One of the most intriguing proposals in RNA evolution, that spliceosomal RNAs derive from a group II intron, invokes an evolutionary splitting of the ancestor (Sharp 1991). Conversely, it is hard to imagine the earliest forms of complex RNAs being as large as their modern counterparts, and fusion of originally separate RNA modules has been proposed for the evolution of the ribosome, self-splicing introns, and even tRNA.
tmRNA presents an especially interesting workshop on both evolutionary splitting and fusion. It has been split at least twice, independently in separate bacterial lineages (Keiler et al. 2000). Both known cases result from gross gene rearrangement, and produce similar two-piece tmRNAs, that share a drastically altered topology compared to the one-piece form. The story comes full circle with an article in this issue by Jacob et al. (2004), who describe a one-piece descendant from one of these two-piece lineages, that is, the resealing of the split RNA. The convergent evolution to a two-piece form suggests that the split benefits tmRNA function; one possible benefit discussed below could even rationalize the instance of resealing.
The large subunit (LSU) rRNA provides a contrasting precedent for splitting and resealing over evolutionary time. Although idiosyncratic breaks are found in LSU rRNAs of diverse bacteria, eukaryotes, and organelles (Gerbi 1986; Burgin et al. 1990), one fragmentation is highly conserved, that producing the 5.8S and 28S pieces in eukaryotes. Microsporidia, however, produce no separate 5.8S molecule (Vossbrinck and Woese 1986). Originally, this was interpreted as retention of the prokaryotic condition in a very early-branching eukaryotic lineage, but because microsporidia are now recognized as highly derived fungi (Keeling et al. 2000), it instead represents the resealing of a split RNA.
This splitting and resealing of the LSU rRNA differs in many ways from that of tmRNA. First, it probably corresponds to the development and subsequent loss of an RNA processing system. Gene rearrangement need not be invoked; the 5.8S sequence matches the 5′ end of both the prokaryotic and microsporidian unitary LSU rRNAs, and the same order of the 5.8S and 28S cognate portions is retained in all these rRNA genes. Second, consequences for ribosome structure are not necessarily drastic. The split ends of 5.8S and 28S remain base-paired together much as the corresponding regions do in the one-piece LSU rRNAs. Third, although the split site can be mapped to an interface of the ribosome (the vicinity of the deacylated end of the E-tRNA), no proposals have been made for functional effects of the split/fused status.
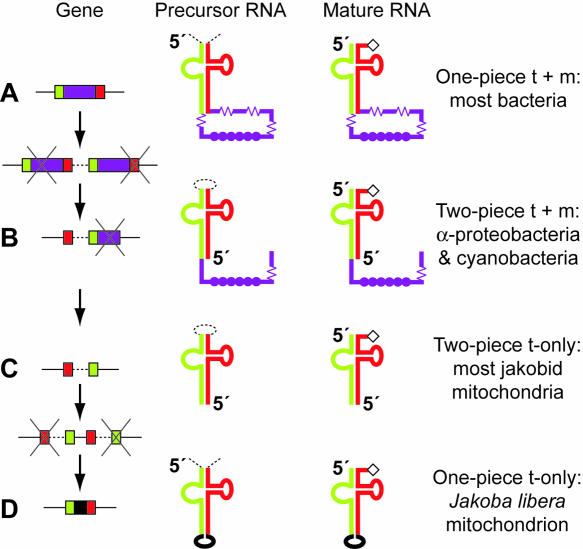
For tmRNA, both splitting and resealing have been accomplished by circular permutation of the gene. Circular permutation is an operation on a string that has the same effect as if the ends were joined to form a circle, and the circle were then broken open at a different point. This operation is followed literally in a useful experimental technique for RNA analysis (Pan 2000). However, for natural DNA sequences, circular permutation is more likely to proceed not literally as above, but rather through recombination to form a tandem repeat that is then resolved (A to B transition in Fig. 1 ▶). Many of us first encountered the principle in certain bacteriophage DNAs; for example, each T4 phage particle contains a linear DNA molecule that is a different permutation of the same circular genome sequence. This permutation results from the processing of long replicative concatemers into pieces slightly larger than the unit genome length.
FIGURE 1.
RNA splitting and resealing through gene permutation and repermutation. (A) Standard tmRNA is produced by tRNA-like end-processing (removing portions marked by dashed lines). The tRNA domain (green and red segments) is charged with alanine (diamond). The mRNA domain (purple) contains the tag reading frame (filled circles) and multiple pseudoknots (springs). (B) Circular gene permutation, probably through tandem duplication with degeneration at the ends of the tandem, has occurred independently in two bacterial lineages. In both lineages, tRNA-like processing at internal sites produces a two-piece tmRNA. (C) The permuted gene from Alphaproteobacteria has been retained in the most primitive of mitochondria, but with loss of the mRNA domain. (D) Jacob et al. (2004) have found a repermuted mitochondrial gene producing a small, one-piece homolog, again lacking the mRNA domain.
Circular permutation of an individual gene is observed when a segment of the gene usually at the upstream end is found instead at the downstream end. A few permuted protein-coding genes have been identified that produce circularly permuted proteins (Lindqvist and Schneider 1997). For RNA genes, the LSU rRNA again provides a precedent. Its gene is permuted in the Tetrahymena pyriformis mitochondrion, with a tRNA spacer positioned between the switched gene segments. Processing at this spacer in the precursor RNA produces yet another split version of LSU rRNA (Heinonen et al. 1987).
tmRNA is named for its modular combination of tRNA and mRNA functions. It is charged with alanine, which it transfers into protein, and it contains a reading frame that is translated. tmRNA helps bacteria deal with a ribosome that has stalled at the 3′ end of an mRNA, through a remarkable interplay with the ribosome (Keiler et al. 1996). Charged tmRNA enters the A site of the stalled ribosome and transfers its alanyl moiety into the nascent protein, without the benefit of a codon:anticodon interaction. Then its reading frame is switched into the decoding center, replacing the stalled mRNA. Translation of tmRNA proceeds to its stop codon, where the ribosome is freed. The resulting protein, a product of two separate mRNAs, receives a tmRNA-encoded peptide tag that is a signal for many proteases to degrade the tagged protein. Thus, tmRNA both rescues stalled ribosomes and provides for decay of the incomplete protein product. Analysis of mutant tmRNAs suggests that the latter, more stiking effect is actually less important for bacterial physiology than the former (Withey and Friedman 2003).
The tRNA and mRNA domains are connected by an elongated analog of the anticodon stem (tmRNA has no true anticodon). In the standard bacterial one-piece tmRNA, this connecting stem seals the mRNA portion into a loop of approximately 300 nt (Fig. 1A ▶). Processing of the precursor is like that for a tRNA; flanking sequences are removed and the RNA is left with a CCA tail, ready to be charged by alanyl-tRNA synthetase. In both two-piece lineages, gene permutation has placed the transcription start site at a position within the mRNA loop, and has left a small intervening segment between the two switched gene portions (Fig. 1B ▶). The intervening segment in the precursor forms an internal loop at the end of the acceptor stem that is removed, again by tRNA-like processing. Although a housekeeping processing system acting at the tRNA domain is what physically splits the tmRNA into two pieces, the effect relative to the one-piece form is to have broken open the looped mRNA domain. This is certainly a drastic topological change.
At first glance the split might seem detrimental to tmRNA function, perhaps destabilizing the RNA. However, the convergent evolution to this altered form, independently in two bacterial lineages, suggests that tmRNA function might, in fact, derive some benefit from the break in the loop. Consider the problem that the unbroken loop presents for translation in the first place. A recent cryo-EM picture of tmRNA partially engaged in the ribosome shows that the loop is relatively small, approximately of a size that it might barely slip onto the neck of the small subunit (Valle et al. 2003). The loop is sealed by extensive base-pairing, both at the acceptor stem and at the long connecting stem, and an abutting pseudoknot is likely to further stabilize the connecting stem through coaxial stacking. Presuming that the loop persists during translation, it is likely to cause difficulties for translation. Usually we tend to ignore the ends of a linear mRNA engaged in translation, but here we must consider the path of the looped mRNA; does it wrap around the outside of the ribosome or double back through the interface? The loop is moreover ratcheting around triplet-by-triplet as tmRNA translation proceeds, which may drag it over inhospitable surfaces and produce stress in the loop. The four to five pseudoknots always found in this loop in bacteria may serve to relieve such stress; unfolding of a single pseudoknot would expand the loop substantially.
In contrast, no such stress can ever develop in two-piece tmRNA, because the mRNA domain loop is permanently opened. Avoidance of stress during translation may be the evolutionary rationale that has driven the permutation of the gene. All tmRNA genes known from the Alphaproteobacteria (Agrobacterium, Rickettsia, etc.) are found in the permuted form. An independently permuted gene is found in a small group of cyanobacteria. Both groups have been shown to produce two-piece tmRNA. The latter case is especially interesting because an unpermuted cyanobacterial gene has been found bearing extremely high sequence homology with the permuted gene (Williams 2002). Taking this as a close approximation to the ancestral sequence shows that the evolution of this two-piece form was somewhat more complicated than the basic permutation model of tandem gene duplication with degeneration of the outermost segments. Correlating with the idea that pseudoknots are an alternative avenue for stress relief, pseudoknot number drops from five in the one-piece cyanobacterial form to one in the two-piece form, and from four to one (or two) in the Alphaproteobacteria (Gaudin et al. 2002; Keiler et al. 2000).
Identifying the tmRNA gene in Alphaproteobacteria inspired a search in their descendants, the mitochondria (Keiler et al. 2000). This failed for most mitochondria; however, a homolog was identified in the most primitive mitochondrion whose complete sequence was available (Lang et al. 1997), that of the jakobid protist Reclinomonas americana. Its permuted gene clearly displays the tRNA domain of tmRNA, but lacks the reading frame (Fig. 1C ▶). Without its “m,” one hesitates to still call it tmRNA. This homolog cannot act identically to tmRNA in bacteria, and its function in mitochondria is therefore unclear.
Jacob et al. (2004) have investigated these mitochondrial tmRNA homologs further, showing that they are indeed expressed, with the expected ends from tRNA-type processing, and are substrates in vitro for (Escherichia coli) RNase P and alanyl-tRNA synthetase. They show that three jakobid genera have the permuted gene, but find a remarkable reversal of gene permutation in the Jakoba libera mitochondrion, which reconfigures the tmRNA homolog back to a one-piece form (Fig. 1D ▶). This molecule, composed of only 102 nt (Fig. 1 ▶ of Jacob et al. 2004), emphasizes the complete loss of the tmRNA reading frame in mitochondria. Although the characteristics of tmRNA are clearly discerned in its tRNA domain, on the whole, it more resembles a tRNA than an entire tmRNA. Indeed, if tmRNA originated by expansion of a tRNA, the Jakoba molecule represents a near return home, but by a very circuitous route.
The new results imply that the mitochondrial homolog has some tRNA-like function, although certainly not the classical one, because it has no anticodon. It may still function in the rescue of stalled ribosomes, even though it cannot reproduce the trick used by authentic tmRNA, which taps into the ordinary mechanism for ribosome release by delivering the ribosome to its stop codon.
Gene repermutation might be rationalized by the earlier loss of the mRNA domain in mitochondria. It was suggested above that the main benefit of the two-piece composition for tmRNA function is to prevent stress during translation. If so, then the mitochondrial homolog, being untranslated, would gain no benefit from being split. For whatever function the tmRNA homolog does have in mitochondria, the one-piece construction might again be more sensible, driving the reversal of gene permutation.
Analysis of the primitive jakobid mitochondria continues to reveal surprises concerning RNA. Earlier it had revealed putative Shine-Dalgarno interactions and bacterial-type RNase P RNA (Lang et al. 1997). A second article in this issue, from the same laboratory (Leigh and Lang 2004), describes an apparently novel system for editing the 3′ ends of tRNAs.
[Commentary on Jacob et al. 2004 (this issue); also refers to Leigh and Lang 2004 (this issue).]
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5274104.
REFERENCES
- Burgin, A.B., Parodos, K., Lane, D.J., and Pace, N.R. 1990. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell 60: 405–414. [DOI] [PubMed] [Google Scholar]
- Gaudin, C., Zhou, X., Williams, K.P., and Felden, B. 2002. Two-piece tmRNA in cyanobacteria and its structural analysis. Nucleic Acids Res. 30: 2018–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi, S.A. 1986. The evolution of eukaryotic ribosomal DNA. Biosystems 19: 247–258. [DOI] [PubMed] [Google Scholar]
- Heinonen, T.Y., Schnare, M.N., Young, P.G., and Gray, M.W. 1987. Rearranged coding segments, separated by a transfer RNA gene, specify the two parts of a discontinuous large subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J. Biol. Chem. 262: 2879–2887. [PubMed] [Google Scholar]
- Jacob, Y., Seif, E., Paquet, P.O., and Lang, B.F. 2004. Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Keeling, P.J., Luker, M.A., and Palmer, J.D. 2000. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 17: 23–31. [DOI] [PubMed] [Google Scholar]
- Keiler, K.C., Waller, P.R., and Sauer, R.T. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993. [DOI] [PubMed] [Google Scholar]
- Keiler, K.C., Shapiro, L., and Williams, K.P. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. 97: 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B.F., Burger, G., O’Kelly, C.J., Cedergren, R., Golding, G.B., Lemieux, C., Sankoff, D., Turmel, M., and Gray, M.W. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387: 493–497. [DOI] [PubMed] [Google Scholar]
- Leigh, J. and Lang, B.F. 2004. Mitochondrial 3′ tRNA editing in the jakobid Seculamonas ecuadoriensis: A novel mechanism and implications for tRNA processing. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Lindqvist, Y. and Schneider, G. 1997. Circular permutations of natural protein sequences: Structural evidence. Curr. Opin. Struct. Biol. 7: 422–427. [DOI] [PubMed] [Google Scholar]
- Pan, T. 2000. Probing RNA structure and function, by circular permutation. Methods Enzymol. 317: 313–330. [DOI] [PubMed] [Google Scholar]
- Valle, M., Gillet, R., Kaur, S., Henne, A., Ramakrishnan, V., and Frank, J. 2003. Visualizing tmRNA entry into a stalled ribosome. Science 300: 127–130. [DOI] [PubMed] [Google Scholar]
- Vossbrinck, C.R. and Woese, C.R. 1986. Eukaryotic ribosomes that lack a 5.8S RNA. Nature 320: 287–288. [DOI] [PubMed] [Google Scholar]
- Williams, K.P. 2002. Descent of a split RNA. Nucleic Acids Res. 30: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withey, J.H. and Friedman, D.I. 2003. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 57: 101–123. [DOI] [PubMed] [Google Scholar]