Abstract
The carboxyl-terminal domain (CTD) of RNA polymerase II (pol II) plays an important role in coupling transcription with precursor messenger RNA (pre-mRNA) processing. Efficient capping, splicing, and 3′-end cleavage of pre-mRNA depend on the CTD. Moreover, specific processing factors are known to associate with this structure. The CTD is therefore thought to act as a platform that facilitates the assembly of complexes required for the processing of nascent transcripts. The mammalian CTD contains 52 tandemly repeated heptapeptides with the consensus sequence YSPTSPS. The C-terminal half of the mammalian CTD contains mostly repeats that diverge from this consensus sequence, whereas the N-terminal half contains mostly repeats that match the consensus sequence. Here, we demonstrate that 22 tandem repeats, from either the conserved or divergent halves of the CTD, are sufficient for approximate wild-type levels of transcription, splicing, and 3′-end cleavage of two different pre-mRNAs, one containing a constitutively spliced intron, and the other containing an intron that depends on an exon enhancer for efficient splicing. In contrast, each block of 22 repeats is not sufficient for efficient inclusion of an alternatively spliced exon in another pre-mRNA. In this case, a longer CTD is important for counteracting the negative effect of a splicing silencer element located within the alternative exon. Our results indicate that the length, rather than the composition of CTD repeats, can be the major determinant in efficient processing of different pre-mRNA substrates. However, the extent of this length requirement depends on specific sequence features within the pre-mRNA substrate.
Keywords: RNA polymerase II, CTD, splicing, 3′-end cleavage, transcription
INTRODUCTION
Protein coding genes, which represent the largest class of genes within eukaryotic genomes, are uniquely transcribed by RNA polymerase II (pol II). Primary pol II transcripts are usually extensively processed before the resulting mature mRNA can be exported to the cytoplasm and translated into protein. The processing steps include the addition of an m7G cap at the 5′-end, splicing of introns, and 3′-end formation by cleavage and polyadenylation. Capping, splicing, and 3′-end cleavage/polyadenylation are closely coupled to transcription by pol II (for review, see Bentley 1999; Hirose and Manley 2000; Cramer et al. 2001; Howe 2002; Neugebauer 2002; Rosonina and Blencowe 2002). The specific mechanisms by which these processing events are coupled are not well understood, although it has been shown that each step is promoted by the carboxyl-terminal domain (CTD) of the largest subunit of pol II. The CTD is a unique feature of RNA pol II.
The CTD consists of tandem repeats of a heptapeptide sequence whose consensus, YSPTSPS, is highly conserved among eukaryotes. The number of repeats varies from species to species. The CTD of budding yeast contains 26 or 27 repeats, whereas there are 42 repeats in Caenorhabditis elegans, 44 in Drosophila melanogaster, and 52 in all mammalian pol IIs that have been characterized. The significance of the variation in repeat number among eukaryotic species is not known, although it is noteworthy that there is a parallel relationship between the number of repeats and the complexity of an organism. Deletion experiments have demonstrated that the CTD is essential in yeast and mammalian cells.
Truncation of the CTD in yeast and mammals reduces activator-dependent stimulation of transcription of some genes (Allison and Ingles 1989; Gerber et al. 1995; Meininghaus and Eick 1999). Moreover, differential phosphorylation of the CTD correlates with steps in the transcription cycle. Pol II that contains a hypophosphorylated CTD (pol IIa) assembles a preinitiation complex together with general transcription factors (Conaway et al. 1992; Koleske et al. 1992; Maxon et al. 1994). Upon phosphorylation of the CTD, pol II converts to a processive, elongating form (pol IIo) that must become dephosphorylated prior to re-incorporation into the preinitiation complex. Differential phosphorylation of CTD repeats at positions Ser2 and Ser5 has been implicated in different stages of pol II elongation (Komarnitsky et al. 2000).
Truncation of the CTD also reduces the efficiency of capping, splicing, and 3′-end formation of pre-mRNA in vivo (Cho et al. 1997; McCracken et al. 1997a,b). Capping enzymes, the cleavage factor CstF, and specific splicing factors and splicing-related proteins have been found to interact with the CTD (for review, see Bentley 1999; Hirose and Manley 2000; Howe 2002; Maniatis and Reed 2002). Purified pol II, or a phosphorylated CTD itself, can stimulate splicing, capping, and 3′-end formation in vitro (Hirose et al. 1999; Ho and Shuman 1999; Zeng and Berget 2000), and is also required for efficient 3′-end formation in nuclear extracts (Hirose and Manley 1998; Ryan et al. 2002). These observations have contributed to a model in which the CTD acts as a molecular “platform” for processing factors, whereby it facilitates efficient cotranscriptional capping, splicing, and 3′-end formation.
Recent findings indicate that the control of alternative splicing may also be coupled to transcription by pol II. For example, the level of inclusion of the alternative fibronectin EDI exon in a minigene reporter changes significantly when transcription is driven by different promoters (Cramer et al. 1999). In a subsequent study by the same investigators, it was demonstrated that factors influencing pol II processivity, including transcriptional activators, influence EDI inclusion levels, apparently by enhancing or delaying the transcription of cis-acting splicing-regulatory sequences that modulate the selection of adjacent splice sites (Kadener et al. 2001). We recently showed that strong transcriptional activators can result in the increased pre-mRNA splicing of constitutive introns, as well as increased cleavage levels (Rosonina et al. 2003). These effects of strong activators were shown to be dependent on the pol II CTD.
Whereas the N-terminal half of the mammalian CTD resembles the yeast CTD, which consists mostly of heptapeptide repeats corresponding to the consensus sequence, the C-terminal half contains repeats that mostly diverge from the consensus, yet nevertheless are conserved among mammals (Fig. 1A ▶; Barron-Casella and Corden 1992). The functional significance of the presence of different CTD repeats is not understood. Recent studies have begun to address this question by examining the role of different blocks of human CTD repeats in transcription and pre-mRNA processing. In transient transfection experiments using primarily reporters derived from human β-globin and β-IFN gene sequences, Bentley and colleagues have provided evidence that a minimum of 15 CTD repeats is necessary for efficient 5′-end cap formation (Fong and Bentley 2001). However, efficient transcription, splicing and 3′-end formation minimally required 25 repeats, from either the conserved or divergent halves of the CTD, together with a 10 amino acid sequence located at the very C-terminus of the CTD (Fig. 1A ▶; Fong et al. 2003). In another recent study, Manley and colleagues (Ryan et al. 2002) demonstrated that efficient 3′-end cleavage of a pre-mRNA substrate derived from the adenovirus L3 pre-mRNA in vitro requires a CTD of at least 26 repeats. The most important determinant in this case was the length of the CTD, rather than the type of CTD repeats present (Ryan et al. 2002). It was not clear from these studies whether the differences in the CTD requirements observed were due to the different assay systems used, or else were a consequence of the unexplored possibility that different pre-mRNAs have different requirements for CTD repeats in order to be transcribed and processed efficiently.
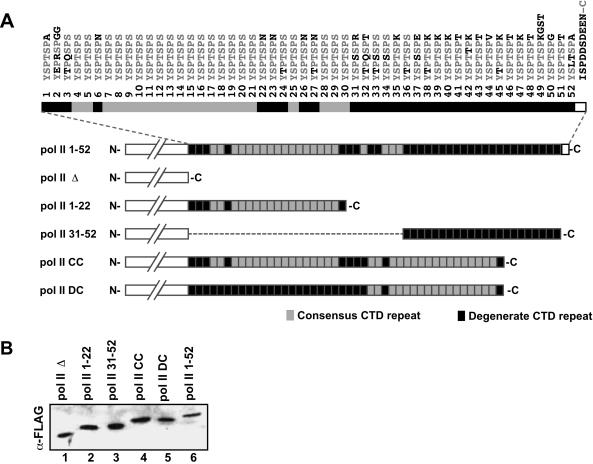
FIGURE 1.
Primary structure of the CTD of pol II. (A) The primary sequence of the 52-repeat human CTD is shown, with the heptapeptide repeat numbers indicated. Residues that do not follow the consensus (YSPTSPS) are highlighted in black. A schematic representation of the CTDs of the different pol II expression plasmids used is also shown, where consensus and degenerate CTD repeats are indicated as light and dark-shaded boxes, respectively. The white box located at the very C-terminus of the CTD represents its terminal 10 amino acid (nonheptapeptide repeat) sequence (see Introduction). The expression plasmids each contain an N-terminal Flag-epitope and an α-amanitin-resistant pol II catalytic domain fused to a CTD with the following heptapeptide repeat composition: 1-52 (pol II 1-52; wild type), repeats 1-22 (pol II 1-22), repeats 31-52 (pol II 31-52), two tandem blocks of repeats 1-22 (pol II CC), or fusion of repeats 31-52 to 1-22 (pol II DC). (B) Expression of mutant pol II derivatives containing different CTDs. Each pol II expression plasmid shown in (A) was transfected into human 293 cells, and the corresponding protein lysates were immunoblotted with the α-Flag antibody.
In the present study, we examined the role of different CTD repeats of mammalian pol II in splicing and 3′-end formation for three different pre-mRNA reporters in vivo. We demonstrate that 22 repeats of the CTD, containing either mostly consensus or entirely degenerate repeats, are sufficient for efficient transcription, splicing, and 3′-end formation of a pre-mRNA containing a constitutive intron. Twenty-two repeats were also found to be sufficient for near wild-type levels of splicing of an exonic splicing enhancer (ESE)-dependent pre-mRNA reporter. However, in a third pre-mRNA reporter, the level of inclusion of an alternatively spliced exon from the CD44 gene was more dependent on the number of pol II CTD repeats. In this case, a CTD of 22 repeats containing mostly consensus or degenerate repeats was not sufficient for efficient inclusion of the alternative exon. Interestingly, deletion of an exonic splicing silencer (ESS) within the CD44 exon reduced the dependence on the CTD for efficient inclusion, indicating that the length of the CTD is important for counteracting the negative effect of an ESS. Our results provide evidence that different pre-mRNAs have different dependencies on the CTD for efficient processing. In each case, different blocks of repeats of the CTD of equal length are functionally equivalent. Thus, for at least for some pre-mRNAs, it is the length rather than the nature of the CTD repeats that is more important for efficient pre-mRNA processing.
RESULTS
Expression of RNA polymerase II mutants containing different CTDs
To investigate whether there are different roles for subdomains of the CTD consisting primarily of either consensus or degenerate heptapeptide repeats, we constructed expression plasmids for Flag epitope-tagged pol II containing the full-length CTD (pol II 1-52), no CTD (pol II Δ), or the “consensus domain” of the CTD (pol II 1-22), or the “degenerate domain” of the CTD (pol II 31-52; Fig. 1A ▶). Specifically, the N-terminal-most 22 CTD repeat segment was designated the consensus domain because it consists of 17 perfect YSPTSPS repeats, and only five nonconsensus repeats. The C-terminal-most 22 repeat segment was designated the degenerate domain, because it contains entirely nonconsensus repeats (refer to Fig. 1A ▶ for sequences of the repeats comprising the consensus and degenerate domains).
To address whether truncation of the CTD, rather than elimination of one of the domains, might alter properties of pol II, we also constructed Flag epitope-tagged pol II expression plasmids containing two tandem copies of the 22-repeat domains, in every combination. Thus, each of these modified pol IIs contained 44 repeats consisting of two fused consensus domains (pol II CC), fused consensus and degenerate domains (pol II CD), fused degenerate domains (pol II DD), or the degenerate domain fused to the consensus domain (pol II DC). In addition to an N-terminal Flag epitope tag, each expression plasmid contained the point mutation Asn792→Asp, which renders the expressed polymerase resistant to the potent inhibitor of pol II, αamanitin (Nguyen et al. 1996). Thus, in the presence of this drug, the function of each mutant pol II can be assayed in the absence of endogenous pol II, which becomes degraded. Each plasmid was transfected into human 293 cells, and the expression levels of the mutant pol IIs were analyzed by immunoblotting with the α-Flag antibody (Fig. 1B ▶). With the exception of pol II CD and pol II DD, which were expressed poorly, the other six pol II derivatives expressed well and at comparable levels (Fig. 1B ▶, cf. lanes 1–6). We therefore limited our study to these six pol II-CTD deletion mutants.
Pol II with a CTD of 22 heptapeptide repeats can transcribe and efficiently process pre-mRNA reporters containing constitutive and ESE-dependent introns
In previous studies, truncation of the CTD to five repeats was shown to significantly reduce the efficiency of transcription, 5′-end capping, splicing, and 3′-end cleavage of pre-mRNA generated from the reporter pGal5HIV2TarCat-2 (McCracken et al. 1997a,b). This reporter consists of the chloramphenical acetyl transferase (CAT) gene upstream of the SV40 t intron and cleavage signal (Fig. 2A ▶, see figure legend for details). To further investigate the requirement of the CTD for processing of this reporter, we tested whether either the 22-repeat consensus or degenerate domains of the CTD are sufficient for its efficient splicing and 3′-end cleavage. Human 293 cells were transfected with expression plasmids for pol II Δ, pol II 1-22, or pol II 31-52; 16 h post-transfection, cells were treated with α-amanitin and re-transfected with the reporter pre-mRNA. Splicing and 3′-end cleavage levels of RNA generated by each pol II mutant was monitored by RNase protection analysis using the 5′-end, splicing and 3′-end cleavage protection probes indicated in Figure 2A ▶. Mapping of the 5′-end of RNAs demonstrated that pol II 1-22, pol II 31-52, and pol II 1-52 all initiated transcription at the expected position. As expected from previous reports (Gerber et al. 1995; Okamoto et al. 1996; Meininghaus and Eick 1999), pol II lacking a CTD (pol II Δ) did not transcribe RNA from this minigene reporter (Fig. 2B ▶, lane 1). Notably, the level of splicing, as determined by quantification of the percent splicing of RNAs, was similar for RNAs generated by pol II 1-22, pol II 31-52, and pol II 1-52 (Fig. 2B ▶ and adjacent bar graph), with splicing efficiency at ~60%–68%. Similarly, there was only a minor difference in the cleavage levels of RNAs generated from these three polymerases, with a small reduction (~10%) in the level of cleavage of pol II 1-22 and pol II 31-52 transcripts, compared to transcripts produced by pol II 1-52. We also determined the level of splicing and 3′-end cleavage of pGal5HIV2TarCat-2 reporter RNAs when transcribed with pol II containing a CTD consisting of heptapeptide repeats 1–15. In this case, both processing activities were reduced by 30%–50% (data not shown; refer to Fig. 2D ▶). By normalizing and summing the signals for processed and unprocessed RNAs, only small differences in the steady-state levels of RNAs generated by pol II 1-22, pol II 31-52, and pol II 1-52 were detected (Fig. 2C ▶). Both pol II 1-22 and pol II 31-52 generated ~80% as much steady-state RNA as did pol II containing a wild-type CTD. Overall, these observations indicate that, although the CTD is required for efficient transcription and pre-mRNA processing, a truncated CTD of 22 heptapeptide repeats is sufficient for generating near wild-type levels of transcription, splicing, and 3′-end cleavage of pGal5HIV2TarCat-2 reporter RNAs.
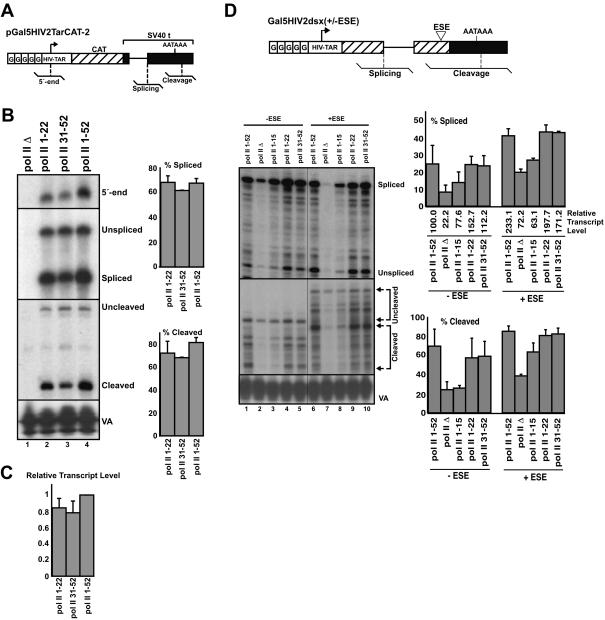
FIGURE 2.
A CTD of 22 repeats is sufficient for efficient transcription, splicing, and 3′-end cleavage of constitutively spliced and ESE-dependent pre-mRNA reporter. (A) Schematic representation of the pGal5HIV2TarCAT-2 pre-mRNA reporter, which contains the HIV-2 promoter and five upstream Gal4 DNA binding sites, the chloramphenical acetyl transferase (CAT) gene, and the SV40 t intron and cleavage/polyadenylation signal. Transcriptional initiation, splicing, and 3′-end cleavage were monitored using the RNase protection probes shown schematically below the reporters. (B) Cells transfected with α-amanitin-resistant pol II deletion mutant expression plasmids, and an expression plasmid for the activator Gal4-VP16, were treated with α-amanitin, then retransfected with the pGal5HIV2TarCAT-2 pre-mRNA reporter and the pol III expression plasmid pSP-VA. RNA was recovered and analyzed by RNase protection using probes for measuring transcriptional initiation (5′-end), splicing, and cleavage levels. Levels of the VA RNA were also determined by RNase protection, allowing for normalization of signals due to variations in transfection efficiency and RNA recovery. Representative analyses from five independent repeat experiments are shown. Quantification of splicing and cleavage efficiencies is shown in the adjacent bar graphs. RNA signals were first corrected for the number of U-residues before calculating the percent spliced and cleaved transcripts. (C) Levels of transcripts shown in (B) were calculated by summing the U-corrected spliced and unspliced signals, and the U-corrected cleaved and uncleaved level signals, and dividing these values by the corresponding VA signals. The resulting values are represented as a fraction of the transcript level resulting from pol II 1-52. (D) Schematic representation of the Gal5HIV2dsx(+/−ESE) reporter, with RNase protection probes for monitoring splicing and cleavage indicated. The (+ESE) version of the reporter contains a synthetic (6xGAA repeat) ESE sequence within the 3′-exon; this ESE stimulates the recognition of the adjacent, suboptimal 3′ splice site. Cells were transfected with the α-amanitin-resistant pol II deletion mutant expression plasmids, activator Gal4-VP16, Gal5HIV2dsx either with (+ESE) or without (−ESE) the ESE, and pSP-VA. Cells were treated with α-amanitin, and recovered RNA was analyzed using the splicing and cleavage probes indicated in the diagram. RNA transcribed by pol II Δ from the Gal5HIV2dsx(+/−ESE) reporter was detectable above background levels, and therefore quantified. An α-amanitin-resistant pol II containing the first 15 CTD heptapeptide repeats, which expresses to comparable levels as pol II with a wild-type CTD (Fong and Bentley 2001), was also included for comparison. The bar graphs indicate quantification of splicing and 3′-cleavage levels from three repeat separate analyses, and were calculated as described in Fig. 1B ▶. Relative transcript levels were calculated as in Fig. 1C ▶, and are adjusted relative to the level for pol II 1-52 (−ESE reporter), which was set to 100.
We also tested the activities of RNA pol II derivatives containing CTD repeats 1-15, 1-22, 31-52, or no CTD, in transcription, splicing, and 3′-end processing of a pre-mRNA reporter derived from the doublesex (dsx) gene of Drosophila (Fig. 2D ▶). This pre-mRNA reporter, which consists of a region spanning exons 3 and 4 of the dsx gene, requires an ESE in exon 4 for efficient splicing and 3′-end cleavage. Insertion of a typical mammalian ESE consisting of GAA repeats in place of the natural dsx ESE results in efficient processing of this substrate in transfected 293 cells (McCracken et al. 2002). We recently showed that the transcriptional activator-dependent stimulation of splicing and 3′-end cleavage of this pre-mRNA reporter is CTD-dependent (Rosonina et al. 2003). Similar to the results described above with the pGal5HIV2TarCat-2 reporter, either block of 22 repeats from the conserved or degenerate halves of the CTD was sufficient for near wild-type levels of transcription, splicing, and 3′-end cleavage of the dsx reporter pre-mRNAs, whether or not they contained the ESE (Fig. 2D ▶, cf. lanes 4,5,9,10 and lanes 1,6, and refer to the adjacent bar graph). Transcription by pol II Δ, which was detectable from this reporter (refer to Fig. 2 ▶ legend), resulted in significantly reduced levels of splicing, 3′-end cleavage, and transcription (Fig. 2D ▶, cf. lanes 2 and 7 with lanes 1,4,5 and 6,9,10, respectively, and refer to the adjacent bar graph). Truncation of the CTD to 15 repeats resulted in an intermediate level of transcription, splicing, and 3′-end cleavage compared to when the reporter was transcribed with pol II containing a CTD of 22 repeats or no CTD. Although the CTD-dependent levels of steady-state transcripts and pre-mRNA processing roughly paralleled each other, we demonstrated previously that the levels of splicing and 3′-end cleavage of the dsx reporter pre-mRNA are controlled independently of the steady-state transcript levels of this reporter (Rosonina et al. 2003; Figs. 2 ▶, 3 ▶). We therefore conclude that for two different pre-mRNA substrates, a pol II with a CTD of 22 repeats from either the conserved or divergent halves of the CTD is sufficient for efficient transcription, splicing, and 3′-end formation, whereas pol II with a CTD of 15 or less repeats is not.
FIGURE 3.
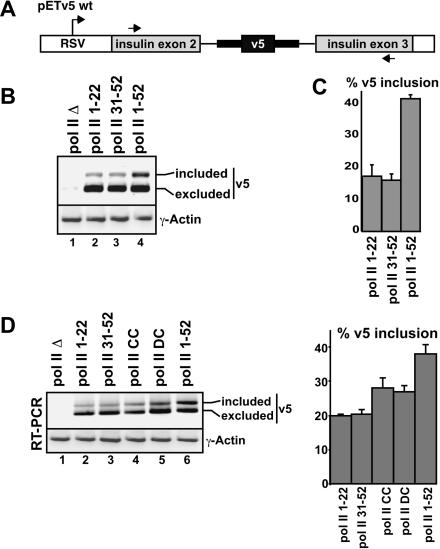
Truncation of the CTD results in reduced inclusion levels of the alternatively spliced CD44 v5 exon. (A) Diagram of the pETv5 wt reporter minigene used to monitor alternative splicing. The variant v5 exon, from the human CD44 gene, is flanked by native CD44 intron sequences (indicated by a bold line) and intron/exon sequences from the constitutively spliced exon 2-intron 2-exon 3 region of the human insulin gene. Inclusion of v5 was detected by RT-PCR using primers specific for the constitutively spliced insulin exons, as indicated (Konig et al. 1998). (B) Human 293 cells were transfected with expression plasmids for the pol II deletion mutants indicated (refer to Fig. 1A ▶ ) and with pETv5 wt. Recovered RNAs were analyzed by RT-PCR. Spliced RNAs containing (included) or lacking v5 (excluded) are indicated. The level of γ-actin RNA was determined by RT-PCR as a control for RNA recovery. (C) The percent v5 inclusion measured from three independent experiments, with the average level and standard deviations shown. For quantification of the percent v5 inclusion, RT-PCR was performed using a radiolabeled PCR primer, as described in Materials and Methods, and signals were quantified by phosphorimager analysis. Linearity of signal detection was confirmed by comparing three different amounts of input RNA in the RT-PCR assays (data not shown). (D) RT-PCR analysis and quantification of the percent v5 inclusion levels in transcripts produced using additional pol II deletion mutants, as indicated. The experiment and analysis were performed as described in B and C.
Truncation of the CTD to 22 repeats results in reduced inclusion of the alternatively spliced v5 exon from the CD44 gene
To investigate whether the CTD is important for modulating the inclusion of an alternative exon, we next asked whether the consensus or degenerate domains are sufficient for full levels of inclusion of an alternatively spliced exon (v5) derived from the CD44 cell surface glycoprotein gene. Figure 3A ▶ shows the structure of the minigene (pETv5) tested, which consists of the CD44 v5 exon and surrounding native intron sequences, flanked by constitutively spliced exons 2 and 3 of the human insulin gene (Konig et al. 1998). The level of inclusion of the v5 exon in mRNA generated from this pre-mRNA reporter is dependent on specific trans-acting factors, which are targets of different signal transduction pathways (Konig et al. 1996, 1998). The pETv5 vector was transfected into α-amanitin-treated human 293 cells expressing α-amanitin-resistant pol II Δ, pol II 1-22, pol II 31-52, or pol II 1-52 (see Materials and Methods). RNA recovered from cells 24 h post-transfection was analyzed by RT-PCR, using the primers indicated in Figure 3A ▶. The level of inclusion of v5 was determined by quantifying the percent of RNA containing or lacking this exon (Fig. 3B ▶). As reported previously, v5 is largely excluded from pETv5 mRNA in 293 cells (Konig et al. 1998). However, a further reduction in the level of inclusion is apparent when the CTD was truncated to 22 repeats (Fig. 3B ▶, cf. lane 4 and lanes 2,3, and quantification in C). Whereas ~40% of transcripts generated by pol II 1-52 contain v5, less than 20% of the transcripts generated by pol II 1-22 and pol II 31-52 contained the variant exon (see quantification in Fig. 3C ▶). Because there was essentially no difference in the level of v5 inclusion for pol II 1-22 and pol II 1-32, it is apparent that the length of the CTD, rather than the nature of the CTD repeats in the transcribing pol II, is more important for efficient inclusion of the CD44 alternative exon.
If v5 inclusion depends primarily on the length of the CTD, then a pol II containing a CTD length that is intermediate between 22 and 52 repeats would be expected to result in an intermediate level of v5 inclusion. To test this, each of the six pol II expression plasmids described in Figure 1A ▶ were transfected into human 293 cells, including pol II CC and pol II DC, which each contain 44 CTD repeats (Fig. 3D ▶). pETv5 was transfected into these cells after the addition of α-amanitin, and RNA was recovered for analysis by RT-PCR. As expected, pol II 1-52 resulted in ~40% inclusion of v5 (RT-PCR, Fig. 3D ▶, lane 6), whereas pol II 1-22 and pol II 31-52 both resulted in ~20% inclusion (Fig. 3D ▶, lanes 2,3, respectively). Pol II CC and pol II DC resulted in an intermediate level of inclusion of v5 (~28% and ~27%, respectively). Thus, a CTD of 44 repeats resulted in about one-third higher levels of v5 inclusion, compared to a CTD of 22 repeats. Together, these results provide strong evidence that the efficiency of v5 inclusion depends on the length of the CTD. More repeats, irrespective of their composition, result in a higher level of v5 inclusion.
The CTD is required to overcome the negative effect of an ESS on v5 inclusion
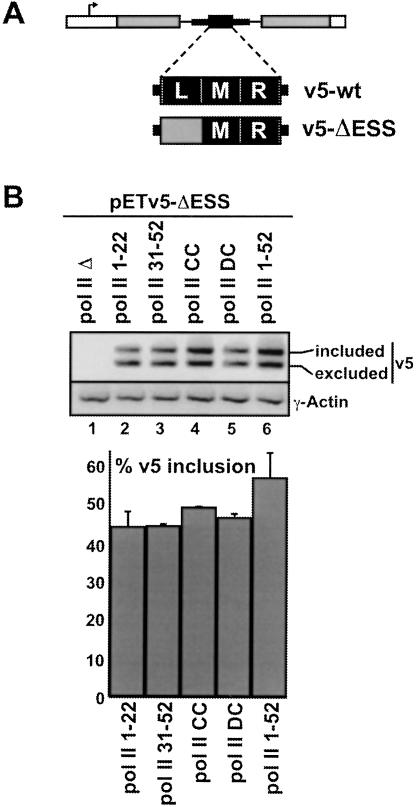
In a previous study, ESE and ESS sequences were identified which influence the level of inclusion of the CD44 v5 exon in pre-mRNA (Konig et al. 1998). Replacement of 40 bp proximal to the 3′ splice site with a sequence from the pBluescript plasmid (creating the pETv5–ΔESS minigene reporter (Fig. 4A ▶; Konig et al. 1998), resulted in a significant increase in inclusion of v5, due to elimination of an ESS. To determine whether differences in the CTD length dependence that we observed for the wild-type form of this reporter are similar in the presence or absence of the ESS, we analyzed the effects of the mutant CTD pol IIs on splicing of pre-mRNA from the pETv5–ΔESS reporter (Fig. 4B ▶). Compared to the pETv5 wt pre-mRNA reporter, an increased inclusion of v5 was observed, regardless of which polymerase transcribed the minigene (cf. Fig. 4B ▶ lanes 2–6 and Fig. 3D ▶ lanes 2–6). For pol II with a wild-type CTD, v5 inclusion was ~57% (see bar graph in Fig. 4B ▶). However, in the absence of the silencer element, the difference in inclusion among the different polymerases was small (between 12% and 18%; compare bar graphs in Figs. 3D ▶ and 4B ▶). This reduction in the dependence on the CTD length for efficient v5 inclusion indicates that the number, rather than the type of CTD repeats, is important for counteracting the negative effects of an ESS that regulates the level of inclusion of an alternative exon.
FIGURE 4.
The CTD-length dependence for efficient inclusion of the CD44 v5 exon is influenced by an exonic splicing enhancer sequence. (A) Location of functionally mapped cis-acting sequences (L, M, R) in the CD44 v5 exon (Konig et al. 1998). L and M contain ESS sequences, whereas R contains an ESE element. Substitution of L with sequences from the pBluescript cloning vector in construct pETv5-ΔESS results in increased inclusion of v5 from this reporter (named pETv5–ΔLblue in Konig et al. 1998). (B) 293 cells were transfected with expression plasmids for each of the pol II deletion mutants, then with pETv5–ΔESS, as described in Fig. 2 ▶. RNA was recovered and analyzed by RT-PCR using the primers indicated in Fig. 3A ▶. The bar graph shows quantification of v5 inclusion levels, as measured in Fig. 3 (B, C) ▶, from three independent experiments.
DISCUSSION
In this study we investigated the requirement for specific pol II CTD heptapeptide repeats for the efficient transcription, splicing, and 3′-end formation of three different pre-mRNAs. We found that N- and C-terminal segments of the CTD consisting, respectively, of 22 repeats that mostly conform to the heptapeptide consensus sequence, or which entirely diverge from the consensus sequence, are sufficient for efficient transcription, splicing, and 3′-end cleavage of a pre-mRNA reporter that contains a constitutive intron, as well as for the transcription and processing of an exon enhancer-dependent pre-mRNA. However, the efficient inclusion of an alternative exon in a third reporter pre-mRNA is reduced when it is transcribed with a pol II containing a CTD of 22 repeats. The inclusion level is reduced to a similar extent whether the block of repeats is derived from the N-terminal-conserved or C-terminal-degenerate halves of the CTD. Together, the results indicate that these two subregions of the CTD are functionally equivalent for different pre-mRNA substrates.
In a previous study by Fong and Bentley (2001), it was concluded that the C-terminal half of the CTD (including 26 heptapeptide repeats) is necessary and sufficient for splicing and 3′-end cleavage, whereas 26 heptapeptide repeats from the N-terminal half of the CTD is not sufficient. Both CTD regions, however, supported efficient capping of pre-mRNAs. The apparent discrepancy between these previous results and our present findings could reflect subtle differences in the nature of the CTD segments that were examined. Alternatively, the pre-mRNAs tested were different, possibly indicating that the CTD requirement for efficient pre-mRNA processing is gene-specific, as is the promoter-specific requirement for the CTD in transcription (Buratowski and Sharp 1990; Gerber et al. 1995; McNeil et al. 1998). This is also suggested by the recent finding that a 10 amino acid sequence (Fig. 1A ▶) at the very carboxyl terminus of the CTD is important for efficient transcription and processing of pre-mRNA reporters derived from β-globin and β-IFN gene sequences (Fong et al. 2003). Because we observed near wild-type levels of transcription, splicing, and 3′-end cleavage with pol IIs containing CTD repeats 1-22 or 31-52, both of which lack this sequence, we conclude that the C-terminal-most 10 amino acid sequence is not generally required for efficient transcription or pre-mRNA processing in vivo. Our results are in agreement with another recent report demonstrating that the length rather than the type of CTD repeats is more important for efficient pre-mRNA processing (Ryan et al. 2002).
That processing of different pre-mRNAs may have different requirements for the pol II CTD is further supported by our observation that 22 CTD repeats is sufficient for efficient splicing of the pGal5HIV2TarCAT-2 and dsx reporters, but not for efficient inclusion of the CD44 v5 exon in the pETv5 reporter. The observation that a CTD containing 44 repeats results in a level of inclusion of this alternative exon that is intermediate between that obtained with a 22-repeat CTD and the wild-type CTD implies that the level of inclusion correlates with the length of the CTD. This dependency on the CTD for efficient inclusion of the CD44 v5 exon was reduced by deletion of an ESS element in the alternative exon, suggesting that an important role for the CTD might be to counteract the negative activity of an ESS.
In conclusion, our results argue that different pre-mRNA substrates depend on the CTD to different extents for efficient processing. In the pre-mRNA substrates that we analyzed, the length of the CTD is more important than the particular segment of heptapeptide repeats, whether derived from the conserved or degenerate CTD subdomains.
MATERIALS AND METHODS
Plasmids
Previously described plasmids include the plasmid for expression of Gal4-VP16 (pSGVPΔ490, Yankulov et al. 1994), control plasmid pSP-VA (pSp65-VA1, Blau et al. 1996), reporters pGal5HIV2TarCAT-2, Gal5HIV2dsx(+/−ESE), and plasmids for generating ribonuclease (RNase) protection probes for these reporters (Yankulov et al. 1994; McCracken et al. 1997a; Rosonina et al. 2003), and minigene reporters pETv5 and pETv5–ΔESS (gift from Harald Konig, Konig et al. 1998). Pol II expression plasmids for pol II Δ, pol II 1-52, pol II 1-22, and pol II 31-52 were described previously (Rosonina et al. 2003), as was the plasmid for pol II 1-15 (Fong and Bentley 2001). Expression plasmids for pol II CC and pol II DC were constructed by PCR amplification of CTD repeats 31-52 with ClaI and NotI terminal restriction sites and subcloning into pol II 1-22 and pol II 31-52, respectively.
Transfections and RNA analysis
Human 293 cells grown on 15-cm plates were transfected using FuGENE 6 (Roche) per the manufacturer’s instructions, with 5 μg of the appropriate α-amanitin-resistant pol II expression plasmid, and in the case of the pGal5HIV2TarCAT-2 and Gal5HIV2dsx(+/−ESE) reporters, 0.5 μg of Gal4-VP16; 16 h later, cells were retransfected with 5 μg reporter, or control plasmid, and 0.5 μg of pSP-VA. RNA was isolated 48 h after the initial transfection as described (Yankulov et al. 1994). For the experiment shown in Figure 2D ▶, all plasmids were transfected simultaneously, 16 h prior to the addition of α-amanitin. pSP-VA encodes a pol III transcript that can be used to compare RNA recovery and transfection efficiency levels between samples, and for normalization of signals.
RNase protection analyses were performed as described (Yankulov et al. 1994). Samples were analyzed by electrophoresis through a 6% polyacrylamide-urea gel, and autoradiography and phosphorimager analysis. Percent splicing and cleavage were calculated by first correcting the appropriate band signals for the number of U residues (α32P-UTP was used in in vitro transcription reactions to generate RNA probes), then dividing the corrected spliced (or cleaved) band signal by the total of the unspliced (or uncleaved) and the spliced (or cleaved) signal, multiplied by 100. Transcript level was determined by summing the corrected spliced and unspliced signal divided by the corrected VA signal, and averaging with the value obtained by summing the corrected cleaved and uncleaved signals. Values were normalized compared to the value obtained for pol II 1-52, and for Figure 2D ▶, values were multiplied by 100. Experiments shown were repeated twice, unless indicated otherwise.
For RT-PCR, 1 μg of total RNA was reverse-transcribed by Superscript II reverse transcriptase (Invitrogen), according to the manufacturer’s specifications, using dT18 as a primer. RT products were resuspended in 40 μL of sterile distilled water, and various dilutions were analyzed by PCR to ensure linearity. One μL of RT product, or 1 μL of dilutions up to 10,000-fold were added to a PCR cocktail containing an end-labeled primer for radioactive analysis. Percent inclusion of v5 exon was calculated by dividing the densitometry signal for the included band by the total signal for the included and excluded signals, times 100. Sequences for the insulin exon-specific oligonucleotide primers for the detection of inclusion levels of the v5 exon from the pETv5 minigene construct were described (Konig et al. 1998). For γ-Actin control RT-PCR, sequence-specific primers were, forward: 5′-ACGTCGCCCTG GACTTCGAGC-3′ and reverse: 5′-CTAGAAGCATTTGCGGTG GAC-3′. All RT-PCRs were repeated using the One-step RT-PCR kit (QIAGEN), per the manufacturer’s specifications, using 1 μg of total RNA, and products were analyzed by ethidium-bromide stain agarose gel electrophoresis. Digital photographs of the gel were obtained by gel documentation, and black-to-white inverted images are shown in Figures 3 ▶ and 4 ▶.
Western blots
Thirty micrograms of protein lysates were analyzed on a 7.5% SDS-polyacrylamide gel by polyacrylamide gel electrophoresis. For Figure 1B ▶, α-Flag (M5; Sigma) was used as a primary antibody at a 1000-fold dilution.
Acknowledgments
We thank J. Ingles, A. Cochrane, M. Bakowski, J. Calarco, S. McCracken, and H. Siu for helpful discussions and comments on the manuscript. D. Bentley, H. Konig, and M. Vigneron generously provided various plasmids and reagents used in these studies. Our work was supported by an operating grant from the National Cancer Institute of Canada (NCIC) to B.J.B. E.R. is a Research Scholar of NCIC, supported with funds provided by the Terry Fox run. B.J.B. is the recipient of an MRC/CIHR Scholarship Award and Premier’s Research Excellence Award.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5207204.
REFERENCES
- Allison, L.A. and Ingles, C.J. 1989. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc. Natl. Acad. Sci. 86: 2794–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron-Casella, E. and Corden, J.L. 1992. Conservation of the mammalian RNA polymerase II largest-subunit C-terminal domain. J. Mol. Evol. 35: 405–410. [DOI] [PubMed] [Google Scholar]
- Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell. Biol. 11: 347–351. [DOI] [PubMed] [Google Scholar]
- Blau, J., Xiao, H., McCracken, S., O’Hare, P., Greenblatt, J., and Bentley, D. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16: 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski, S. and Sharp, P.A. 1990. Transcription initiation complexes and upstream activation with RNA polymerase II lacking the C-terminal domain of the largest subunit. Mol. Cell. Biol. 10: 5562–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, E.J., Takagi, T., Moore, C.R., and Buratowski, S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 11: 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway, R.C., Bradsher, J.N., and Conaway, J.W. 1992. Mechanism of assembly of the RNA polymerase II preinitiation complex. J. Biol. Chem. 267: 8464–8467. [PubMed] [Google Scholar]
- Cramer, P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E., and Kornblihtt, A.R. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4: 251–258. [DOI] [PubMed] [Google Scholar]
- Cramer, P., Srebrow, A., Kadener, S., Werbajh, S., de la Mata, M., Melen, G., Nogues, G., and Kornblihtt, A.R. 2001. Coordination between transcription and pre-mRNA processing. FEBS Lett. 498: 179–182. [DOI] [PubMed] [Google Scholar]
- Fong, N. and Bentley, D.L. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 15: 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, N., Bird, G., Vigneron, M., and Bentley, D.L. 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 22: 4274–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, H.P., Hagmann, M., Seipel, K., Georgiev, O., West, M.A., Litingtung, Y., Schaffner, W., and Corden, J.L. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374: 660–662. [DOI] [PubMed] [Google Scholar]
- Hirose, Y. and Manley, J.L. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96. [DOI] [PubMed] [Google Scholar]
- ———. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Hirose, Y., Tacke, R., and Manley, J.L. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 13: 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C.K. and Shuman, S. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3: 405–411. [DOI] [PubMed] [Google Scholar]
- Howe, K.J. 2002. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta 1577: 308–324. [DOI] [PubMed] [Google Scholar]
- Kadener, S., Cramer, P., Nogues, G., Cazalla, D., de la Mata, M., Fededa, J.P., Werbajh, S.E., Srebrow, A., and Kornblihtt, A.R. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20: 5759–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske, A.J., Buratowski, S., Nonet, M., and Young, R.A. 1992. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell 69: 883–894. [DOI] [PubMed] [Google Scholar]
- Komarnitsky, P., Cho, E.J., and Buratowski, S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig, H., Moll, J., Ponta, H., and Herrlich, P. 1996. Trans-acting factors regulate the expression of CD44 splice variants. EMBO J. 15: 4030–4039. [PMC free article] [PubMed] [Google Scholar]
- Konig, H., Ponta, H., and Herrlich, P. 1998. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 17: 2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Maxon, M.E., Goodrich, J.A., and Tjian, R. 1994. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: A model for promoter clearance. Genes & Dev. 8: 515–524. [DOI] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S., Wickens, M., and Bentley, D.L. 1997a. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S., and Bentley, D.L. 1997b. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 11: 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Lambermon, M., and Blencowe, B.J. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, J.B., Agah, H., and Bentley, D. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes & Dev. 12: 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meininghaus, M. and Eick, D. 1999. Requirement of the carboxy-terminal domain of RNA polymerase II for the transcriptional activation of chromosomal c-fos and hsp70A genes. FEBS Lett. 446: 173–176. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being cotranscriptional. J. Cell Sci. 115: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.T., Giannoni, F., Dubois, M.F., Seo, S.J., Vigneron, M., Kedinger, C., and Bensaude, O. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res. 24: 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, H., Sheline, C.T., Corden, J.L., Jones, K.A., and Peterlin, B.M. 1996. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. 93: 11575–11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina, E. and Blencowe, B.J. 2002. Gene expression: The close coupling of transcription and splicing. Curr. Biol. 12: R319– 321. [DOI] [PubMed] [Google Scholar]
- Rosonina, E., Bakowski, M.A., McCracken, S., and Blencowe, B.J. 2003. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 278: 43034–43040. [DOI] [PubMed] [Google Scholar]
- Ryan, K., Murthy, K.G., Kaneko, S., and Manley, J.L. 2002. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol. 22: 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov, K., Blau, J., Purton, T., Roberts, S., and Bentley, D.L. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77: 749–759. [DOI] [PubMed] [Google Scholar]
- Zeng, C. and Berget, S.M. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20: 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]