Abstract
The ribosome is a two-subunit molecular machine, sporting a working cycle that involves coordinated movements of the subunits. Recent structural studies of the 70S ribosome describe a rather large number of intersubunit contacts, some of which are dynamic during translocation. We set out to determine which intersubunit contacts are functionally indispensable for the association of ribosome subunits by using a modification interference approach. Modification of the N-1 position of A715, A1912, or A1918 in Escherichia coli 50S subunits is strongly detrimental to 70S ribosome formation. This result points to 23S rRNA helices 34 and 69, and thus bridges B2a and B4, as essential for ensuring stability of the 70S ribosome.
Keywords: ribosome, 23S rRNA, intersubunit bridges, modification interference
INTRODUCTION
All ribosomes consist of two unequal subunits. Although the large subunit catalyses the peptidyl transferase reaction and the small subunit catalyses decoding of mRNA codons, the two-subunit organization itself is a prerequisite for dynamic ribosome functioning, for example, translocation (Spirin 2002). The ribosome stays together by virtue of tRNAs that interconnect the subunits and a number of intersubunit bridges. Nevertheless, tRNAs are not needed for subunit association per se (e.g., Blaha et al. 2002).
The presence of various rRNA regions in intersubunit contacts was determined by the use of footprinting (Chapman and Noller 1977; Herr and Noller 1979) and modification interference technologies (Herr et al. 1979). The first unambiguously located intersubunit contact was placed by chemical cross-linking between 23S rRNA helix 69 and 16S rRNA helices 44 and 45 (Mitchell et al. 1992). More recent footprinting studies determined a number of rRNA positions that participate in intersubunit contacts (Merryman et al. 1999a,b). Recent medium-resolution X-ray crystallography (Yusupov et al. 2001) and cryo-EM-based models of the 70S ribosome (Gabashvili et al. 2000; Gao et al. 2003) define, in addition to RNA–RNA bridges, several protein–protein and protein–RNA intersubunit bridges. Generally, intersubunit bridges as defined by structural studies are in good agreement with data obtained by chemical methods. The Yusupov et al. (2001) model incorporates 12 bridges, which translate into more than 30 individual interactions between 50S and 30S subunits. These bridges seem to be largely conserved between the three kingdoms of life (Spahn et al. 2001; Gao et al. 2003). Notably, the more central positions (as defined by proximity to peptidyl transferase [PT] and decoding centers) in both 30S and 50S are occupied by bridges, consisting entirely of RNA (bridges B2a, B2b, B2c, B3, and B7a), whereas protein–protein and protein–RNA bridges (B1a, B1b, B7b, and B8, plus elements of B4, B5, and B6) are more peripheral (Yusupov et al. 2001; Gao et al. 2003). Centrally located RNA–RNA bridges contribute more than 80% of the intersubunit contacts (Gao et al. 2003). Recently, it has been shown that central RNA–RNA bridges are static during EF-G-GTP binding whereas some peripheral protein-containing bridges change conformation, resulting in a major ratchetlike movement of ribosomal subunits (Gao et al. 2003; Valle et al. 2003). This implies that the core interactions between ribosomal subunits in the 70S ribosome might consist of the centrally located RNA–RNA bridges as opposed to a more regulatory role for the peripheral bridges.
We have used chemical modification of 23S rRNA inside the 50S subunits in combination with selection of functional 50S to determine the nucleotides in 23S rRNA that are functionally important for the stability of the 70S ribosome. We found that dimethyl sulfate (DMS) modifications of N-1 positions of adenines 715 in helix 34 and 1912 as well as 1918 in helix 69 interfere with 70S ribosome formation. These results point to two intersubunit bridges, B2a and B4, as essential for the stability of ribosomal subunit interaction.
RESULTS AND DISCUSSION
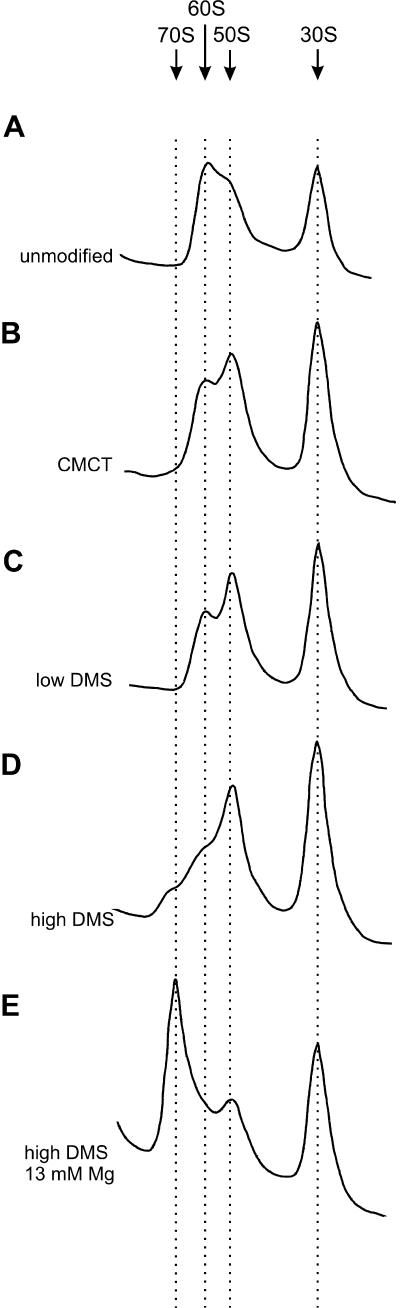
Our intention was to chemically modify 50S ribosomal subunits at a low level, so that the majority of the 50S population would retain its ability to bind 30S subunits. After modification with DMS or 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT), 50S subunits were reassociated with unmodified 30S subunits to determine which modified positions are excluded from the 70S ribosomes. Ribosomal subunit reassociating and subsequent sucrose gradient centrifugation was carried out in 6 mM MgCl2 to obtain partial 70S formation. Under these conditions, native subunits reassociate with 50%–60% efficiency (Fig. 1A ▶). All reassociation experiments that were performed in 6 mM MgCl2 exhibited ribosome sedimentation rates of ~60S (Fig. 1A–D ▶), in accordance with Blaha et al. (2002). To avoid overmodification, we used two concentrations of DMS, 17 mM and 85 mM. This corresponds to DMS/50S molar ratios of 8.5 × 103 and 4.25 × 104. 50S subunits, modified with the lower concentration of DMS or with CMCT, reassociated with 30%–40% efficiency in 6 mM MgCl2 and sedimented with velocities similar to unmodified ribosomes (Fig. 1B,C ▶). Therefore, modification of 50S subunits with CMCT or with 17 mM DMS reduces its reassociation efficiency by 20%–50%. Such an intermediate level of counterselection makes it likely that (1) a specific interference pattern can be found and (2) positions that are counterselected in our assay are indeed relevant as major contributors to 70S stability. 50S subunits that were modified with 85 mM DMS and reassociated in 6 mM MgCl2 produced two partially overlapping peaks in the 70S region (Fig. 1D ▶). Therefore, we also reassociated these 50S subunits in 13 mM MgCl2, where 70S ribosomes sedimented as one peak (Fig. 1E ▶). We could not perform the selection experiment by including tRNA because under stable tRNA binding conditions, for example, 13 mM MgCl2 (Schilling-Bartetzko et al. 1992), all modified 50S reassociates into 70S ribosome (data not shown). Hence no selection was possible.
FIGURE 1.
Sucrose gradient patterns of modified and subsequently reassociated 50S subunits. Ribosomal particles were separated by sucrose gradient centrifugation. (A) unmodified 50S reassociated in 6 mM MgCl2. (B) 50S, modified with CMCT were reassociated in 6 mM MgCl2. (C) 50S, modified with 17 mM of DMS, were reassociated in 6 mM MgCl2. (D) 50S, modified with 85 mM DMS, were reassociated in 6 mM MgCl2. (E) 50S, modified with 85 mM DMS, were reassociated in 13 mM MgCl2.
Scanning of the domains II–V of 23S rRNA by reverse transcriptase-directed primer extension revealed that modifications at two regions were strongly counterselected in reassociated 70S subunits.
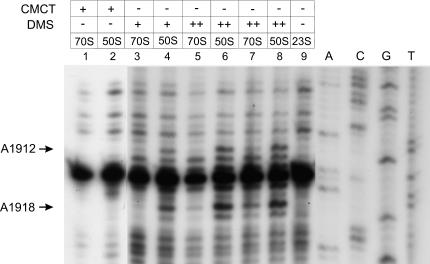
First, modification of A1912 and A1918 at their N-1 positions by DMS strongly interfered with 70S ribosome formation (Fig. 2 ▶). The interference was more pronounced if 70S ribosomes were formed in 6 mM MgCl2 (Fig. 2 ▶, lanes 3–6). When 13 mM MgCl2 was used for 70S reassociation, somewhat less interference was observed (Fig. 2 ▶, lanes 7,8). A1912 and A1918 are a part of the loop of 23S rRNA helix 69, contributing to the intersubunit bridge B2a.
FIGURE 2.
Reverse transcriptase analysis of the positions of the DMS and CMCT modifications in the 23S rRNA. Positions A1912 and A1918, whose modification interfere with 50S reassociation, are denoted by arrows. The dideoxy sequencing lanes are indicated by A, C, G, and T. (+) 17 mM DMS; (++) 85 mM DMS. (Lanes 1–6) selection experiments done in 6 mM MgCl2; (lanes 7,8) selection experiments done in 13 mM MgCl2.
Second, DMS modification of A715 conferred strong interference to 70S reassociation at 6 mM MgCl2, but did not interfere at 13 mM MgCl2 (Fig. 3 ▶). A715 is a part of the loop of helix 34 and intersubunit bridge B4.
FIGURE 3.
Reverse transcriptase analysis of the positions of the DMS and CMCT modifications in the 23S rRNA. Position A715, whose modification interferes with 50S reassociation, is denoted by an arrow. The dideoxy sequencing lanes are indicated by A, C, G, and T. (+) 17 mM DMS; (++) 85 mM DMS. (Lanes 1–3) selection experiments done in 6 mM MgCl2; (lanes 4,5) selection experiments done in 13 mM MgCl2.
No CMCT-specific interferences were found in 23S rRNA, although a number of CMCT-modified uridines were detected. The number of positions accessible to DMS and CMCT modification in 50S subunits was similar to the results summarized in Egjeberg et al. (1990).
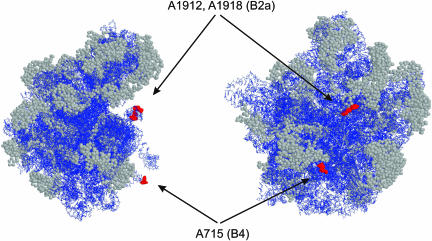
We have determined three positions in 23S rRNA that are functionally indispensable for 70S ribosome formation. The interfering positions, being a subset of modifiable positions at or near intersubunit contact areas, are located at two known intersubunit bridges, B2a and B4. This implicates bridges B2a and B4 as having a central role in the stability of 70S ribosomes (rather than a dynamic or regulatory role). This is in accordance with their recent assignment as immobile in the large-scale intersubunit rearrangement that accompanies ribosomal translocation (Gao et al. 2003; Valle et al. 2003). Although bridge B2a is one of the central RNA-only bridges (Yusupov et al. 2001), bridge B4 is more peripheral and, in addition to RNA (helix 34 of 23S rRNA and helix 20 of 16S rRNA), has a protein component in the 30S side (positions 59–63 and 86–87 of S15; Culver et al. 1999; Gao et al. 2003). However, what they have in common is that interfering positions at both helices 69 and 34 protrude from the main body of 50S subunit (Fig. 4 ▶).
FIGURE 4.
Modeling of interfering positions into the crystal structure of Deinococcus radiodurans 50S subunits (Harms et al. 2001; PDB accession code 1KC9). The left view is from the L1 side and the right view is from the 30S side of the 50S subunit. Arrows indicate interfering positions (E. coli numbering) and corresponding bridges. RasWin molecular graphics was used to highlight interfering positions in red spacefill. The rest of the RNA is blue and α-carbons of r-proteins are in gray spacefill.
In the medium-resolution X-ray crystallography-based model of Thermus thermophilus 70S ribosome, nucleotides A1913, C1914, and A1918 are in contact with the 16S rRNA helix 44 (Yusupov et al. 2001) whereas cryo-EM studies of Escherichia coli 70S ribosome point to A1912 and A1913 as the 50S component of B2a (Gao et al. 2003). We have found that N-1 modifications of A1912 and A1918 cause disruption of intersubunit association whereas modification of A1913 does not (Fig. 2 ▶). N-1 positions of adenine have been shown to be involved in minor groove RNA interactions (Nissen et al. 2001). Indeed, bridge B2a, which is made of the loop of helix 69 (23S rRNA positions 1912–1918) on the 50S side, has been found to dock into helix 44 of 16S rRNA by minor groove–minor groove interactions (Yusupov et al. 2001). Therefore, we have good reason to believe that observed interferences with 70S ribosome formation are caused by disruption of specific interactions of A1912 and A1918 with 16S rRNA helix 44. Accordingly, N-1 positions of A1912 and A1918 are likely to be involved in B2a formation via minor groove interaction with helix 44 of 16S rRNA.
Interestingly, helix 69 contains three pseudouridines (Ψ1911, mΨ1915, and Ψ1917; Ofengand 2002). Modified nucleosides are often found in functionally important regions of rRNA (Ofengand 2002). The fact that Ψ1915 and Ψ1917 are well conserved in all three kingdoms stresses the importance of the helix-loop 69 in ribosome functioning. Indeed, helix 69 contacts tRNAs at both the A site and the P site (Yusupov et al. 2001; Stark et al. 2002; Bashan et al. 2003). This makes it both a central structural component of the 70S ribosome and a good candidate for a mediator of signals between 50S and 30S subunits during the elongation cycle. Mutations in the helix 69 have been found to affect translational fidelity (O’Connor and Dahlberg 1995), supporting its role in coordinating crosstalk between two ribosomal subunits. Also noteworthy is the observation, based on hydroxyl-radical footprinting, that the ribosome anti-association factor IF3 and 23S rRNA helix 69 appears to contact overlapping areas in the 30S subunit (Dallas and Noller 2001). This suggests that disrupting bridge 2a may also be the way to ribosomal subunit dissociation in situ. Conformational flexibility of helix 69 (Ban et al. 2000; Harms et al. 2001; Yusupov et al. 2001) might enable it to carry the A site peptidyl-tRNA acceptor stem during translocation, making it effectively a molecular crane (Bashan et al. 2003). Nevertheless, our results clearly show that helix 69 can productively participate in intersubunit contacts in the absence of tRNAs. Thus, the essentially static structural role of helix 69 in ensuring the stability of the 70S ribosome and the aforementioned more dynamic tRNA-dependent roles appear to be separable.
MATERIALS AND METHODS
50S subunits were dissociated from tight-coupled 70S ribosomes by sucrose gradient centrifugation in 1 mM MgCl2 (Bommer et al. 1996). Modification of 50S subunits was conducted as in Stern et al. (1988). Eight A260 units of 50S and 1.2 μL or 6 μL of DMS stock (4 μL DMS, 16 μL ethanol) were used per 150 μL DMS reaction (17 mM and 85 mM DMS, respectively) and 75 μL of CMCT stock (126 mg/mL) per 150 μL CMCT reaction (149 mM CMCT). Modification reactions commenced for 5 min at 37°C and were stopped by addition of 15 μL 0.1% adenine on an ice bath. Stern et al. (1988) state that under conditions similar to our low DMS (17 mM) or CMCT modification protocols, no more than a few modifications per 23S rRNA molecule occur. Ribosomes were further purified from modifying agents by Sephacryl S400 spin columns (Amersham Pharmacia), equilibrated in buffer M6 (6 mM MgCl2, 60 mM NH4Cl, 30 mM Tris-HCl at pH 7.5, 60 mM KCl, 5 mM 2-mercapto ethanol) or M13 (same as M6, except with 13 mM MgCl2; Maiväli et al. 2002). 70S ribosomes were reassociated in buffer M6 or M13 by adding 8 A260 units of unmodified 30S subunits to modified 50S subunits in 1 mL final volume. Reassociation was carried out for 30 min at 37°C. 70S ribosomes and 50S subunits were fractionated by 10%–20% sucrose gradient centrifugation in M6 or M13 (Beckmann rotor SW 28, 20.4k rpm, 17 h) and rRNA was purified by silica binding as in Maiväli et al. (2002). Primer extension was done as in Stern et al. (1988). 23S rRNA domains II–V were scanned for modifications. Domains I and VI are not suspect as intersubunit contact areas (Yusupov et al. 2001). The identity of interfering positions was ascertained by at least five independent replications of the modification–selection experiment. Band densitometric analysis and quantification of observed interferences at 6 mM MgCl2 revealed a 70%–90% reduction of band intensities in reassociated 70S ribosomes if compared with the 50S fractions (data not shown).
Acknowledgments
We thank Dr. Tanel Tenson and M.Sc. Silja Kuusk (both University of Tartu, Estonia) for helpful comments on the manuscript. The research was supported by Estonian Science Foundation Grant 4425, EU framework V Grant No. QLK2-CT-2000-00935, and HHMI International Research Grant No. 55000332.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5220504.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Agmon, I, Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H.A., et al. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11: 91–102. [DOI] [PubMed] [Google Scholar]
- Blaha, G., Burkhardt, N., and Nierhaus, K.H. 2002. Formation of 70S ribosomes: Large activation energy is required for the adaptation of exclusively the small ribosomal subunit. Biophys. Chem. 96: 153–161. [DOI] [PubMed] [Google Scholar]
- Bommer, U., Burkhardt, N., Jünemann, R., Spahn, C.M.T., Triana-Alonso, F.J., and Nierhaus, K.H. 1996. Ribosomes and polysomes. In Subcellular fractionation. A practical approach (eds. J. Graham and D. Rickwoods), pp. 271–301. Oxford University Press, Oxford, UK.
- Chapman, N.M. and Noller, H.F. 1977. Protection of specific sites in 16 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J. Mol. Biol. 109: 131–149. [DOI] [PubMed] [Google Scholar]
- Culver, G.M., Cate, J.H., Yusupova, G.Z., Yusupov, M.M., and Noller, H.F. 1999. Identification of an RNA–protein bridge spanning the ribosomal subunit interface. Science 285: 2133–2136. [DOI] [PubMed] [Google Scholar]
- Dallas, A. and Noller, H.F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8: 855–864. [DOI] [PubMed] [Google Scholar]
- Egjeberg, J., Larsen, N., and Garrett, R.A. 1990. Structural map of 23S rRNA. In The ribosome: Structure, function and evolution (eds. W.E. Hill et al.), pp. 168–179. American Society for Microbiology, Washington, DC.
- Gabashvili, I.S., Agrawal, R.K., Spahn, C.M., Grassucci, R.A., Svergun, D.I., Frank, J., and Penczek, P. 2000. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100: 537–549. [DOI] [PubMed] [Google Scholar]
- Gao, H., Sengupta, J., Valle, M., Korostelev, A., Eswar, N., Stagg, S., Roey, P., Agrawal, R.K., Harvey, S.C., Sali, A., et al. 2003. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801. [DOI] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- Herr, W. and Noller, H.F. 1979. Protection of specific sites in 23 S and 5 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J. Mol. Biol. 130: 421–432. [DOI] [PubMed] [Google Scholar]
- Herr, W., Chapman, N.M., and Noller, H.F. 1979. Mechanism of ribosomal subunit association: Discrimination of specific sites in 16 S RNA essential for association activity. J. Mol. Biol. 130: 433–449. [DOI] [PubMed] [Google Scholar]
- Maiväli, Ü., Pulk, A., Loogväli, E., and Remme, J. 2002. Accessibility of phosphates in domain I of 23 S rRNA in the ribosomal 50 S subunit as detected by RP phosphorothioates. Biochim. Biophys. Acta 1579: 1–7. [DOI] [PubMed] [Google Scholar]
- Merryman, C., Moazed, D., Daubresse, G., and Noller, H.F. 1999a. Nucleotides in 23S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285: 107–113. [DOI] [PubMed] [Google Scholar]
- Merryman, C., Moazed, D., McWhirter, J., and Noller, H.F. 1999b. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285: 97–105. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Osswald, M., and Brimacombe, R. 1992. Identification of intermolecular RNA cross-links at the subunit interface of the Escherichia coli ribosome. Biochemistry 31: 3004–3011. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., and Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor, M. and Dahlberg, A.E. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 254: 838–847. [DOI] [PubMed] [Google Scholar]
- Ofengand, J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514: 17–25. [DOI] [PubMed] [Google Scholar]
- Schilling-Bartetzko, S., Bartetzko, A., and Nierhaus, K.H. 1992. Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. J. Biol. Chem. 267: 4703–4712. [PubMed] [Google Scholar]
- Spahn, C.M.T., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Spirin, A.S. 2002. Ribosome as a molecular machine. FEBS Lett. 514: 2–10. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.J., Zemlin, F., Wintermeyer, W., and van Heel, M. 2002. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 9: 849–854. [DOI] [PubMed] [Google Scholar]
- Stern, S., Moazed, D., and Noller, H.F. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164: 481–489. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M., and Frank, J. 2003. Locking and unlocking ribosomal motions. Cell 114: 123–134. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]