Abstract
Introduction of a premature termination codon (PTC) into an exon of a gene can lead to nonsense-mediated decay of the mRNA, which is best characterized as a cytoplasmic event. However, increasing evidence has suggested that PTCs may also influence the nuclear processing of an RNA transcript, leading to models of nuclear surveillance perhaps involving translating nuclear ribosomes. We used quantitative RT-PCR to measure the in vivo steady-state levels of every exon-intron junction in wild-type, PTC-containing, and missense-containing precursor mRNAs of both the nonrearranging dihydrofolate reductase (DHFR) and the somatically rearranging Ig-μ genes. We find that each exon-intron junction’s abundance and, therefore, the rate of intron removal, is not significantly affected by the presence of a PTC in a neighboring exon in either the DHFR or Ig-μ pre-mRNA. Similarly, the abundance of the uncleaved Ig-μ polyadenylation sites does not differ between wild-type and PTC-containing Ig-μ pre-mRNAs. Our Ig-μ data were confirmed by RNase protection analyses, and multiple cell isolates were examined to resolve differences with previously published data on steady-state pre-mRNA levels. We conclude that the presence of a PTC affects the rate of neither splicing nor the cleavage step of 3′ end formation during pre-mRNA processing in the nucleus. Our results are discussed with respect to existing evidence for nuclear surveillance mechanisms.
Keywords: nonsense, nonsense-mediated decay, pre-mRNA splicing, quantitative RT-PCR, RNase protection, DHFR, Ig-μ
INTRODUCTION
Introduction of a premature termination codon (PTC) into the open reading frame (ORF) of a protein-coding gene can lead to nonsense-mediated mRNA decay (NMD). NMD prevents the translation of truncated, functionally defective proteins. Recent advances have greatly illuminated the molecular mechanisms underlying how mammalian cells know that an mRNA harbors a PTC. However, whether cells can recognize the presence of a PTC before the mRNA has been processed and exported from the nucleus has remained controversial.
The importance of introns to nonsense-mediated decay is demonstrated by observations that intronless transcripts are not susceptible to NMD (Maquat and Li 2001) and that insertion of an intron into the 3′ untranslated region of an mRNA causes the normal termination codon to behave like a PTC (Carter et al. 1996; Thermann et al. 1998). The presence of a PTC can also cause changes in the splicing pattern of a pre-mRNA. Exon skipping is common under the selective pressure of a PTC (Dietz et al. 1993; Dietz and Kendzior 1994; Gersappe et al. 1999; Mendell and Dietz 2001; Caputi et al. 2002; Cartegni et al. 2002; Mendell et al. 2002; Wang et al. 2002a,b). Such nonsense-mediated alternative splicing (NAS) has been traced in some cases to the disruption of an exonic splicing enhancer or of important RNA secondary structure (Caputi et al. 2002; Cartegni et al. 2002). In other cases, NAS appears to be a direct consequence of an in-frame PTC (Li et al. 2002; Mendell et al. 2002). Furthermore, PTCs have been reported to inhibit pre-mRNA splicing in an ORF-dependent manner both in vivo (Naeger et al. 1992; Lozano et al. 1994; Gersappe and Pintel 1999; Gersappe et al. 1999) and in vitro (Aoufouchi et al. 1996). Mühlemann et al. (2001) studied two rearranging genes, TCR-β and Ig-μ, and observed that PTCs caused both higher levels and accumulation of unspliced precursor mRNAs at the site of transcription, suggesting that pre-mRNA processing efficiency is affected by the presence of a PTC.
Because the position of the PTC relative to the downstream intron is critical for NMD, a mechanism for marking the former site of introns on a mature mRNA was predicted (Maquat 1995, 2002). Subsequently, evidence has accumulated that the act of splicing deposits a multiprotein assembly, the exon-junction complex (EJC; Zhang et al. 1998a,b; Le Hir et al. 2000, 2001), on the nascent mRNA. When a translating ribosome then encounters a PTC upstream of an EJC, the mRNA is targeted for degradation (Le Hir et al. 2000, 2001). Yet, most PTC-containing mRNAs appear to be degraded while in association with the nucleus (Urlaub et al. 1989; Baserga and Benz 1992; Cheng and Maquat 1993; Belgrader and Maquat 1994; Belgrader et al. 1994; Lozano et al. 1994; Simpson and Stoltzfus 1994; Aoufouchi et al. 1996; Carter et al. 1996; Kessler and Chasin 1996; Li et al. 1997; Brogna 1999; Bühler et al. 2002). These apparently conflicting findings are consistent with two models: cotranslational export and/or nuclear translation. During cotranslational export, the mRNA is read by cytoplasmic ribosomes as it exits the nucleus, generating a decrease in the level of mRNA associated with the nuclear fraction. Considerable evidence for this “pioneer” round of translation has been reported by Maquat and colleagues (Ishigaki et al. 2001; Lejeune et al. 2002). Alternatively, if the mRNA were proofread by nuclear translation, the simplest explanation would be that PTCs are detected by ribosomes, each of whose subunits, as well as translation factors and charged tRNAs, reside in the nucleus for at least a short while before export to the cytoplasm (Lejbkowicz et al. 1992; Arts et al. 1998; Lund and Dahlberg 1998; Pederson and Politz 2000; Bohnsack et al. 2002).
The “nuclear scanning” model for detecting PTCs before an mRNA leaves the nucleus, originally proposed by Chasin and colleagues (Urlaub et al. 1989), is supported by accruing evidence for translation within the nucleus (for critical review, see Hentze 2001; Maquat 2002; Dahlberg et al. 2003). Mangiarotti (1999) provided evidence for the coupling of transcription and translation in Dictyostelium nuclei. Iborra et al. (2001) reported that nascent polypeptides accumulate in the nucleus of HeLa cells, also concluding that transcription and translation are coupled. However, Nathanson et al. (2003) have argued that the observations of Iborra et al. (2001) might have been due to cytoplasmic contamination and overpermeabilization of the cells.
If RNA transcripts are scrutinized by nuclear translation before or during splicing, then the presence of a PTC might be expected to affect the relative rates of intron removal. We therefore examined the in vivo abundance of 5′ exon-intron junctions in wild type, PTC-containing, and corresponding missense-containing precursor mRNAs of both the nonrearranging dihydrofolate reductase (DHFR) and the somatically rearranging Ig-μ genes, using quantitative reverse transcription-polymerase chain reaction (QRT-PCR). We observe that the relative rates of 5′ splice site cleavage are not affected by the presence of PTCs in various exons of either the DHFR or the Ig-μ pre-mRNAs. The Ig-μ results were confirmed by RNase protection assays, and the abundance of the Ig-μ uncleaved polyadenylation sites were also analyzed by QRT-PCR. Our results lead to two conclusions. First, the presence of a PTC alters neither the rate nor order of intron removal or the rate of cleavage/polyadenylation of a pre-mRNA. Second, careful comparison of the same cell lines examined by Mühlemann et al. (2001) reveal twofold rather than fivefold higher levels of Ig-μ pre-mRNA in the presence of a PTC.
RESULTS
Introns in DHFR pre-mRNAs are spliced at the same rate with or without a PTC in a neighboring exon
Previously, a method was devised to determine the relative rates of intron removal from a particular transcript by examining the abundance of unspliced exon-intron junctions within a steady-state population of partially processed cellular pre-mRNAs (Patel et al. 2002). QRT-PCR is used to determine the amount of each unspliced 5′ splice site, which is inversely proportional to the relative rate of removal of that intron. QRT-PCR is a highly sensitive method, allowing even pre-mRNAs of very low abundance to be analyzed. We applied this method to pre-mRNAs containing PTCs to establish whether the rate or order of removal of the introns neighboring the PTC-containing exon might be altered.
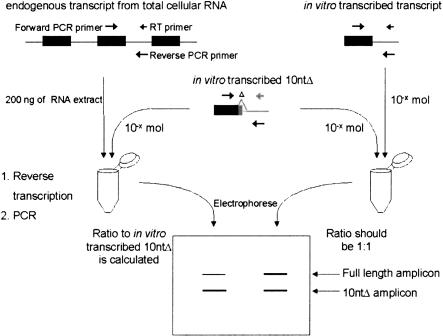
The procedure begins with the extraction of total RNA from cells containing the wild-type gene or from cells containing a PTC or a missense mutation in the gene. At the same time, two control RNAs are in vitro transcribed and quantified (Fig. 1 ▶). The first control RNA contains ~500 nucleotides surrounding the 5′ exon-intron junction that will be amplified, and the second control RNA contains the same sequence, but 10 nucleotides defining the 5′ splice site have been deleted. In order to quantify the amount of PCR product produced by the RT-PCR reaction, a known amount of the 10ntΔ control RNA is added to either an equal amount of the full-length control RNA or to a known amount of RNA purified from cell extract. The two RNA mixtures are reverse-transcribed, and then the cDNAs are amplified by PCR to generate products of ~100–150 nucleotides. The 10-nucleotide difference in length between the two PCR products allows their separation when electrophoresed through a denaturing polyacrylamide gel; the ratio of these bands is calculated on a phosphorimager. The internal standard RNA (10ntΔ) thus controls for possible variation in the amplification efficiency of sequences corresponding to different 5′ splice sites due to differences in sequence or secondary structure.
FIGURE 1.
Diagram of the quantitative RT-PCR assay. First, a titration is performed for each PCR amplicon to determine how many moles of an in vitro-transcribed RNA, containing the region to be amplified with a 10-nt deletion, should be added competitively to 200 ng of total extract RNA. The RNA mixture then undergoes RT using a primer specific for each exon-intron junction to be examined; 3/100 of the RT reaction is next amplified by PCR using primers specific to each exon-intron junction, with the forward primer being spiked with radiolabel. The PCR products are electrophoresed on a denaturing 10% polyacrylamide gel, and the ratio of the full-length product to the 10-nt-shorter product is determined on a phosphorimager. For example, if the full-length product were twice as intense as the shorter control product, then the exon-intron abundance would be calculated as 2 × 10−x moles/200 ng of extract = 10 × 10−x/μg of extract. At the same time, a reaction with no extract RNA and containing only in vitro-transcribed full-length and 10ntΔ control RNA is subjected to the same RT and PCR amplification steps. The ratio of these two PCR products should be 1:1, thereby serving as a control for validating the analysis (see Materials and Methods for more detail).
We first tested the QRT-PCR assay for its ability to distinguish less than 10-fold differences in 5′ splice site abundance. The standard QRT-PCR assay used throughout this report analyzes 200 ng of extract RNA. Titrations of the 10ntΔ control are routinely performed to determine how much to add to the reactions to obtain ratios that are ~1:1. However, to test the sensitivity of this assay in detecting less than 10-fold differences in RNA abundance, we began by adding 20, 100, 200, 400, and 1000 ng of extract RNA (corresponding to 0.1, 0.5, 1, 2, and 5 times the amount of pre-mRNA) to the same amount of 10ntΔ control. Reverse transcription and PCR were performed on these dilutions for exon-intron junctions 2, 3, and 5 of the Ig-μ pre-mRNA, described in more detail later. As seen in Table 1 ▶, the experimental results were as expected. When 1/10 the amount of pre-RNA (20 ng) was added to the reaction, the experimental result was ~1/10 for all three exon-intron regions assayed; when 5 times the pre-mRNA (1000 ng) was added, the resulting ratio was five times the normal ratio of 1:1. Thus, the observed ratios conform closely to the expected outcomes.
TABLE 1.
Quantitative RT-PCR is sensitive to less than 10-fold changes in RNA levels.
| Experimental ratio | ||||
| Dilution factor | Amount of extract RNA | μE/I2 | μE/I3 | μE/I5 |
| 0.1 | 20 ng | −0.01 | 0.11 | 0.13 |
| 0.5 | 100 ng | 0.36 | 0.58 | 0.50 |
| 1 | 200 ng | 1 | 1 | 1 |
| 2 | 400 ng | 2.0 | 1.9 | 1.8 |
| 5 | 1000 ng | 4.2 | 3.8 | 4.0 |
To test whether QRT-PCR can distinguish small differences in RNA levels in total RNA isolated from cell extracts, a titration experiment was performed. Ig-μ exon 2-intron 2 (μE/I2), exon 3-intron 3 (μE/I3), and exon 5-intron 5 (μE/I5) were analyzed by quantitative RT-PCR. Normally, the reaction uses 200 ng of extract RNA and an experimentally determined amount of 10ntΔ control RNA. In five different reactions, 1/10 (20ng), 1/2 (100ng), 1 times (200ng), 2× (400ng), and 5× (1000 ng) the amount of extract RNA was added to the same quantity of 10ntΔ control RNA. The experimental ratios should reflect the amount of extract RNA added; for example, when one-half the normal amount of extract RNA is added (100 ng), then the resulting experimental ratio should be 0.5. The actual ratios calculated from the QRT-PCR reactions were 0.36 for μE/I2, 0.58 for μE/I3, and 0.50 for μE/I5, closely corresponding to the expected value.
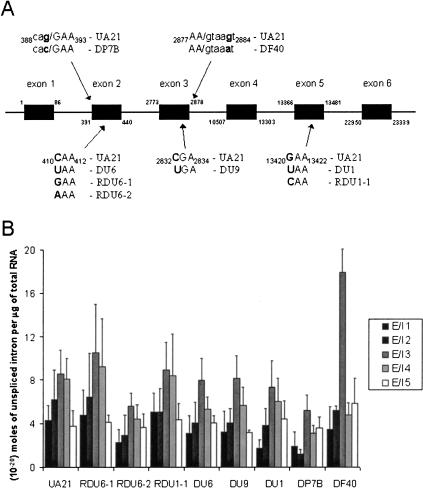
The nonrearranging DHFR gene was chosen for initial analysis. Use of this endogenous gene, with well studied PTC and missense mutations (Carothers et al. 1983, 1993; Urlaub et al. 1989; Kessler et al. 1993; Kessler and Chasin 1996), avoids any artifacts that might be caused by exogenous expression of a transfected construct. The DHFR pre-mRNA contains six exons and five introns (Fig. 2A ▶). We examined three PTC-containing cell lines with mutations in exons 2, 3, and 5, named DU6, DU9, and DU1, along with the parental cell line containing the hemizygous wild-type DHFR, UA21. Three missense mutants at positions corresponding to the PTCs in exons 2 and 5 were also tested: cell lines RDU6-1 and RDU6-2, containing two different missense codons in the same position as the PTC in exon 2, and RDU1-1, with a missense codon in the same position as the PTC in exon 5. The missense mutants control for any changes in the rate or order of intron removal from the DHFR transcript generated by alterations in exonic sequences (e.g., an exonic splicing enhancer).
FIGURE 2.
The abundance of Chinese hamster ovary DHFR exon-intron junctions in the cellular RNA population. (A) The ~24-kb CHO DHFR pre-mRNA (not to scale) contains six exons (black boxes) and five introns (thin black lines between boxes). The nucleotide at the start and end of each exon is numbered above or below. Seven different CHO cell lines were used. Each is hemizygous for the DHFR locus and was created from the same wild-type cell line (UA21). Three nonsense mutants with premature termination codons (PTCs) in exons 2, 3, and 5 (DU6, DU9, and DU1) and three missense mutants, in which two of the nonsense mutations had reverted to sense (RDU6-1, RDU6-2, and RDU1-1), were examined. Two splicing mutants were also assayed: DP7B contains a mutation in the 3′ splice site between intron 1 and exon 2, and DF40 has a mutation in the 5′ splice site between exon 3 and intron 3. The arrows indicate the approximate location of each PTC, missense, or splice site mutation on the DHFR pre-mRNA, and the sequence is provided with base numbers beside the name of each mutant cell line. Exonic sequences are capitalized, intronic sequences are in lowercase, and the boundary is shown as a “/”. (B) The five exon-intron junctions of DHFR pre-mRNA from the wild-type and missense and nonsense mutant cell lines were quantitated by QRT-PCR of total RNA. Assays were performed with various control and cellular RNA preparations 4–8 times. The QRT-PCR assays of each of the exon-intron junctions (E/I) from DP7B strain were repeated 3–6 times; those for DF40 were repeated 2–4 times. Bars show the number of moles of each DHFR exon-intron junction in 1 μg of total RNA. Cell lines are named on the x-axis. The error bars show the standard deviation (S.D.).
RNA purified from extracts of Chinese hamster ovary (CHO) cells hemizygous for the DHFR gene and various control RNA transcript preparations were subjected to QRT-PCR (Fig. 2B ▶). Each QRT-PCR assay was performed 4–8 times. The data reveal that the abundance of the 5′ exon-intron boundaries of the five DHFR introns is very similar from cell line to cell line, all on the order of 10−20 moles per microgram of extract RNA. Because the transcription rate of one nonsense mutant does not differ from wild type (Urlaub et al. 1989), the introns must be removed at roughly the same rate from the DHFR pre-mRNA in the wild-type, missense- and nonsense-containing cell lines. Introns 1 and 5 appear to be removed somewhat faster and intron 3 more slowly. The nonsense cell lines that contain PTCs in exon 2, 3, or 5 exhibit no significant differences in 5′ exon-intron abundance relative to wild type. We conclude that the rates of removal of DHFR introns are not affected by PTCs in neighboring exons.
DHFR splice site mutants provided positive controls establishing that changes in 5′ exon-intron boundary abundance can indeed be detected by QRT-PCR. Each QRT-PCR assay for the splicing mutants was performed 2–6 times. DP7B has a point mutation (nt 390 G→C) in the 3′ splice site between intron 1 and exon 2 (Fig. 2A ▶; Carothers et al. 1993). By QRT-PCR, DP7B has levels of exon-intron junctions 1, 3, 4, and 5 that are not significantly different from those of wild type (UA21), but the abundance of the intron 2 5′ splice site is the lowest of any strain examined (see Fig. 2B ▶). DF40 has a point mutation (nt 2883 G→A) in the 5′ splice site between exon 3 and intron 3 (Carothers et al. 1993). We observe that the abundance of exon-intron junction 3 in mutant DF40 DHFR pre-mRNA is twofold higher than in wild-type pre-mRNA (Fig. 2B ▶). The significant differences in exon-intron boundary abundance in these splicing mutants provide evidence that changes in intron removal can indeed be detected by QRT-PCR analysis of the cellular RNA population.
The relative rates of removal of Ig-μ introns are also unaffected by PTCs in neighboring exons
The Ig-μ mRNA encodes the immunoglobulin μ heavy chain (Ig-μ). Ig-μ pre-mRNAs containing PTCs have been reported to accumulate at or near the site of transcription (Mühlemann et al. 2001). Accumulation could result from alterations in mRNA processing, preventing the mature mRNA from leaving the site of transcription. Because one such processing event is splicing, we used the QRT-PCR assay to analyze the Ig-μ PTC-containing mutants that display pre-mRNA accumulation.
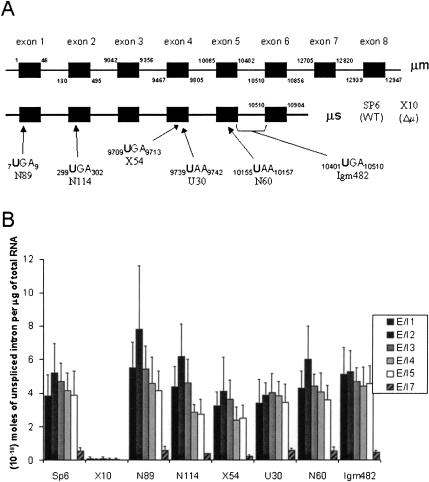
The cell lines used by Mühlemann et al. (2001) contain mutations in the endogenous Ig-μ gene of mouse B cells (Connor et al. 1993, 1994). Two different forms of μ mRNA are produced in B cells depending on whether the first or second poly(A) site is used: a 2.4-kb secreted form (μs) and a 2.7-kb membranous form (μm; Fig. 3A ▶; Connor et al. 1993, 1994). The relative amounts depend on the state of B-cell ontogeny; the hybridomas used here produce 90% secreted form. Secreted Ig-μ pre-mRNA, like DHFR pre-mRNA, contains six exons and five introns. The membranous form has a shortened exon 6 and two additional 3′ exons. Six different PTC-containing mutants, along with the wild-type hybridoma, were studied (the set used by Mühlemann et al. 2001 plus two additional cell lines; Connor et al. 1993, 1994). Unfortunately, no corresponding missense mutants were available for the Ig-μ cell lines.
FIGURE 3.
The abundance of mouse B-cell hybridoma Ig-μ exon-intron junctions in the cellular RNA population. (A) The Ig-μ pre-mRNA is shown (not to scale) with introns indicated by thin black lines between the exons (black boxes), with the start and end of each exon numbered. Ig-μ mRNA has two polyadenylation sites: Use of the first leads to a larger exon 6 in a 2.4-kb mRNA for the secreted form (μs), and use of the second creates a smaller exon 6 joined to exons 7 and 8 in a 2.7-kb membranous form (μm) of the mRNA. Ninety percent of the Ig-μ mRNA in these cell lines is Ig-μs. Wild type (Sp6), a cell line deleted for Ig-μ (X10), and six cell lines containing PTCs in Ig-μ exons (N89, N114, X54, U30, N60, and Igm482) were assayed. Arrows depict the approximate location of each PTC, and the sequence on the Ig-μ pre-mRNA with base numbers is given. For those mutants in which the PTC was created by deletion of one or more nucleotides, the base numbering remains the same as that for wild-type Ig-μ pre-mRNA, causing some PTCs to appear to have four or more nucleotides. The PTC of Igm482 is formed only when exons 5 and 6 are spliced together. Even though N89 and Igm482 mutants contain PTCs, they produce nearly wild-type levels of Ig-μ mRNA (Connor et al. 1994; Mühlemann et al. 2001). (B) Total RNA was extracted from the various mouse B-cell hybridoma lines. QRT-PCR was used to determine the number of moles of each exon-intron junction per μg of total RNA. QRT-PCR was performed for exon-intron junctions 1–5 of Ig-μ cell lines a minimum of six times, with the majority being repeated 8–10 times. Cell lines are named on the x-axis. The error bars represent the standard deviation (S.D.).
The eight cell lines examined were Sp6 (wild type), X10 (deletion of Ig-μ), N89 (PTC in exon 1), N114 (PTC in exon 2), X54 (PTC in exon 4), U30 (PTC in exon 4), N60 (PTC in exon 5), and Igm482 (PTC in exons 5/6; Fig. 3A ▶). All of the mutant cell lines were derived from the same parental cell line, the wild-type Sp6. The PTC in Igm482 is formed only upon splicing together exons 5 and 6. As expected, nonsense mRNAs from N114, X54, U30, and N60 are almost completely degraded in the cell (see Fig. 5B,D ▶; Connor et al. 1993). However, two of the mutants, N89 and Igm482, generate significant levels of mRNA. N89, with a premature stop at the third codon in the mRNA, produces ~25% wild-type levels of mRNA (Connor et al. 1994). It is not understood why the N89 message is not degraded completely, because it appears that translation does not reinitiate downstream of the initial start codon. Interestingly, in the case of Igm482, membranous Ig-μ mRNA undergoes nonsense-mediated decay, whereas the secreted form does not. Here, the shorter mRNA presumably escapes degradation because its PTC is in the last exon of the secreted form, whereas the PTC is upstream of two introns in the membranous form.
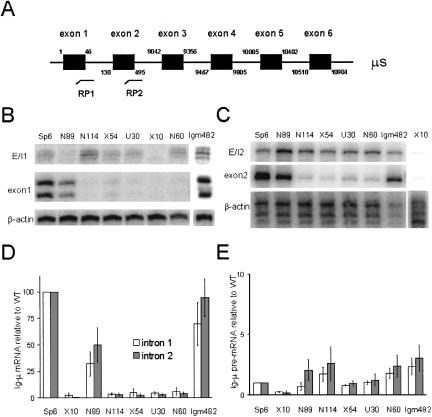
FIGURE 5.
The levels of spliced and unspliced Ig-μ mRNA as determined by RNase protection. (A) The Ig-μ secreted pre-mRNA is shown (not to scale) with introns indicated by thin black lines between the exons (black boxes), with the start and end of each exon numbered. The two probes used for RNase protection, RP1 and RP2, are shown below in the approximate locations where they hybridize. RP1 spans exon 1-intron 1 and RP2 spans exon 2-intron 2. (B) Mouse B-cell hybridoma RNAs were hybridized to RP1. Top panel: The region with triplet bands (due to frayed ends) corresponding to exon 1-intron 1 (E/I1), assessing the level of Ig-μ pre-mRNA. Middle panel: A doublet with size predicted for protection of exon 1, corresponding to the spliced form of Ig-μ mRNA. Bottom panel: A protected band corresponding to β-actin mRNA in the extract, to which all of the Ig-μ data were normalized. The Igm482 analysis was a separate experiment. All were run on 6% denaturing polyacrylamide gels. (C) Mouse B-cell hybridoma RNAs were subjected to RNase protection using the RP2 probe. Top panel: Protection of the unspliced exon 2-intron 2 boundary (E/I2); middle: the protected spliced mRNA; bottom: a triplet representing β-actin mRNA to which the Ig-μ data were normalized. A different β-actin probe was used here because the former probe protected a region similar in size to that protected by the RP2 probe. The X10 data were obtained in a separate experiment. All were electrophoresed on 6% denaturing polyacrylamide gels. (D) The data from three or four RNase protection assays using RP1 and five using RP2 were averaged and are presented in the graph. The levels of Ig-μ spliced mRNA for each cell line are plotted relative to those for Sp6 (wild type, WT) set at 100%, as done by Mühlemann et al. (2001). The error bars represent the standard deviation (S.D.). (E) The levels of Ig-μ pre-mRNA are plotted relative to Sp6 (WT) set at 1, as done by Mühlemann et al. (2001). The assays were repeated three or four times for RP1 and five times for RP2. The error bars represent the standard deviation (S.D.).
RNA extracted from each of these eight cell lines was subjected to QRT-PCR for exon-intron junctions 1–5 of Ig-μ, 6–10 times (Fig. 3B ▶). Exon-intron junctions 1–5 exist at ~10−18 moles per μg of RNA extract. Like the pattern seen for DHFR, the relative Ig-μ exon-intron junction levels do not differ significantly between wild-type and nonsense-containing pre-mRNAs. Introns 1–5 are found in both forms of Ig-μ, whereas intron 7 is present only in the mRNA of the membranous Ig-μ. As can be seen in Figure 3B ▶, the exon-intron 7 junction is indeed an order of magnitude less abundant than the first five junctions, providing an excellent positive control for the assay. The deletion mutant, X10, contains no Ig-μ introns, and none are detected by the QRT-PCR assay, providing a negative control (Fig. 3B ▶). Because run-on analyses (Mühlemann et al. 2001) have previously shown that the transcription rate of the majority of these cell lines is similar, we conclude that the rates of removal of Ig-μ introns are unaffected by PTCs in neighboring exons.
Polyadenylation is also unaffected by the presence of a PTC in the Ig-μ pre-mRNA
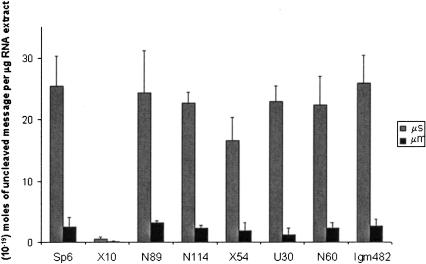
Even though splicing is unaffected by the presence of a PTC in the message, it remained possible that some other pre-mRNA processing step might be perturbed by the presence of a nonsense codon. We therefore tested 3′ end cleavage leading to polyadenylation. Using the same QRT-PCR assay, primers were designed to flank the Ig-μs and Ig-μm poly(A) sites. Again, the levels of uncleaved pre-mRNA should be inversely proportional to the rates of cleavage and polyadenylation. The data in Figure 4 ▶ reveal that the level of the membranous Ig-μm poly(A) site is about 10-fold less than that of the secreted Ig-μs, as expected. Again, the presence of a PTC in exon 1, 2, 4, or 5 does not significantly alter the abundance of either the uncleaved secreted or membranous poly(A) sites.
FIGURE 4.
The abundance of Ig-μ secreted (μs) and membranous (μm) poly(A) sites. The levels of the uncleaved poly(A) sites in secreted and membranous Ig-μ pre-mRNA were quantified by QRT-PCR. These QRT-PCR assays were repeated 4–9 times. Note that the uncleaved polyadenylation site of the mRNA for the secreted form is 10-fold more abundant than that of the membranous form. Cell lines are named on the x-axis. The error bars represent the standard deviation (S.D.).
RNase protection assays confirm the quantitative RT-PCR results
The results shown in Figure 3B ▶ revealed no significant differences in 5′ exon-intron junction abundance between the wild type and the PTC-containing mutants of Ig-μ when assessed by QRT-PCR. In contrast, Mühlemann et al. (2001) detected the accumulation of Ig-μ pre-mRNA in the same PTC-containing cell lines by an RNase protection assay. We therefore applied RNase protection to examine the levels of spliced and unspliced Ig-μ mRNA in the mouse B-cell lines. We used two probes, one of which spanned a portion of the exon 1-intron 1 junction (RP1) and the other a portion of the exon 2-intron 2 junction (RP2; Fig. 5A ▶). When these probes hybridize to spliced mRNA, only the exon portion of each probe would be protected from RNase digestion; when the probes bind unspliced pre-mRNA, the longer exon-intron portion would be protected.
The amount of spliced Ig-μ mRNA calculated to exist in the various cell lines (Fig. 5D ▶) based on the RNase protection data (Fig. 5B,C ▶) agrees well with previously published data (Connor et al. 1994; Mühlemann et al. 2001). The cell lines N114, X54, U30, and N60 all contain PTCs, and the Ig-μ mRNA undergoes nonsense-mediated decay. Accordingly, the levels determined by RNase protection in these cell lines are 6% or less relative to the wild-type Sp6 strain. The two cell lines with nonsense mutations that are not subject to NMD, N89 and Igm482, likewise exhibit levels that agree with those reported previously (Connor et al. 1994; Mühlemann et al. 2001). We also performed Northern analyses and obtained further confirmation of the Ig-μ mRNA levels (data not shown).
On the other hand, in contrast to the conclusions of Mühlemann et al. (2001), we observed no significant increase in the unspliced pre-mRNA levels in the RNA populations of PTC-containing mutants compared to wild type (Fig. 5E ▶). These results from RNase protection agreed well with our QRT-PCR results in that no significant accumulation of pre-mRNA was observed in the PTC-containing mutants relative to wild type (Fig. 3B ▶). Moreover, the relative levels of pre-mRNA quantified by RNase protection corresponded very well with the levels determined by QRT-PCR. The cell lines exhibited at most a threefold difference from the wild type, but the observed standard deviation makes that difference not significant.
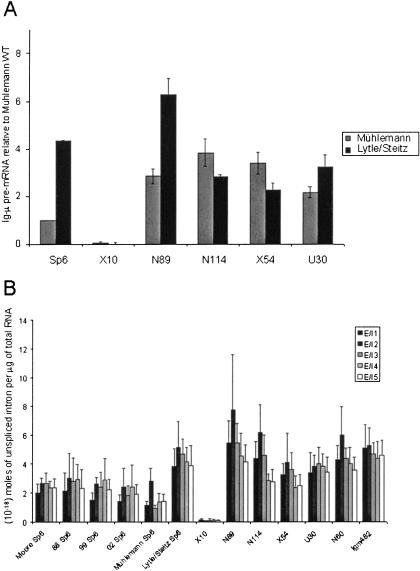
The contrast between our Ig-μ data, which do not show enhanced levels of PTC-containing versus wild-type pre-mRNA, and the published results of Mühlemann et al. (2001) was puzzling. To investigate this difference, RNA extracts from six of the Ig-μ cell lines were obtained directly from the laboratory of Dr. Oliver Mühlemann. QRT-PCR was performed on each of the extracts. The QRT-PCR result of the exon 4-intron 4 junction was arbitrarily chosen to represent the amount of pre-mRNA in each cell line, and the relative amount in each cell line versus the Mühlemann wild type was calculated (Fig. 6A ▶). The results obtained for these 12 RNA extracts (six from Mühlemann and six from Lytle/Steitz) were replicated by another method in the Mühlemann laboratory (O. Mühlemann, pers. comm.). Significantly, the Mühlemann Sp6 RNA extract had less than one-fourth the amount of pre-mRNA than the Lytle/Steitz Sp6 extract, causing all other cell lines to appear to have a significantly greater amount of pre-mRNA. Thus, the difference in pre-mRNA levels in the Sp6 strains can account for the contrasting conclusions of Mühlemann et al. (2001) versus ours, even though both of these Sp6 cell lines were obtained from the laboratory of Dr. Marc Shulman (Connor et al. 1993, 1994).
FIGURE 6.
Ig-μ RNA levels are different in different Sp6 cell lines. (A) The relative levels of Ig-μ pre-mRNA in RNA populations extracted from the wild-type Sp6 and mutant cell lines in the Lytle/Steitz lab or in the Mühlemann lab were quantified by QRT-PCR. The exon 4-intron 4 junction was arbitrarily chosen to represent the level of pre-mRNA. The level in the wild-type extract from Mühlemann was set at 1, and the relative levels in all other extracts are shown. Note that the Lytle/Steitz Sp6 levels are ~four times the Mühlemann Sp6 level. Both sets of experiments were repeated twice. The error bars represent the standard deviation (S.D.). (B) The exon-intron junction levels for Ig-μ cell lines grown in the Lytle/Steitz lab are reproduced (from Fig. 3B ▶) for comparison with the exon-intron junction levels in Sp6 cell lines obtained from the Moore or Shulman (88, 99, 02) labs and grown in the Lytle/Steitz lab; Mühlemann lab Sp6 RNA is also included. The results, determined by QRT-PCR, are shown in the graph. Determination of the exon-intron junction levels in the Lytle/Steitz Sp6 cells were repeated 8–10 times; those of the other Sp6 cells were repeated four times. The Mühlemann Sp6 RNA was assayed twice. The error bars represent the standard deviation (S.D.).
To test whether variation in isolates of cell lines or growth conditions might account for the differences in pre-mRNA levels, additional isolates of the same Sp6 cell line were examined. Three more Sp6 isolates were obtained from the Shulman lab: from vials frozen in 1988, 1999, and 2002 (88 Sp6, 99 Sp6, and 02 Sp6, respectively). Also, the Sp6 cells used by the laboratory of Dr. Melissa Moore were obtained. Total RNA was extracted from these four additional cell lines and assayed by QRT-PCR at least four times. The results are presented in Figure 6B ▶ along with the previously determined exon-intron boundary levels for the other cell lines (see Fig. 3B ▶). These Sp6 cell lines have ~twofold lower levels than the original Lytle/Steitz Sp6 cell line and ~twofold higher levels than the Mühlemann Sp6 extract RNA. We conclude that the calculated levels of PTC-containing Ig-μ pre-mRNAs depend on which wild-type Sp6 cell lines are used for comparison, but on average appear to be ~twofold enhanced.
DISCUSSION
We embarked on this study with the premise that a PTC might affect the rate or order of intron removal if premRNAs are indeed surveyed by nuclear translation. We analyzed two endogenous genes: the nonrearranging DHFR gene in Chinese hamster ovary cells and the somatically rearranging Ig-μ gene in mouse B cells. We monitored the rate of each intron’s removal relative to all other introns within these pre-mRNAs by QRT-PCR. Our finding that all of the exon-intron boundaries in the PTC-containing pre-mRNAs were of similar abundance to those in wild-type or missense pre-mRNAs allows us to conclude that introns neighboring exons that contain a PTC in either the DHFR or Ig-μ pre-mRNA are excised neither more slowly, nor more quickly, than introns in the wild-type transcripts. We also analyzed and found the rates of cleavage at polyadenylation sites in the Ig-μ transcripts to be unaffected by the presence of a PTC.
QRT-PCR is a sensitive technique that can be used to quantify even low-abundance RNAs within cellular populations. We provide both positive and negative controls to demonstrate the validity of our conclusions. The DHFR splice site mutants, DF40 and DP7B, did show alterations in the abundance of exon-intron junctions 3 and 2, respectively (Fig. 2 ▶), as expected from prior analyses (Carothers et al. 1993). In contrast, point mutations creating missense codons did not affect the abundance of exon-intron boundaries in the DHFR pre-mRNA population. The Ig-μ exon-intron 7 junction and the μm polyadenylation site were both detected at an order of magnitude lower abundance compared to the other Ig-μ amplified regions (Figs. 3 ▶, 4 ▶), as expected for the membrane-bound versus the secreted form of the mRNA.
Our QRT-PCR data (Figs. 2 ▶, 3 ▶) show that the splicing rates of the various introns within either the DHFR or Ig-μ pre-mRNA do not differ greatly. The less than twofold differences contrast with the up to sevenfold differences that can be detected for U12-type relative to U2-type intron removal from a single transcript (Patel et al. 2002). Whereas we find that intron 1 appears to be removed most quickly from the CHO DHFR pre-mRNA (Fig. 2B ▶), Kessler et al. (1993) suggested that intron 1 is the last to be removed. They performed RT-PCR on cellular RNA using forward primers annealing to the various introns and a reverse primer complementary to the last exon; because only one intron-containing PCR product was detected, they concluded that intron 1 was removed last. We repeated RT-PCR assays with similar primers and were able to detect multiple intron-containing PCR products (data not shown). Thus, we suspect that the sensitivity of the assays can explain this discrepancy with the conclusions of Kessler et al. (1993).
In addition to measuring the relative rates of RNA processing events occurring on a specific pre-mRNA, the QRT-PCR technique enabled us to assess the absolute amount of each exon-intron boundary inside cells. Compared to the wild-type control, we observed no difference in the levels of PTC-containing pre-mRNA for DHFR. This lack of effect of PTCs on pre-mRNA levels agrees with data published for other nonrearranging genes. When the amounts of nonsense or wild-type β-globin pre-mRNA in erythroid cells were measured by S1 nuclease mapping and RNA blotting, no difference was observed upon insertion of a PTC (Maquat et al. 1981; Lim et al. 1992). Similarly, in nonerythroid cells, Baserga and Benz (1992) found by S1 nuclease analysis that the level of nuclear PTC-containing β-globin RNA was indistinguishable from that of wild type, in accord with the conclusions of others (Humphries et al. 1984; Kugler et al. 1995). Cheng and Maquat (1993) demonstrated by RT-PCR that triosephosphate isomerase pre-mRNA levels likewise do not change upon insertion of a PTC. Gaspar et al. (1991) found by RT-PCR that the steady-state precursor RNA levels of androgen receptor do not vary with or without a PTC. Finally, Kessler and Chasin (1996) by RT-PCR observed no effect of PTCs on pre-mRNA levels for the APRT gene.
In the case of Ig-μ, our initial results likewise showed no significant difference in the levels of any of the PTC-containing pre-mRNAs relative to wild type. Our inability to reproduce the PTC-dependent accumulation of pre-mRNA reported by Mühlemann et al. (2001) we then found to be due to the fact that the wild-type cell lines used by Drs. Mühlemann and Moore versus those used in our laboratory differed in their levels of Ig-μ pre-mRNA. Because all other cell lines were normalized to wild type by Mühlemann et al. (2001), the enhancement of nonsense pre-mRNA levels was directly attributable to the lower wild-type levels they detected. When we assayed additional Sp6 cell lines, including theirs (Fig. 6B ▶), we observed that the levels of Ig-μ pre-mRNA vary from cell line to cell line. Perhaps the environment of the cells in transit, the culture conditions, or the passage number of the cells affects the growth and pre-mRNA levels of these B-cell hybridomas. This natural variability makes definitive conclusions about pre-mRNA accumulation difficult. On average, we observed an approximate twofold increase in pre-mRNA levels in the PTC-containing cell lines relative to the wild type, not as dramatic as the fivefold difference in pre-mRNA levels reported by Mühlemann et al. (2001).
The only genes that previously have been reported to show an increase in PTC-containing pre-mRNA and intron levels are a viral gene from minute virus of mice (Gersappe et al. 1999; Gersappe and Pintel 1999) and the somatically rearranging and hypermutating genes, Ig-μ and TCR-β (Mühlemann et al. 2001). The latter two genes undergo programmed DNA rearrangements to juxtapose the variable and constant regions, and approximately two-thirds of these recombination events lead to the formation of a PTC (Kronenberg et al. 1986; Fang et al. 1996). It has therefore been proposed that these cells have developed special mechanisms leading to nonsense pre-mRNA accumulation for these hypermutating genes. The reported accumulation of TCR-β pre-mRNA for stably and transiently transfected PTC-containing TCR-β mini-genes was not as dramatic as for the Ig-μ pre-mRNA (Mühlemann et al. 2001), more similar to our results for Ig-μ. Perhaps the integration of these genes at nonnative loci in the HeLa genome (or lack of integration in the case of transiently transfected constructs) affects the fate of their pre-mRNAs.
In summary, our finding that the presence of a PTC does not affect the rates of two pre-mRNA processing events, splicing and cleavage/polyadenylation, could be taken as evidence against the nuclear surveillance of pre-mRNA by translating ribosomes. On the other hand, it is possible that the progression of ribosomes along a nuclear transcript, even if it occurs, simply does not detectably alter the rates of these processes. Transcription rate, in contrast, has been observed to affect alternative splicing (de la Mata et al. 2003), and the accumulation of PTC-containing transcripts at sites of transcription could be due to altered nuclear trafficking rather than a perturbation of RNA processing events. One wonders whether the several Ig-μ wild-type cell isolates examined here would all exhibit lack of pre-mRNA accumulation at the site of transcription relative to PTC-containing Ig-μ mutants if examined under the microscope. In addition, it will be interesting to learn whether the phenomenon of PTC-induced pre-mRNA accumulation at the site of transcription is specific to rearranging genes, or can be seen for endogenous nonrearranging genes, such as DHFR, as well.
MATERIALS AND METHODS
Preparation of RNA standards
To create transcription templates for RNA standards for the DHFR analyses, DNA segments containing 600–750 bases flanking each of the five exon-intron junctions were PCR-amplified from either the plasmids pMB5, pB6-14, and pB6-1 (gift from Dr. Lawrence Chasin; Carothers et al. 1983) or from genomic DNA extracted by Trizol (Invitrogen). To create templates for the Ig-μ analyses, DNA segments flanking exon-intron junctions 1–5 and 7 and the poly(A) sites for secreted and membranous Ig-μ were PCR-amplified from the plasmid pRSp6 (gift from Dr. Marc Shulman, containing the Ig-μ genomic sequence inserted into pSV2neo). XhoI and NotI restriction sites, introduced by the PCR primers, were used to insert the amplified segments into pBluescript SK+ vectors (Stratagene) downstream of a T7 promoter. Ten-nucleotide deletions were introduced either by PCR mutagenesis or by using the QuikChange Mutagenesis system (Stratagene). RNA standards were prepared by in vitro transcription from the T7 promoter essentially as described by Tarn and Steitz (1996), internally trace-labeling with [α-32P] UTP. Gel-purified transcripts were quantified by liquid scintillation counting and stored at −80°C in aqueous solution containing 20 μg/mL yeast total carrier RNA.
Quantitative RT-PCR analyses
Total RNAs for DHFR or Ig-μ analyses were prepared using Trizol (Invitrogen) according to the manufacturer’s directions. The CHO cells were grown to 70%–90% confluence in F12 medium (Gibco-BRL) supplemented with 5% fetal calf serum (Gemini BioProducts) and 30 μM thymidine (Sigma). The mouse B-cell hybridomas were grown in suspension in Dulbecco’s modified Eagle’s medium (Gibco-BRL) supplemented with 12% newborn calf serum (Gemini BioProducts), 110 mg/L sodium pyruvate (Gibco-BRL), and 10−5 M β-mercaptoethanol (Gibco-BRL). Contaminating genomic DNA was removed by treating with RQ1 DNase (Promega) in 5 mM MgCl2 and 50 mM Tris pH 8.0 at 37°C for 1 h, followed by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. Reverse transcription was then performed using Thermoscript RT (Invitrogen) according to the manufacturer’s directions, with 0.83 μM gene-specific RT primers and 200 ng total cellular RNA, and various concentrations (from 8.3 × 10−22 moles/μL for DHFR exon-intron boundaries 1–5, 8.3× 10−20 for Ig-μ introns 1–5, 8.3 × 10−21 for Ig-μ intron 7, 3.3 × 10−20 for Ig-μ secreted poly(A) site, and 4.2 × 10−21 for Ig-μ membranous poly(A) site) of in vitro-transcribed RNA standards. RT reactions (10 μL) were incubated at 37°C for 15 min, then 55°C for 30 min, then 65°C for 30 min, and finally 85°C for 10 min. Escherichia coli RNase H (0.1 U/μL; Invitrogen) was added and the samples incubated at 37°C for 30 min. PCR was then performed using Platinum Taq polymerase (Invitrogen) in the buffer supplied adding 1.5 mM MgCl2, 0.4 mM dNTPs, 200 nM unlabeled primers doped with 5′-radiolabeled forward primer (to label only one strand of the amplicon), and 7.5% (by volume) of reverse-transcribed cDNA. The PCR primers were complementary to sequences equidistant from and flanking each exon-intron boundary to yield PCR products of ~150 bp. The RT primer was specific for each exon-intron junction and contained the first 16 bases of the reverse PCR primer designed for that junction. All primer sequences are available upon request. Between 21 and 27 cycles of PCR were performed with 2-min extension times and annealing temperatures of 64°C for all samples (based on lowest predicted Tm). A PCR reaction with no cDNA was performed as a negative control for each experiment. Amplicons were separated on a 10% polyacrylamide sequencing gel and quantitated using a Molecular Dynamics PhosphorImager. The control ratios for each QRT-PCR reaction were generally close to 1:1. If the ratios differed by more than 2.5-fold, then the control RNA transcripts were discarded and remade. For DHFR, the average control ratio for the exon 1-intron 1 junction (E/I1) was 0.83, for E/I2 0.81, for E/I3 0.90, for E/I4 0.96, and for E/I5 0.97. For Ig-μ, the average control ratio for E/I1 was 1.09, for E/I2 0.84, for E/I3 0.84, for E/I4 0.91, for E/I5 0.74, and for the μs polyadenylation site 0.99. For the Ig-μ controls used at 10−21 moles/μL, the ratios were more difficult to maintain at 1:1. The average control ratio for Ig-μ E/I7 was 0.45 and for the μm polyadenylation site was 1.71.
RNase protection assays
The RNase protection protocol was adapted from several published earlier (Dumas et al. 1996; Lindsey and Wilkinson 1996; Mühlemann et al. 2001). The probes for the Ig-μ exon 1-intron 1 (RP1) and exon 2-intron 2 junctions (RP2) were PCR-amplified from the pRSp6 plasmid described above. The subclones were inserted into the bluescript plasmid, pBSCSK+ (Stratagene) and used as templates for transcription. RP1 was 158 nt containing 29 bases of nonhybridizing plasmid sequence, 48 bases of antisense to exon 1, and 81 bases of antisense to intron 1. RP2 was 484 nt containing 29 bases of nonhybridizing plasmid sequence, 290 bases of antisense to exon 2, and 165 bases of antisense to intron 2. The β-actin cDNA was cloned from RNA extracted from the Sp6 cell line by Trizol (Invitrogen) and placed behind a T3 promoter in the bluescript plasmid, pBSCSK+ (Stratagene). Two different β-actin probes were necessary in order to avoid protecting regions similar in size to those protected by either RP1 or RP2 for Ig-μ. The β-actin plasmid was cleaved with XhoI or XbaI to create templates for transcription that produce RNAs of 306 and 178 nt, respectively. The RNAs were transcribed at the following specific activities: RP1 at 11,100 cpm/fmole, RP2 at 17,100 cpm/fmole, β-actin XhoI at 129 cpm/fmole, and β-actin XbaI at 412 cpm/fmole. Twelve fmole of RP1 or RP2 and 10 fmole of β-actin XbaI or 5 fmole of β-actin XhoI were added to tubes alone (undigested controls) or to tubes containing 10 μg of extract RNA. Samples were dried and resuspended in 7.5 μL of hybridization buffer (40 mM PIPES, 400 mM NaCl, 1 mM EDTA, and 80% formamide). The samples were heated to 85°C for 5 min and incubated at 65°C overnight. The next day, 87.5 μL of digestion buffer [10 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM EDTA, 4.66 μg pancreatic RNase A, and 1090 Units of RNase T1 (Ambion)] was added to the samples, which were heated at 30°C for 30 min. The undigested controls were resuspended in the same buffer without RNase and treated the same as the RNased samples. Then, 10 μL of 20% SDS and 2.5 μL of proteinase K (20 mg/mL) were added. The samples were heated at 37°C for 15 min, phenol/chloroform/isoamyl alcohol-extracted, and ethanol-precipitated. The samples were then resuspended in formamide loading buffer and electrophoresed on a 10% denaturing polyacrylamide gel.
Acknowledgments
We thank Dr. Lawrence Chasin (Columbia Univ.) for his gift of CHO cell lines containing wild-type, mutant, and revertant DHFR and additional plasmids; Dr. Marc Shulman (Univ. of Toronto) for several deliveries of mouse hybridoma cell lines containing wild-type, deletion, and mutant Ig-μ and additional plasmids; Dr. Melissa Moore (Brandeis Univ.) for N89 and Sp6 cell lines; Dr. Oliver Mühlemann (Univ. of Bern) for his RNA extracts of several of the Ig-μ cell lines; and Rachel Mitten-Fry and Voula Mili for critical evaluation of this manuscript. This work was supported by a Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship (F32 GM67478-01) and a grant from the NIH (GM26154). J.A.S. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5241404.
REFERENCES
- Aoufouchi, S., Yelamos, J., and Milstein, C. 1996. Nonsense mutations inhibit RNA splicing in a cell-free system: Recognition of mutant codon is independent of protein synthesis. Cell 85: 415–422. [DOI] [PubMed] [Google Scholar]
- Arts, G.J., Kuersten, S., Romby, P., Ehresmann, B., and Mattaj, I. W. 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17: 7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga, S.J. and Benz Jr., E.J. 1992. Beta-globin nonsense mutation: Deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc. Natl. Acad. Sci. 89: 2935–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader, P. and Maquat, L.E. 1994. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol. Cell. Biol. 14: 6326–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader, P., Cheng, J., Zhou, X., Stephenson, L.S., and Maquat, L.E. 1994. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol. Cell. Biol. 14: 8219–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack, M.T., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E., and Gorlich, D. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21: 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna, S. 1999. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5: 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler, M., Wilkinson, M.F., and Mühlemann, O. 2002. Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep. 3: 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Kendzior Jr., R.J., and Beemon, K.L. 2002. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes & Dev. 16: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers, A.M., Urlaub, G., Ellis, N., Chasin, L.A. 1983. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 11: 1997–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers, A.M., Urlaub, G., Grunberger, D., and Chasin, L.A. 1993. Splicing mutants and their second site suppressors at the dihydrofolate reductase locus in chinese hamster ovary cells. Mol. Cell. Biol. 13: 5085–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding the nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Carter, M.S., Li, S., and Wilkinson, M.F. 1996. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 15: 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. and Maquat, L.E. 1993. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol. Cell. Biol. 13: 1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor, A., Collins, C., Jiang, L., McMaster, M., and Shulman, M.J. 1993. Isolation of new nonsense and frameshift mutants in the immunoglobulin μ heavy-chain of hybridoma cells. Som. Cell. Mol. Genet. 19: 313–320. [DOI] [PubMed] [Google Scholar]
- Connor, A., Wiersma, E., and Shulman, M.J. 1994. On the linkage between RNA processing and RNA translatability. J. Biol. Chem. 269: 25178–25184. [PubMed] [Google Scholar]
- Dahlberg, J.E., Lund, E., and Goodwin, E.B. 2003. Nuclear translation: What is the evidence? RNA 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata, M., Alonso, C.R., Kadener, S., Fededa, J.P., Blaustein, M., Pelisch, F., Cramer, P., Bentley, D., and Kornblihtt, A.R. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- Dietz, H.C. and Kendzior Jr., R.J. 1994. Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat. Genet. 8: 183–188. [DOI] [PubMed] [Google Scholar]
- Dietz, H.C., Valle, D., Francomano, C.A., Kendzior Jr., R.J., Pyertiz, R.E., and Cutting, G.R. 1993. The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259: 680–683. [DOI] [PubMed] [Google Scholar]
- Dumas, S., Le Hir, H., Bodeau-Pean, S., Hirsch, E., Thermes, C., and Mallet, J. 1996. New species of human tyrosine hydroxylase mRNA are produced in variable amounts in adrenal medulla and are overexpressed in progressive supranuclear palsy. J. Neurochem. 67: 19–25. [DOI] [PubMed] [Google Scholar]
- Fang, W., Mueller, D.L., Pennell, C.A., Rivard, J.J., Li, Y.S., Hardy, R.R., Schlissel, M.S., and Behrens, T.W. 1996. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity 4: 291–299. [DOI] [PubMed] [Google Scholar]
- Gaspar, M.L., Meo, T., Bourgarel, P., Guenet, J.L., and Tosi, M. 1991. A single base deletion in the Tfm androgen receptor gene creates a short-lived messenger RNA that directs internal translation initiation. Proc. Natl. Acad. Sci. 88: 8606–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersappe, A. and Pintel, D.J. 1999. A premature termination codon interferes with the nuclear function of an exon splicing enhancer in an open reading frame-dependent manner. Mol. Cell. Biol. 19: 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersappe, A., Burger, L., and Pintel, D.J. 1999. A premature termination codon in either exon of minute virus of mice P4 promoter-generated pre-mRNA can inhibit nuclear splicing of the intervening intron in an open reading frame-dependent manner. J. Biol. Chem. 274: 22452–22458. [DOI] [PubMed] [Google Scholar]
- Hentze, M.W. 2001. Protein synthesis: Believe it or not-translation in the nucleus. Science 293: 1058–1059. [DOI] [PubMed] [Google Scholar]
- Humphries, R.K., Ley, T.J., Anagnou, N.P., Baur, A.W., Neinhuis, A.W. 1984. Beta0–39 thalassemia gene: A premature termination codon causes β-mRNA deficiency without affecting cytoplasmic β-mRNA stability. Blood 64: 23–32 [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293: 1139–1142. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Kessler, O. and Chasin, L.A. 1996. Effects of nonsense mutations on nuclear and cytoplasmic adenine phophoribosyltransferase RNA. Mol. Cell. Biol. 16: 4426–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, O., Jiang, Y., and Chasin, L.A. 1993. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol. 13: 6211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg, M., Siu, G., Hood, L.E., and Shastri, N. 1986. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Ann. Rev. Immunol. 4: 529–591. [DOI] [PubMed] [Google Scholar]
- Kugler, W., Enssle, J., Hentze, M.W., and Kulozik, A.E. 1995. Nuclear degradation of nonsense mutated β-globin mRNA: A post-transcriptional mechanism to protect heterozygotes from severe clinical manifestations of β-thalassemia? Nucleic Acids Res. 23: 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz, F., Goyer, C., Darveau, A., Neron, S., Lemieux, R., and Sonenberg, N. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. 89: 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Leonard, D., and Wilkinson, M.F. 1997. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: A nuclear-associated translation-like mechanism. J. Exp. Med. 185: 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Wachtel, C., Miriami, E., Yahalom, G., Friedlander, G., Sharon, G., Sperling, R., and Sperling, J. 2002. Stop codons affect 5′ splice site selection by surveillance of splicing. Proc. Natl. Acad. Sci. 99: 5277–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.K., Sigmund, C.D., Gross, K.W., and Maquat L.E. 1992. Nonsense codons in human β-globin mRNA result in the production of mRNA degradation products. Mol. Cell. Biol. 12: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey, J.S. and Wilkinson, M.F. 1996. Pem: A testosterone- and LH-regulated homeobox gene expressed in mouse Sertolli cells and epididymis. Dev. Biol. 179: 471–484. [DOI] [PubMed] [Google Scholar]
- Lozano, F., Maertzdorf, B., Pannell, R., and Milstein, C. 1994. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 13: 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E. and Dahlberg, J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Mangiarotti, G. 1999. Coupling of transcription and translation in Dictyostelium discoideum nuclei. Biochemistry 38: 3996–4000. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 1995. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1: 453–465. [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. 2002. Nonsense-mediated mRNA decay. Curr. Biol. 12: R196–197. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. and Li, X. 2001. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA 7: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E., Kinniburgh, A.J., Rachmilewitz, E.A., and Ross, J. 1981. Unstable β-globin mRNA in mRNA-deficient β0 thalassemia. Cell 27: 543–553. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T. and Dietz, H.C. 2001. When the message goes awry: Disease-producing mutations that influence mRNA content and performance. Cell 107: 411–414. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., ap Rhys, C.M.J., and Dietz, H.C. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422. [DOI] [PubMed] [Google Scholar]
- Mühlemann, O., Mock-Casagrande, C.S., Wang, J., Shulin, L., Custódio, N., Carmo-Fonseca, M., Wilkinson, M.F., and Moore, M.J. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8: 33–44. [DOI] [PubMed] [Google Scholar]
- Naeger, L.K., Schoborg, R.V., Zhao, Q., Tullis, G.E., and Pintel, D.J. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes & Dev. 6: 1107–1119. [DOI] [PubMed] [Google Scholar]
- Nathanson, L., Xia, T., and Deutscher, M.P. 2003. Nuclear protein synthesis: A re-evaluation. RNA 9: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A., McCarthy, M., and Steitz, J.A. 2002. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 21: 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. and Politz, J.C. 2000. The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 148: 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, S.B. and Stoltzfus, C.M. 1994. Frameshift mutations in the v-src gene of avian sarcoma virus act in cis to specifically reduce v-src mRNA levels. Mol. Cell. Biol. 14: 1835–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn, W.Y. and Steitz, J.A. 1996. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84: 801–811. [DOI] [PubMed] [Google Scholar]
- Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., Hentze, M.W., and Kulozik, A.E. 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 17: 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub, G., Mitchell, P.J., Ciudad, C.J., and Chasin, L.A. 1989. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol. Cell. Biol. 9: 2868–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Chang, Y-.F., Hamilton, J.I., and Wilkinson, M.F. 2002a. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hamilton, J.I., Carter, M.S., Li, S., and Wilkinson, M.F. 2002b. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science 297: 108–110. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Sun, X., Qian, Y., LaDuca, J.P., and Maquat, L.E. 1998a. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: A possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 18: 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Sun, X., Qian, Y., and Maquat, L.E. 1998b. Intron function in the nonsense-mediated decay of beta-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4: 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]