently shown to be resistant to nonionic detergents (Shi et al. 2003). We treated the isolated RNP complexes with both nonionic and anionic detergents to determine if HCV NS proteins would remain intact in the complexes. Our results show that the HCV RNA complexes are resistant to 2% NP-40, whereas SDS treatment destroyed the HCV RNP complexes (Fig. 5B ▶, lane 5). The Western blot analysis demonstrated the presence of a representative NS5a protein in the RNP complexes treated with NP-40 (Fig. 5B ▶, lanes 3,4). Treatment of FCA4 lysates with 1% SDS, however, completely disrupted the RNP complexes, indicating that these complexes are assembled as membrane-bound RNA–protein complexes (lane 5).
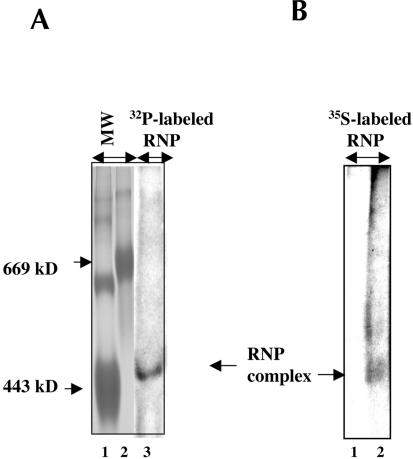
Next, we attempted to determine an approximate molecular mass of the HCV RNP complex using the nondenaturing polyacrylamide gel electrophoresis (PAGE) system that has been previously described for estimating the molecular mass of spliceosome RNP complexes (Smith et al. 1991). The HCV RNP complexes were radiolabeled using the cell-free replication scheme recently described by us (Ali et al. 2002). The HCV RNA replication was carried out in the presence of [α-32P]CTP. The FCA4 cells metabolically labeled in the presence of 35S-methionine were also used as a source of RNP complexes in this analysis. Radiolabeled RNP complexes were purified by the monomeric avidin affinity procedure as described in Materials and Methods. The samples were treated with 1% NP-40 and electrophoresed on 5% nondenaturing PAGE for 24 h. An approximate molecular mass of the HCV RNP complexes was estimated to be ~450 kD (Fig. 6A ▶, lane 3; Fig. 6B ▶, lane 2). This analysis demonstrates that RNP complexes containing either 32P-labeled RNA or 35S-methionine-labeled proteins migrate with similar molecular mass (Fig. 6A ▶, lane 3; Fig. 6B ▶, lane 2). The Huh7 cellular lysate did not contain such complexes (Fig. 6B ▶, lane 1).
FIGURE 6.
Determination of molecular mass of HCV RNP complex. (A) The RNA in the HCV RNP complexes was 32P-radiolabeled in the presence of [α-32P]CTP by a cell-free replication assay as described previously (Ali et al. 2002). (B) The proteins in the HCV RNP complexes were metabolically labeled with 35S-methionine. The RNP complexes derived from FCA4 or Huh7 cellular lysates were purified by avidin-agarose affinity purification (Fig. 1 ▶) and treated with 1% NP-40 for 20 min at 4°C and electrophoresed on 5% native PAGE at 100 V for 24 h at room temperature. RNP complexes were detected by autoradiography (A, lane 3; B, lane 2). (B, lane 1) Huh7 lysates were used as the control. (A, lanes 1,2) Molecular weight markers including apoferretin (443 kD) and thyroglobulin (669 kD; Sigma) were stained with Coomassie brilliant blue.
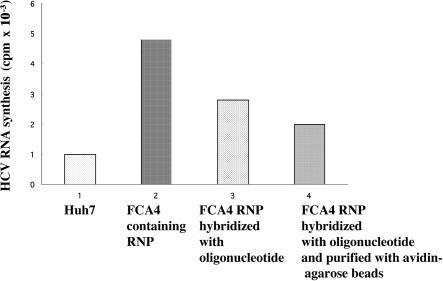
The FCA4 lysates prepared by our method as previously described (Ali et al. 2002) are competent in replication. Once the oligonucleotide anneals to the RNA in the RNP complex, these RNP complexes are unable to complete the synthesis of RNA because of interference in the progression of viral replicase on the template with an annealed oligonucleotide. The RNA products synthesized after the RNP complex hybridized with the oligonucleotide produce a smear of radiolabeled RNA bands (data not shown). Hence, we present the incorporation of radioactivity in these reactions. FCA4 RNP complexes were purified by the avidin-agarose method and subjected to RNA synthesis as described above. Incorporation of [32P]CTP into newly synthesized RNA is presented in Figure 7 ▶. As can be seen, FCA4 cellular lysates subjected to RNA synthesis show significant incorporation of TCA-precipitable 32P counts (bar 2). FCA4 RNP complexes subjected to affinity purification followed by the RNA synthesis protocol also show RNA synthesis but at a lower level (bar 4). Reduction in the level of RNA synthesis reflects the incomplete nature of RNA synthesis caused by interference by the oligonucleotide annealed to the template RNA. When HCV RNP complexes were annealed to oligonucleotides and directly subjected to RNA synthesis, a similar reduction in RNA synthesis was observed (bar 3). These data clearly imply that although purified RNP complexes are replication-competent, annealed oligonucleotides offer interference to the progression of RNA synthesis. Huh-7 cellular lysates subjected to affinity purification and RNA synthesis show background levels of [32P]CTP incorporation.
FIGURE 7.
HCV replication assay. Huh7 (bar 1) and FCA4 lysates containing RNP complexes hybridized with oligonucleotides and followed by the avidin-agarose affinity purification step were incubated with [32P]CTP for 90 min during an RNA synthesis reaction, as described in Materials and Methods (bar 4). The incorporation of 32P radioactivity in the FCA4 lysates directly subjected to RNA synthesis (bar 2) or annealed to the neomycin oligonucleotide followed by RNA synthesis (bar 3) is presented. The radiolabeled lysates (1 μL) were spotted on DE81, ion exchange filters (Whatman). The filters were dried and washed extensively with 100 mM sodium phosphate buffer (pH 8.0). The 32P-labeled RNA products bound to the filters were measured by scintillation counting.
DISCUSSION
In this study, we used a two-step purification scheme as outlined in Figure 1 ▶ to enrich and specifically isolate the HCV NS proteins that are associated with replication-competent RNP complexes. Using this strategy, we demonstrated the association of all the NS proteins that are encoded by the HCV subgenomic replicon with the RNP complex. The fact that all the HCV NS proteins are associated with the RNP complex is not entirely surprising because the initial selection of the subgenomic replicon from liver cells infected with HCV relied on the inclusion of minimal NS proteins in the generation of autonomous replicons (Lohmann et al. 1999). Spontaneous appearance of subgenomic replicons has been previously observed in pestiviruses and flaviviruses (Khromykh and Westaway 1997; Behrens et al. 1998).
Positive-strand RNA viruses replicate their genomic RNA in membrane-bound multiprotein complexes (Lai 1998; Ahlquist 2002). Association of host factors in concert with the viral proteins during RNA synthesis by these viruses has been demonstrated, but very little, if anything, is known about the nature of interactions and their functional role during replication. Cellular proteins are known to either directly interact with the 3′ end of the RNA genome or bind to the RdRp holoenzyme RNP complex (Lai 1998). Previous work on the association of various cellular proteins in the viral RNP complexes has revealed the presence of host translation factors, proteins involved in Pol III transcription (La antigen), and those associated with the hnRNP complex such as PTB, poly(rC)-binding protein, and the like (Lai 1998). These proteins have been shown to interact with the 5′ and 3′ NCRs of the viral genomic RNA (Lai 1998). In the case of HCV, several HCV NS proteins have been shown to associate with each other (Dimitrova et al. 2003).
Translation and replication functions of positive-strand RNA viruses are considered to be linked (Novak and Kirkegaard 1994). First, the genomic RNA is translated, and after several rounds of translation, a molecular switch is believed to direct the initiation of RNA replication within a membrane-bound RNP complex (Novak and Kirkegaard 1994). Circumstantial evidence indicates that the HCV RNA may follow the same strategy. A recent report demonstrated dramatic structural alterations in the cellular membranes resembling membranous web-like structures in cells expressing HCV NS proteins (Gosert et al. 2003). Shi et al. (2003) demonstrated the association of detergent-resistant (1% NP-40) HCV RNP complexes with lipid rafts. In the present analysis, we found RNP complexes to be resistant to even higher concentrations of NP-40 (up to 2%; Fig. 5B ▶). The level of HCV gene expression may vary in different stages of liver disease. It is very likely that varying levels of HCV gene expression are dictated by the host/viral factors constituting the RNP complex. Depending on the state of hepatocytic growth, different host factors may be involved in the formation of the HCV RNP complex. The contents of the RNP complex may also be modified indirectly by signal transduction pathways involving cytokines and protein kinases at a given stage of hepatocytic growth. In addition, the HCV translation/replication activities, which occur in the ER membrane and induce an ER stress response (Tardif et al. 2002), may also impinge their influence on intracellular events that may in turn impact on the HCV gene expression. We have recently reported that HCV NS5a induces oxidative stress via Ca2+ signaling and activates NF-κB by an alternate pathway, which does not involve serine phosphorylation of IκBα and its degradation by proteasomes (Gong et al. 2001; Waris et al. 2003). NS5a alone or in the context of the subgenomic replicon activates a cellular tyrosine kinase(s), which phosphorylates IκBα at Tyr 42 and 305 residues and targets its degradation by calpain proteases (Waris et al. 2003). Activation of cellular tyrosine kinase(s) and subsequent activation of transcription factors such as NF-κB, STAT-3, and others is likely to affect the cellular gene expression with a direct impact on HCV translation/replication. Such changes in the HCV-infected hepatocytes may directly influence the repertoire of host factors available for the HCV RNP complex assembly and its functions. The various levels and the nature of host/viral proteins in the RNP complexes are relevant to the state of liver disease associated with HCV infection. For instance, during acute and chronic hepatitis and cirrhosis, the replication functions of RNP complexes may be dictated by the composition of host/viral proteins. Our future efforts will be directed toward identifying the host factors associated with the HCV RNP complex in the liver tissues infected with HCV. The role of host factors in HCV translation and replication is directly relevant to chronic liver disease associated with the viral infection.
MATERIALS AND METHODS
Cell cultures
Huh7 and FCA4 human hepatoma cells were routinely grown in Dulbecco’s MEM media (GIBCO BRL) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin sulfate, and 10% fetal calf serum. The FCA4 cell line stably expresses an HCV subgenomic replicon with a single adaptive mutation and a deletion of a serine residue at position 1176 (Guo et al. 2001). FCA4 cells were maintained in G418-containing medium in a subconfluent state to ensure optimal cell growth and hence efficient replicon replication.
Reagents and antibodies
The 5′ end biotin-tagged oligonucleotide complementary to the neomycin phosphotransferase gene, both in sense and anti-sense polarities, was synthesized by Invitrogen-LifeTechnology. A digoxigenin-ddUTP oligonucleotide 3′ end labeling kit and anti-digoxigenin monoclonal antibody were purchased from Roche Molecular Biochemicals. Monomeric avidin-agarose beads were purchased from Pierce.
The polyclonal antibody against HCV C-100 peptide was a gift from Dr. M. Houghton (Chiron, Inc). The C-100 peptide represented amino acids between 1569 and 1931, which includes NS3 (1026–1657), NS4a (1658–1711), and NS4b (1712–1972). This antibody recognizes NS3, NS4a, and NS4b. NS4a/b antibody was purchased from Virogen, Inc. Anti-NS5a was a gift from C. Cameron (Pennsylvania State University), and NS5b was a gift from Marc Collett (ViroPharma, Inc.).
Preparation of replication lysates
The cytoplasmic fractions of FCA4 and Huh7 cells were prepared by a protocol described previously (Ali et al. 2002). Huh7 cells were transiently transfected with in vitro synthesized BM4–5 subgenomic replicon RNA as described previously (Ali et al. 2002). Briefly, each plate with a subconfluent culture was washed gently with cold wash buffer (150 mM sucrose, 30 mM HEPES at pH 7.4, 33 mM ammonium chloride, 7 mM KCl, and 4.5 mM magnesium acetate), followed by the addition of lysolecithin buffer (250 μg/mL lysolecithin in sterile wash buffer) for 1–2 min. To each plate containing lysolecithin solution, 3 mL of wash buffer was added, mixed, and aspirated. After all the buffer was removed, cells were scraped from each plate in 120 μL of incomplete reaction buffer (100 mM HEPES at pH 8.5; 50 mM ammonium chloride; 7 mM KCl; 1 mM spermidine; 1 mM each ATP, GTP, and UTP; 10 μM CTP; 1 mM DTT; and 10% glycerol), then transferred to Eppendorf tubes and lysed gently by pipetting 15 times. The cell suspension was centrifuged for 5 min at 1600 rpm in a microfuge at 4°C. The cytoplasmic fraction (supernatant) was aliquoted and stored at −70°C.
For the HCV replication assay, the cytoplasmic extract was incubated with [α-32P]CTP (30 μCi; 800 Ci/mmole) for 1.5 h at 30°C. The replication reaction was terminated by adding SDS-containing STE buffer (10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.5% SDS), followed by extraction with phenol–chloroform–isoamyl alcohol (25:24:1) and water-saturated ether. The RNAs were precipitated in ethanol. The centrifuged pellet was washed with 70% ethanol and resuspended in RNase-free water. The replication products were analyzed by native agarose gel electrophoresis.
Isolation of HCV ribonucleoprotein complex
The biotinylated oligonucleotide containing the following nucleotide sequence: 5′-BioTEGGCCCGTCGTGGCCAGCCACGATAGCC GCGCTGCCTCGTCCTGCAGTTCATTCAGGGC-3′ was synthesized complementary to the neomycin phosphotransferase gene sequence. The anti-sense oligonucleotides and unrelated (nonspecific) oligonucleotides were also synthesized containing biotin tag at the 5′ end. Digoxigenin molecules were added at the 3′ ends by terminal transferase in the presence of digoxigenin-ddUTP as described previously (Schmitz et al. 1991). Equal amounts of FCA4 and Huh7 replication lysates were incubated with 50 pmoles of biotin and digoxigenin-tagged neomycin oligonucleotides for 2 h at 30°C to produce neomycin-oligonucleotide:RNP complex hybrids. The hybrid complexes were allowed to bind monomeric avidin-agarose beads overnight at 4°C. Avidin beads were washed several times; first with DT300 buffer (buffer D containing 0.02% Tween-20 and 300 mM KCl) and then with ETO buffer (20% glycerol, 1 mM HEPES at pH 7.9, 0.25 mM EDTA, 0.5 mM DDT, and 0.02% Tween-20). Each wash was followed by centrifugation. The bound complexes were eluted with 2 mM d-biotin for 20 min at room temperature. The eluate was diluted to 500 μL with buffer D (20% [v/v] glycerol, 20 mM HEPES at pH 7.9, 100 mM KCl, 0.25 mM EDTA, and 0.5 mM DDT) and immunoprecipitated with anti-digoxigenin monoclonal antibody (10 μg) overnight at 4°C. The immune complexes were incubated with protein G-Sepharose for 1 h at 4°C. The bound complexes were washed with buffers D and ETO, respectively, and subjected to SDS-PAGE.
Western blot analysis
The protein samples were fractionated by 10% SDS/PAGE. Gels were electroblotted onto nitrocellulose membranes in transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol). Membranes were treated overnight in blocking buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.3% polyvinlypyrrolidone, 0.5% Tween-20 [w/v]) and subsequently incubated with antisera against HCV proteins. Immunoreactive proteins were visualized using the ECL kit (Amersham).
Nondenaturing polyacrylamide gel electrophoresis
FCA4 lysates (200 μL) were incubated with 1 mM each ATP, GTP, and UTP; 10 μM CTP, [α-32P]CTP (30 μCi; 800 Ci/mmole) for 1.5 h at 30°C. The reaction was terminated by adding STE buffer (10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 150 mM NaCl). The [α-32P]CTP-labeled RNP complexes were hybridized with biotin-tagged oligonucleotides as described above. The hybrid complexes were allowed to bind overnight at 4°C with avidin beads. Avidin-agarose beads were washed with DT300 and ETO buffers. The bound complexes were eluted with 2 mM d-biotin at room temperature. The eluate was treated with 1% NP-40 for 20 min at 4°C and electrophoresed by 5% native PAGE at 100 V for 24 h at room temperature. The ribonucleoprotein complexes were also metabolically labeled in the presence of 35S-methionine. Briefly, cells were incubated for 30 min in methionine- and cysteine-free DMEM supplemented with 2 mM glutamine, antibiotics, and 10% FBS. Cells were labeled for 1 h in the presence of 200 μCi of Trans35S-label (1175 Ci/mmole; ICN) per milliliter of the above media. Cellular lysates were prepared as described above and hybridized with biotin-tagged oligonucleotides as described above.
Acknowledgments
This work is supported by a grant from NIH (DK61566). We thank C. Seegar (Fox Chase Cancer Institute, Philadelphia) for the generous gift of FCA4 and BM4–5.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5124404.
REFERENCES
- Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273. [DOI] [PubMed] [Google Scholar]
- Ali, N. and Siddiqui, A. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirements in internal initiation of translation. J. Virol. 69: 6367–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. 94: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, N., Tardif, K.D., and Siddiqui, A. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76: 12001–12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager, R. and Lohmann, V. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81: 1631–1648. [DOI] [PubMed] [Google Scholar]
- Behrens, S.E., Grassmann, C.W., Thiel, H.J., Meyers, G., and Tautz, N. 1998. Characterization of an autonomous subgenomic pestivirus RNA replication J. Virol. 72: 2364–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight, K.J., Kolykhalov, A.A., and Rice, C.M. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290: 1972–1974. [DOI] [PubMed] [Google Scholar]
- Di Bishceglie, A.M. 1997. Hepatitis C and hepatocellular carcinoma. Hepatology 26: 34S–38S. [DOI] [PubMed] [Google Scholar]
- Dimitrova, M., Imbert, I., Kieny, M.P., and Schuster, C. 2003. Protein–protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77: 5401–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson, J., Penin, F., and Moradpour, D. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12: 517–523. [DOI] [PubMed] [Google Scholar]
- Gale Jr., M., Korth, M.J., Tang, N.M., Tan, S.L., Hopkins, D.A., Dever, T.E., Polyak, S.J., Gretch, D.R., and Katze, M.G. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural protein 5A. Virology 230: 217–227. [DOI] [PubMed] [Google Scholar]
- Gong, G., Waris, G., Tanveer, R., and Siddiqui, A. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induce oxidative stress, and activates STAT-3 and NF-κB. Proc. Natl. Acad. Sci. 98: 9599–9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert, R., Egger, D., Lohmann, V., Bartenshlager, R., Blum, H.E., Bienz, K., and Moradpour, D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77: 5487–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.-T., Bichko, V.V., and Seeger, C. 2001. Effect of α interferon on the hepatitis C virus replicon. J. Virol. 75: 8516–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata, M., Mizushima, H., Akagi, T., Mori, S., Kakiuchi, N., Kato, N., Tanaka, N., Kimura, K., and Shimotohno, K. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural proteins of hepatitis C virus. J. Virol. 67: 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T. and Lai, M.M.C. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71: 8698–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh, A.A. and Westaway, E.G. 1997. Subgenomic replicons of the flavivirus Kunjin: Construction and applications. J. Virol. 71: 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M.M.C. 1998. Cellular factors in the transcription and replication of viral RNA genomes: A parallel to DNA-dependent RNA transcription. Virology 244: 1–12. [DOI] [PubMed] [Google Scholar]
- Lohmann, V., Korner, F., Koch, J.O., Herian, U., Theilmann, L., and Bartenschlager, R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285: 110–113. [DOI] [PubMed] [Google Scholar]
- Murphy, F.A., Fauquet, C.M., Bishop, D.H.L., Ghabrial, S.A., Jarvis, A.W., Martelli, G.P., Mayo, M.A., and Summers, M.D. 1995. Virus taxonomy: Sixth report of the International Committee on Taxonomy of Viruses, pp. 424–426. Spring Verlag, Vienna.
- Novak, J.E. and Kirkegaard, K. 1994. Coupling between genome translation and replication in an RNA virus. Genes & Dev. 8: 1726–1737. [DOI] [PubMed] [Google Scholar]
- Reed, K.E. and Rice, C.M. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242: 55–88. [DOI] [PubMed] [Google Scholar]
- Reyes, G.R. 2002. The nonstructural 5A protein of hepatitis C virus: An expanding, multifunctional role in enhancing hepatitis C virus pathogenesis. J. Biomed. Sci. 9: 187–197. [DOI] [PubMed] [Google Scholar]
- Rijnbrand, R.C.A. and Lemon, S.M. 1999. The hepatitis C viruses (eds. C.H. Hegedorn and C.M. Rice). Spring-Verlag, Berlin.
- Schmitz, G.G., Walter, T., Seibl, R., and Kessler, C. 1991. Nonradioactive labeling of oligonucleotides in vitro with the hapten digoxigenin by tailing with terminal transferase. Anal. Biochem. 192: 222–231. [DOI] [PubMed] [Google Scholar]
- Schwartz, M., Chen, J., Janda, M., Sullivan, M., Boon, J., and Ahlquist, P. 2002. A positive-strand RNA virus replication complex parallels forms and function of retrovirus capsids. Mol. Cell 9: 505–514. [DOI] [PubMed] [Google Scholar]
- Shi, S.T., Lee, K.J., Aizaki, H., Hwang, S.B., and Lai, M.M.C. 2003. Hepatitis C virus RNA replication occurs on a detergent resistant membrane that cofractionates with caveolin-2. J. Virol. 77: 4160–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.O., Tabiti, K., Schaffaner, G., Soldati, D., Albrecht, U., and Birnstiel, M.L. 1991. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligonucleotides. Proc. Natl. Acad. Sci. 88: 9784–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangberg, K., Goober-Larsson, L., Wahren-Herlenius, M., and Schwartz, S. 1999. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated protein. J. Hum. Virol. 2: 296–307. [PubMed] [Google Scholar]
- Tardif, K.D., Mori, K., and Siddiqui, A. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76: 7453–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama, K.K., Lizuka, N., Kohara, M., and Nomoto, A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66: 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walewski, J.L., Keller, T.R., Stump, D., and Branch, A.D. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7: 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Sarnow, P., and Siddiqui, A. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67: 3338–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris, G., Livolsi, A., Imbert, V., Peyron, J.F., and Siddiqui, A. 2003. Hepatitis C virus NS5A activates NF-κB via tyrosine phosphorylation of IκBα and its degradation by calpain protease. J. Biol. Chem. 278: 40778–40787. [DOI] [PubMed] [Google Scholar]
- Xu, Z., Choi, J.T., Yen, T.S., Lu, W., Strohecker, A., Govindrajan, S., Chien, D., Selby, M., and Ou, J. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20: 3840–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]