Abstract
The dicistronic assay for internal ribosome entry site (IRES) activity is the most widely used method for testing putative sequences that may drive cap-independent translation initiation. This assay typically involves the transfection of cells with dicistronic DNA test constructs. Many of the reports describing eukaryotic IRES elements have been criticized for the use of inadequate methods for the detection of aberrant RNAs that may form in transfected cells using this assay. Here we propose the combined use of a new RNAi-based method together with RT-PCR to effectively identify aberrant RNAs. We illustrate the use of these methods for analysis of RNAs generated in cells transfected with dicistronic test DNAs containing either the hepatitis C virus (HCV) IRES or the X-linked inhibitor of apoptosis (XIAP) cellular IRES. Both analyses indicated aberrantly spliced transcripts occurred in cells transfected with the XIAP dicistronic DNA construct. This contributed to the unusually high levels of apparent IRES activity exhibited by the XIAP 5′ UTR in vivo. Cells transfected directly with dicistronic RNA exhibited much lower levels of XIAP IRES activity, resembling the lower levels observed after translation of dicistronic RNA in rabbit reticulocyte lysates. No aberrantly spliced transcripts could be detected following direct RNA transfection of cells. Interestingly, transfection of dicistronic DNA or RNA containing the HCV IRES did not form aberrantly spliced transcripts. These observations stress the importance of using alternative test procedures (e.g., direct RNA transfection) in conjunction with a combination of sensitive RNA analyses for discerning IRES-containing sequences in eukaryotic mRNAs.
Keywords: eukaryotic translation, internal ribosome entry, RNA splicing, XIAP
INTRODUCTION
The initiation of translation of most cellular mRNAs is believed to occur via a cap-dependent ribosome scanning mechanism in which eIF4F (eIF4G, eIF4E, and eIF4A) initially binds the m7G cap of mRNA via the eIF4E component (Kozak 1999). eIF4F then serves to recruit the 43S pre-initiation complex (40S ribosomal subunit, eIF3, eIF1A, and eIF2-GTP-Met-tRNAi), which scans from the 5′ end of the mRNA until the Met-tRNAi interacts with the initiator AUG codon. Translation proceeds following the joining of the pre-initiation complex with the 60S ribosomal subunit and elongation factors and with the dissociation of some initiation factors (for review, see Pestova et al. 2001). Although scanning proves a valid mechanism for the initiation of translation of most cellular mRNAs, it may be complicated when mRNAs possess unusually long 5′ UTRs (Falcone and Andrews 1991) that form thermodynamically stable, GC-rich, secondary structures and/or contain upstream AUG codons or ORFs. Such features have been demonstrated to be inhibitory to canonical ribosome scanning (Geballe and Morris 1994; Jackson and Kaminski 1995).
Internal ribosome entry site (IRES) elements were first demonstrated within the 5′ UTRs of poliovirus (Pelletier and Sonenberg 1988) and encephalomyocarditis virus (EMCV; Jang et al. 1988; Chen and Sarnow 1995) viral RNAs. IRES-mediated translation has since been identified as a common feature of Picornaviridae, Flaviviridae (Pestivirus, hepatitis C virus [HCV]), and Retroviridae (Lentivirus) RNAs (Gale et al. 2000; Buck et al. 2001; Hellen and Sarnow 2001). Translation studies in vitro and in vivo demonstrated that the poliovirus UTR could mediate cap-independent translation through internal ribosome entry following virus-mediated cleavage of eIF4G (Hambidge and Sarnow 1992) by recruitment of some of the canonical eIFs (for review, see Ehrenfeld 1996). In addition, it was demonstrated that several host-cell proteins, including La (Meerovitch et al. 1993), PTB (Hellen et al. 1994), and PCBP2 (Blyn et al. 1997), could stimulate IRES-mediated translation of the poliovirus RNA. Such factors have been referred to as IRES-transactivating factors (ITAFs; Stoneley et al. 2000).
Initiation of cap-independent translation of a eukaryotic mRNA was first reported over a decade ago for the immunoglobulin heavy-chain binding protein, BiP (Macejak and Sarnow 1991). The number of mammalian mRNAs reported to contain IRES elements has expanded and includes transcripts capable of mediating internal initiation during the inhibition of cap-dependent translation that occurs during mitosis and apoptosis. These include mRNAs encoding vascular endothelial growth factor (VEGF; Stein et al. 1998), c-myc (Stoneley et al. 1998), apoptosis activating factor 1 (Apaf-1; Coldwell et al. 2000), fibroblast growth factor (FGF-2; Vagner et al. 1995), cellular-inhibitor of apoptosis 1 (c-IAP1; Van Eden et al. 2004), and X-linked inhibitor of apoptosis (XIAP; Holcik et al. 1999). The mechanisms by which these reported IRES elements function are not yet completely understood but may involve the participation of certain RNA structural motifs as well as cellular ITAFs (Holcik and Korneluk 2000; Mitchell et al. 2001; Holcik et al. 2003).
Recently, the concept of IRES-mediated translation initiation in eukaryotes was challenged on the basis of the methods typically used for IRES identification (Kozak 2001, 2003). One commonly used method is the dicistronic test for IRES activity that relies on an IRES directing the translation of a downstream ORF when positioned between two cistrons in a dicistronic mRNA. However, there is the possibility that cells transfected with dicistronic DNA test constructs may give rise to transcripts that are aberrantly processed either by the presence of a cryptic promoter or by RNA splicing. This may lead to a misinterpretation of the IRES activity for a given test sequence. On this basis, many reports describing eukaryotic IRESs were criticized for using inadequate methods (e.g., Northern blotting) for the detection of such aberrant transcripts (Kozak 2001, 2003). Here we demonstrate the combined use of RNAi and RT-PCR as alternative highly sensitive methods for the assessment of transcript integrity in cells transfected with dicistronic test DNAs containing either HCV or XIAP IRES elements. These assays readily detected aberrantly spliced transcripts in cells transfected with the XIAP dicistronic construct but not with the HCV dicstronic construct. We found that aberrant splicing contributed to the unusually high levels of apparent IRES activity exhibited by the XIAP 5′ UTR in vivo. However, the XIAP 5′ UTR exhibited low IRES activity when RNAs were translated in rabbit reticulocyte lysates or when cells were transfected directly with dicistronic RNAs. These observations stress the importance of using alternative test procedures (e.g., direct RNA transfection) and sensitive RNA analyses for the identification/characterization of putative IRES elements in eukaryotic mRNAs.
RESULTS AND DISCUSSION
IRES-mediated translation in vitro and in transfected cells
The dicistronic assay for IRES activity is the most commonly used test for cap-independent translation initiation in eukaryotic mRNAs. However, this test has been criticized on the basis that even low amounts of alternative mRNA transcripts may not be dicistronic, which may result in a misinterpretation of IRES activity for a given test sequence. To better address these criticisms, we demonstrate here the use of two highly sensitive methods to assess the integrity and identity of RNAs produced in cells transfected with dicistronic DNA test vectors. We propose these methods should be used in conjunction with previously reported methods (i.e., promoter-less test constructs, Northern blotting) to effectively ascertain the presence and identity of aberrant RNAs should they form via cryptic promoters and/or RNA splicing.
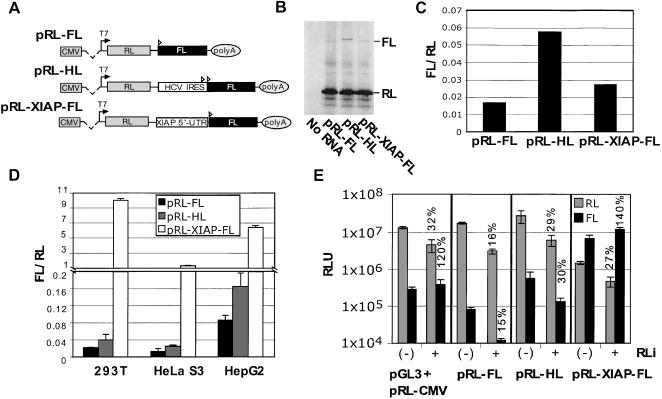
To illustrate the use of these methods, we first assayed cap-independent translation initiation as mediated by HCV and XIAP IRES elements. Dicistronic DNA constructs contained an upstream Renilla luciferase (RL) cistron, an intercistronic region, and a downstream firefly luciferase (FL) cistron. Translation of RL from the dicistronic RNA occurs via cap-dependent scanning, whereas translation of FL depends on the presence of an IRES in the intercistronic region. These intercistronic regions consisted of either a 27-nt spacer in pRL-FL, a 493-nt insert containing the HCV IRES in pRL-HL, or a 495-nt sequence containing the reported XIAP IRES (Holcik et al. 1999) in pRL-XIAP-FL (Fig. 1A ▶). Figure 1B ▶ shows an autoradiogram of proteins translated in RRL from capped dicistronic RNAs. Both HCV- and XIAP-derived sequences could mediate weak translation of the downstream cistron at levels higher than that of the control (i.e., pRL-FL). Ratios of FL-to-RL activity were calculated to correct for variations in RNA levels and were quantitated from the autoradiogram by densitometry (Fig. 1C ▶). These data were corroborated by luciferase assays (data not shown) and were consistent with previous reports that demonstrated the HCV IRES and the XIAP 5′ UTR could support cap-independent translation initiation in vitro (Tsukiyama-Kohara et al. 1992; Holcik and Korneluk 2000). Studies of this kind, although suggestive of IRES activity, also need to be performed in transfected cells because it is well known that translation in RRL is not strictly cap dependent and/or the lysate may lack ITAFs required by some IRESs (Meerovitch et al. 1993). Therefore, a putative IRES element is typically tested in vivo by transiently transfecting cell monolayers with dicistronic DNA constructs. Accordingly, the dicistronic DNAs in Figure 1A ▶ were transfected into 293T, HepG2, and HeLa S3 cells. Twenty hours following transfection, cells were lysed and the RL and FL activities in the lysates determined. Figure 1D ▶ shows the relative IRES activity (FL/RL) for each of the test constructs to be significantly greater than that of the control and to vary with each cell type. In contrast to the data in Figure 1C ▶, the apparent IRES activity for pRL-XIAP-FL was much higher in cells (~250-fold greater than that of pRL-HL in the lysates of 293T cells) and was consistently 20- to 50-fold more active than any other viral or cellular IRES we have tested (data not shown). In contrast to pRL-XIAP-FL, the FL/RL ratios of pRL-FL and pRL-HL were similar in both transfected cells and RRLs. Interestingly, RL activities in the lysates of cells transfected with pRL-XIAP-FL were consistently repressed 90%–96% compared with cells transfected with pRL-FL and pRL-HL, despite transfection of equivalent amounts of DNA (Fig. 1E ▶). This suggested that insertion of the XIAP 5′ UTR negatively affected expression of the upstream RL cistron either by competing for components of the translation apparatus or by compromising the integrity of the dicistronic RNA.
FIGURE 1.
The XIAP 5′ UTR exhibits activity inconsistent with IRES-mediated translation from an intact dicistronic RNA template. (A) A schematic of the DNA constructs used for this study. The locations of CMV and T7 promoters are indicated. The dashed line denotes the position of a chimeric intron normally removed by RNA splicing. Arrowheads are translation initiation codons for the FL ORF, and Poly-A is the polyadenylation signal from the bovine growth hormone gene. (B) Capped RNAs were transcribed from the indicated DNA constructs as detailed in the Materials and Methods. Equal amounts of RNA were used to program RRL supplemented with [35S]-Met/Cys. Radiolabeled protein was detected by SDS-PAGE/autoradiography. The mobilities of RL and FL proteins are indicated on the right. (C) The autoradiogram reproduced in B was scanned to determine the intensity of RL and FL bands, and the FL/RL ratios (i.e., FL normalized to RL) were graphed. (D) pRL-FL, pRL-HL, or pRL-XIAP-FL was transfected into the indicated cell types in quadruplicate. Twenty hours post-transfection, cells were harvested and cell lysates assayed for FL and RL. FL activity was normalized to RL and plotted as the mean ± SD. (E) Indicated DNA plasmids were cotransfected with pSV-βGal and either pBS-U6RLi, an siRNA expression vector targeting RL (+), or pBS-U6ApaI, the cognate vector control (−). 293T cell lysates were assayed for FL and RL activity 48 h post-transfection, and data were normalized to β-Gal activity (see Van Eden et al. 2004) as a control for transfection efficiency. Data are plotted on a logarithmic scale, and in each case the percentage of remaining RL or FL activity, following RL knock-down, is indicted where appropriate. Data are plotted as the mean ± SD from a single experiment performed in triplicate and were reproducible.
We reasoned the unusual behavior of XIAP-containing constructs may have resulted from cellular cap-dependent translation of “altered” transcripts that were functionally monocistronic and contained only the FL ORF. Such transcripts could arise from the presence of a cryptic promoter within the 5′ UTR of XIAP and/or by aberrant mRNA splicing. The latter has been suggested on the basis of the XIAP sequence containing a possible splice-junction motif (Yn closely followed by AG) near the 3′ end of its reported IRES element (Kozak 2003).
We developed a RNAi-based method to analyze the integrity of dicistronic mRNAs transcribed in cells transfected with dicistronic DNA test constructs. For these experiments, DNA constructs were cotransfected with a plasmid encoding a short RNA hairpin transcribed via the murine U6 promoter (pBS-U6RLi; Sui et al. 2002). In turn, this siRNA would activate the RNA induced silencing complex (for review, see Hannon 2002), resulting in the complete degradation of RNAs containing the RL sequence and a corresponding inhibition of RL protein synthesis in the cell. If RL and FL are translated exclusively from an intact dicistronic mRNA, both RL and FL activities should be proportionally reduced following a RL-directed RNAi response. However, if a message is functionally monocistronic, encoding only FL, then FL activity should be unaffected by the presence of RL siRNA. The results of this experiment conducted in 293T cells transfected with dicistronic DNA test constructs are shown in Figure 1E ▶. In each case, the parent plasmid (pBS-U6ApaI) was separately cotransfected as an empty vector control. As expected, RL siRNA significantly reduced RL but not FL activity in cells cotransfected with individual monocistronic FL (pGL3) and RL (pRL-CMV) reporter constructs (cf. pGL3/pCMV-RL + and [−]). Conversely, RNAi decreased RL and FL activities equivalently in cells transfected with either pRL-FL or pRL-HL. This indicated that these dicistronic RNAs were intact. However, in the case of pRL-XIAP-FL, the RL and FL activities were not repressed equivalently following RNAi. FL activity actually increased despite a significant knockdown of RL activity. This indicated that some amount of the mRNA transcribed from pRL-XIAP-FL lacked the upstream RL ORF. These results were reproduced in HepG2 cells (data not shown). Intriguingly, the RL activity produced from pRL-XIAP-FL was much lower than was the activity from any other dicistronic vector, even after RNAi knockdown. These results indicate that alternate transcripts exist in cells transfected with pRL-XIAP-FL that repress RL expression and unlink RL and FL expression. Although this assay was used to indicate the presence of altered transcripts, it does not elucidate the mechanism of their formation (i.e., via a cryptic promoter and/or unexpected RNA splicing).
Analysis of cryptic promoters and RNA splicing
Production of aberrant RNAs in cells transfected with dicistronic DNA constructs has been previously reported (Han and Zhang 2002). In this case, the intercistronic sequence encoded a “cryptic promoter” that resulted in transcription of a mRNA that contained only the downstream ORF. To test for the presence of cryptic promoters within the XIAP 5′ UTR sequence, we removed the CMV promoter from pRL-XIAP-FL and compared FL levels in cells transfected with parental and deletion constructs. In this test, the XIAP sequence contained no detectable promoter activity (data not shown), consistent with previous reports (Holcik et al. 1999). However, this assay cannot be used to identify alternatively spliced RNAs in cells transfected with dicistronic DNAs.
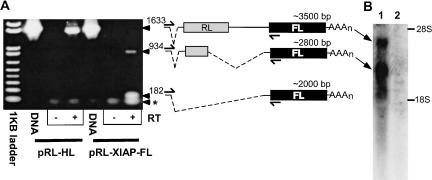
Thus, to detect possible splice variants, total RNA from transfected 293T cells was subjected to RT-PCR as described in the Materials and Methods. PCR products were separated in a 1% (w/v) agarose gel and visualized by ethidium bromide (EtBr) staining (Fig. 2A ▶). Transient transfection with pRL-HL resulted in a single PCR product (1715 base pairs [bp]) indicative of the full-length dicistronic mRNA transcript having spliced-out the upstream chimeric intron. This was verified by sequencing the PCR product. However, transfection with pRL-XIAP-FL resulted in the production of some full-length transcript in addition to abundant amounts of smaller, alternative transcripts. A PCR product (1633 bp) corresponding to full-length transcript (~3500 bp) was visible only upon overexposure of the gel. Smaller PCR products (182 bp and 934 bp) were isolated and sequenced, and their corresponding transcripts are indicated as schematics in the figure. Similar RT-PCR results were obtained following transfection of HepG2 cells (data not shown). The splice-acceptor site within the XIAP-derived sequence was shared in these smaller transcripts (i.e., 5′•••AGÅAAGGUG•••3′). Interestingly, RNA splicing at this position likely functions in the maturation of endogenous XIAP mRNAs. Searches of an EST database indicated multiple XIAP mRNAs exist in which this splice-junction is conserved, despite varying upstream sequences. The location of this splice-junction was conserved in human, mouse, and rat ESTs (e.g., human: BQ423165, AL713196; mouse: U88990, U36842; rat: AB033366, AF304334).
FIGURE 2.
The XIAP 5′ UTR encodes a RNA splice-acceptor site. (A) 293T cells were transfected with either pRL-HL or pRL-XIAP-FL, and RNA was harvested 24 h post-transfection. Total RNA was subjected to RT-PCR, separated in a 1% (w/v) agarose/EtBr gel, and visualized by UV illumination. PCR products from the HCV and XIAP DNA constructs were included as size controls (DNA). (+) and (−) indicate the inclusion or exclusion of RT in the reaction mixtures, respectively. PCR products were excised from the gel for sequencing and identification. The identities and sizes (bp) of each PCR product and their corresponding mRNA are diagrammed on the right. 5′ and 3′ PCR primers are indicated by split-arrows above and below each diagram, respectively. Dashed lines indicate the positions of RNA splice junctions. The asterisk denotes nonelongated oligonucleotide primers. (B) Northern blot of RNA from 293T cells transfected with pRL-XIAP-FL DNA (lane 1) or a control plasmid (lane 2). Blotting was performed as detailed in Materials and Methods. As a loading control, 18S and 28S rRNA bands were visualized by methylene blue staining of the nitrocellulose blot and shown to be equivalent (data not shown).
The 182-bp band corresponded to a singly spliced monocistronic mRNA that possessed a short 5′ UTR followed by the FL ORF. The 934-bp band corresponded to a transcript that had been spliced twice, having had the chimeric intron in addition to the majority of the RL ORF and XIAP-derived sequence removed. Translation of this transcript should result in the formation of a RL-FL fusion protein that lacks RL activity but likely exhibits FL activity (Olsson et al. 1989; Eu and Andrade 2001). Both smaller mRNAs should facilitate efficient cap-dependent translation of FL. The production of these mRNAs is consistent with the observed repression of RL activity and the resistance of FL to RNAi-mediated knockdown that were observed.
We performed Northern blotting to quantitate the amounts of each transcript produced in 293T cells transfected with pRL-XIAP-FL DNA. Figure 2B ▶ shows multiple transcripts were detected following transfection with this DNA (lane 1) but not control DNA vector (lane 2). Two mRNAs were readily detectable and corresponded in size to the intact dicistronic (~3500 bp) and the double-spliced (~2800 bp) transcripts. The double-spliced transcript was the most abundant transcript formed in 293T cells. However, the single-spliced (~2000 bp) transcript that was detected by RT-PCR was not readily apparent by Northern blotting. These results do not corroborate previously published Northern blot data (Holcik et al. 2003).
Collectively, these data indicate that multiple sensitive methods of RNA analysis need to be used when assessing the integrity and identity of transcripts produced in cells transfected with dicistronic DNA test constructs. For instance, the single-spliced (~2000 bp) transcript was readily detectable by RT-PCR but not by Northern blotting. Thus, RT-PCR, although not quantitative, provides a highly sensitive technique for the detection of alternatively spliced mRNAs. Here, Northern blotting, although not sensitive enough to detect all transcripts, can be used as a quantitative assessment of the most prevalent mRNA species. In addition, it is likely that aberrant splicing in cells transfected with dicistronic DNA reporter constructs confounded prior assessments of IRES activity within the XIAP 5′ UTR (Holcik et al. 1999, 2003; Holcik and Korneluk 2000; Henis-Korenblit et al. 2002; Nevines et al. 2003; Warnakulasuriyarachchi et al. 2003). It is possible that other 5′ UTR sequences reported to contain IRES elements may also facilitate RNA splicing. This is of concern because many putative ITAFs reported to stimulate activity of these IRES sequences are known RNA splicing factors (e.g., PTB [for review, see Wagner and Garcia-Blanco 2001] and hnRNP-C [Choi et al. 1986]).
The influence of RNA splicing on apparent IRES-mediated translation
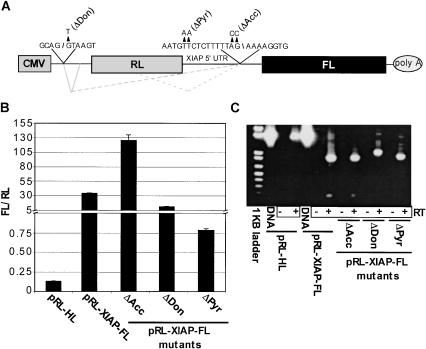
To obtain an accurate assessment of IRES activity for a proposed sequence, the formation of aberrant transcripts resulting from cryptic promoter activity and/or RNA splicing must be eliminated. However, these phenomena may be inseparable, because sequence changes designed to abrogate promoter activity and/or splicing within a test sequence may also adversely affect IRES function. To address this, pRL-XIAP-FL DNA was mutated at the sites corresponding to the splice-donor (GT→CC [pRL-XIAP-FLΔDon]) and the identified splice-acceptor (AG→TT [pRL-XIAP-FLΔAcc]) in an attempt to abrogate RNA splicing and determine its contribution to apparent XIAP IRES activity (Fig. 3A ▶). The splice-acceptor site was located within the XIAP 5′ UTR sequence but was not in the “minimal IRES” region that was previously mapped (Holcik et al. 1999). In addition, we introduced a TT→AA change within the polypyrimidine tract of the UTR at nucleotides −35 to −44 (relative to the XIAP translation initiation codon) that was previously shown to abrogate apparent XIAP IRES activity (Holcik et al. 1999). 293T cells were transfected with these dicistronic mutant constructs in triplicate (as described in the Materials and Methods), and the FL/RL activity ratios were determined and plotted (Fig. 3A ▶). In addition, total RNA was isolated from transfected cells for RT-PCR analysis (Fig. 3B ▶). We found that altering the splice-acceptor site (i.e., pRL-XIAP-FLΔAcc) increased the FL/RL ratio with no apparent change in the RNA splicing profile. However, sequencing of the smallest PCR product revealed the splice-junction had shifted to an AG 5 nt downstream of its original location, which may account for the increase in relative FL activity (Fig. 3A ▶). The increase in FL/RL could have resulted from formation of a shorter leader on the transcript and/or resulted from more efficient RNA splicing (this can not be ruled out because the RT-PCR was not quantitative). Alternatively, mutating the upstream splice-donor site (pRL-XIAP-FLΔDon), not located in the XIAP sequence itself, reduced the FL/RL ratio by ~80%. Interestingly, splicing was not prevented but was altered by this mutation. Higher levels of full-length transcripts were observed, but both full length and spliced products retained the upstream chimeric intron, resulting in slower migration in the gel (Fig. 3C ▶; confirmed by sequencing). This demonstrated the mutation blocked the splicing of the chimeric intron as intended and prevented formation of the smallest splice product. However, despite absence of upstream splicing and a functional 5′ intron, it is important to note that alternate splicing was still mediated by the strong splice acceptor in the XIAP sequence.
FIGURE 3.
Mutation of nucleotides in pRL-XIAP-FL reduces apparent IRES activity and affects RNA splicing in transfected cells. (A) Schematic depicting the pRL-XIAP-FL DNA construct. Gray solid lines indicate splicing of chimeric introns. Dashed lines indicate splicing associated with XIAP sequences. Mutations to pRL-XIAP-FL are indicted by arrowheads and identified in parentheses. Splice-donor and -acceptor locations are denoted by forward and backward slash marks, respectively. (B) Indicated DNA constructs were transfected into 293T cells and lysates generated 24 h post-transfection. FL and RL activities were determined and FL/RL plotted for each construct. (C) RNA was harvested from cells in one well for each of the transfections shown in B and subjected to RT-PCR (as indicated in Materials and Methods). Products were separated in a 1% (w/v) agarose gel and visualized by EtBr staining/UV. PCR products from the HCV and XIAP DNA constructs were included as size controls (DNA). RT was included (+) or excluded (−) from each reaction as indicated.
Polypyrimidine tracts have been implicated in IRES function (Blyn et al. 1997; Faustino and Cooper 2003) but have also been shown to be involved in RNA splicing. Accordingly, the polypyrimidine tract was changed in pRL-XIAP-FL (pRL-XIAP-FLΔPyr). Mutating this sequence correlated with a dramatic 30-fold reduction in IRES activity (i.e., the FL/RL activity ratio), although some activity was still present compared with the HCV IRES. RNA splicing still occurred with transfection of this construct. The polypyrimidine tract was previously reported to be necessary for the formation of a RNP complex containing La that is believed to facilitate XIAP IRES-mediated translation (Holcik and Korneluk 2000). However, we found that the reduction in FL/RL activity with pRL-XIAP-FLΔPyr correlated well with changes in RNA splicing (i.e., the formation of lower amounts of the smaller RNAs). These data indicate that splicing and IRES activity cannot be distinguished simply by mutating DNA constructs. Therefore, IRES activity must be assayed using a method in which splicing does not occur.
Direct transfection of cells with RNA as an assay for IRES activity
Several groups have used direct RNA transfection to assay IRES activity in cells (Koev et al. 2002; Thompson and Sarnow 2003). In cases in which promoter and/or splicing activity is present, this may be the most effective method for assaying IRES activity in cells. However, direct RNA transfection may prevent the opportunity for an IRES to properly associate with certain nuclear ITAFs that may be required for RNP-complex formation and IRES activity. This was previously referred to as a nuclear event that is required for the activation of certain IRESs (Stoneley et al. 2000). However, in the case of XIAP, nuclear-RNA splicing likely contributed to the unusually high levels of apparent IRES activity that was previously reported (Holcik et al. 1999).
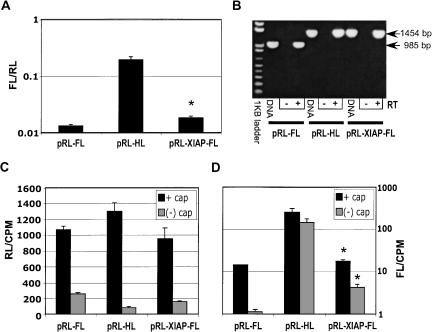
293T cells were directly transfected with dicistronic test RNAs to assay the IRES activity of the XIAP 5′ UTR. RNAs from pRL-FL and pRL-HL were included as negative and positive controls, respectively. Transcripts for the assays were synthesized in vitro in the presence of 32P-CTP, with or without m7GpppG cap. Each runoff transcript also encoded a 35-nt poly(A) tail. Following transcription, RNA was treated with DNase I, phenol-chloroform extracted, then passed through a gel filtration column prior to transfection. This insured transfection mixes lacked contaminating DNA, proteins, and nucleotides (including nonincorporated m7GpppG). Cells were lysed 9 h post-transfection when rates of luciferase synthesis were maximal (Van Eden et al. 2004). Figure 4A ▶ shows the FL/RL activity ratio for each of the indicated capped dicistronic RNAs. The XIAP 5′ UTR exhibited a FL/RL ratio that was marginally, yet statistically, greater (approximately twofold) than that of the empty vector (pRL-FL). The HCV IRES exhibited a FL/RL ratio that was nearly 15-fold higher than the negative control. RNA was also harvested from transfected cells for RT-PCR as a test for RNA splicing (Fig. 4B ▶). The results of this analysis indicated no splicing occurred with any of the transfected dicistronic RNAs. It can also be concluded that the lack of any product from the RT(−) controls demonstrated that DNA from in vitro transcription reactions was not carried over into the transfections. Also, the 95% repression of RL activity in 293T cells transfected with pRL-XIAP-FL DNA (Fig. 1E ▶) did not occur with direct RNA transfection of these cells (Fig. 4C ▶). Similar results were obtained with transfection of HepG2 and HeLa S3 monolayers. However, in these cells the XIAP 5′ UTR failed to demonstrate statistically significant IRES activity (data not shown).
FIGURE 4.
The XIAP 5′ UTR exhibits low IRES activity in cells transfected with RNA. (A) RNA was transcribed in vitro from each of the indicated DNA plasmids in the presence of m7GpppG and [α32P]-CTP. RNA was then treated with DNase I, phenol-chloroform extracted, and purified via gel filtration chromatography. Nine hours post-transfection of RNA, 293 cells were lysed and assayed for FL and RL activity. FL/RL ratios were plotted for each transfection on a logarithmic scale. Asterisk indicates a statistically significant difference between pRL-XIAP-FL and pRL-FL (P < 0.05). (B) RNA was extracted from cells transfected with the indicted RNAs and used for RT-PCR. PCR products were isolated in a 1% (w/v) agarose gel and visualized by EtBr staining/UV. PCR products from the indicated DNA constructs were included as size controls (DNA). (+) and (−) denote the inclusion or exclusion of RT from the reaction, respectively. The arrow to the right indicates PCR products correlating to intact dicistronic RNAs. (C,D) Cells were transfected as above with capped or uncapped 32P-labeled RNA transcribed from the indicated plasmids. Cells were lysed, 9 h post-transfection, and FL and RL activities were determined. Remaining transfected RNA was estimated via scintillation counting of TriZol-extracted RNA. RL (C) and FL (D) were plotted after normalizing to the total amount of each labeled RNA. (D) Data were plotted on a logarithmic scale. Asterisk indicates significant pRL-XIAP-FL IRES activity compared with pRL-FL (P < 0.05). Results are representative of three separate experiments.
Unlike mRNA produced in cells transfected with DNA expression vectors, RNAs transcribed in vitro are not capped with absolute efficiency. Because the dicistronic assay has FL activity normalized to cap-dependent RL activity, FL/RL can vary with capping efficiency. To address this, we compared RL and FL translation from capped versus uncapped dicistronic RNAs in transfected 293T cells. Figure 4C ▶ shows the effect of RNA capping on RL activity, and Figure 4D ▶ shows the effect of capping on FL activity. In both cases, values were normalized based on the amount of radiolabeled RNA remaining in cells 9 h post-transfection, although normalization to the amount of radiolabeled input RNA resulted in identical patterns (data not shown). Surprisingly, the absence of a cap did not significantly decrease the stability of transfected radiolabeled RNA assessed in denaturing gel electrophoresis/autoradiography (Van Eden et al. 2004). Although the lack of cap greatly reduced the translation of FL from pRL-FL RNA, there was much less of an effect on RNAs containing either the HCV IRES or XIAP 5′ UTR sequences (Fig. 4D ▶). These data support the notion that a low level of FL translation from pRL-FL RNA is mediated via ribosome reinitiation and is cap dependent. Conversely, both XIAP and HCV test RNAs were less affected by the presence of an upstream cap because ribosomes may have been directly recruited to their IRES elements. These data provide further evidence that both HCV- and XIAP-derived test sequences could support cap-independent translation initiation in 293T cells. In addition, these data indicate that both sequences do not simply enhance reinitiation but rather mediate direct recruitment of ribosomes. Thus, transfection of both capped and uncapped dicistronic test RNAs may provide a means for discerning downstream FL cistron synthesis resulting from ribosome reinitiation versus the presence of an IRES.
Furthermore, the HCV IRES exhibited similar levels of IRES activity (compared with pRL-FL) in vitro and in cells transfected with either DNA or RNA dicistronic constructs. Thus, nuclear events may not be essential for HCV IRES activity. Collectively, these results support transfection of cells with dicistronic RNA test constructs as a valid method for assaying IRES activity while simultaneously eliminating the possibility of aberrant RNAs formed from cryptic promoters and/or RNA splicing.
Concluding remarks
Methods other than the dicistronic test have also been used for the identification of IRESs. The HIV IRES was identified by its ability to continue directing translation in a monocistronic mRNA during poliovirus infection and its ensuing cap-dependent translation shutoff (Buck et al. 2001). Similarly, the ORF-embedded eIF4GI IRES was first identified by its mRNA maintaining association with polysomes during poliovirus infection. Likewise, transcripts encoding c-myc and BiP were found associated with polysomes during these conditions (Johannes and Sarnow 1998). However, these methods can only aid in the identification of potential IRES sequences that function in the absence of intact eIF4G. The most definitive test of IRES activity was demonstrated in a circular RNA containing the EMCV 5′ UTR (Chen and Sarnow 1995). This demonstrated that neither the cap nor a free 5′-phosphate residue was required for the initiation of translation. However, this technique is limited in that the IRES must be devoid of stop codons and able to direct the translation of a run-on protein.
To date, the dicistronic DNA assay is the predominant method used for demonstrating internal ribosome entry sites in eukaryotic mRNAs. However, precautions must be taken to ensure that translation of the downstream cistron results from IRES activity and not from aberrantly formed RNAs. Northern blotting has frequently been used to assay RNA integrity in cells transfected with dicistronic DNA test constructs, but we have had difficulty detecting some smaller spliced-RNAs by this method. However, by using a RNAi-based technique as well as RT-PCR analysis, we were able to detect aberrant RNA processing in cells transfected with a dicistronic DNA containing the XIAP 5′ UTR. Splicing was anticipated because translation of upstream RL was typically repressed ~20-fold in constructs containing the XIAP 5′ UTR (data not shown). Thus, repression of first cistron expression may also be an indication of aberrant RNA splicing. In addition, we have presented data that show the XIAP 5′ UTR-derived splice elements cannot be easily eliminated by point mutations. RNA transfection was used to assay IRES activity because splicing and IRES activities could not be differentiated in transfections conducted with DNA constructs.
There is evidence to suggest post-transcriptional regulation of cellular XIAP expression occurs (Holcik et al. 1999; Tamm et al. 2000). It was reported that XIAP is translated preferentially during apoptosis when cap-dependent translation is down-regulated (Holcik et al. 1999). We transfected cells directly with dicistronic RNA to assess XIAP IRES activity in the absence of aberrant RNA formation and demonstrated the XIAP 5′ UTR could mediate cap-independent translation, albeit at levels far lower than previously reported (Holcik et al. 1999, 2003; Holcik and Korneluk 2000; Henis-Korenblit et al. 2002; Nevines et al. 2003; Warnakulasuriyarachchi et al. 2003). Further work is required to determine if the weak XIAP IRES activity detected here is stimulated by cell stress or apoptosis.
The data presented here suggest the need for a re-evaluation of proposed ITAFs in the IRES-mediated translation process. In the case of the XIAP IRES, the involvement of La and hnRNP-C requires particular clarification because these factors are known to be associated with the RNA splicing apparatus (Choi et al. 1986; Fouraux et al. 2002). We propose that the methods presented here will complement those currently in use and contribute to the accurate identification/characterization of viral and cellular IRESs.
MATERIALS AND METHODS
Materials
Fetal bovine serum (FBS) was purchased from Hyclone Laboratories, and m7G(5′)ppp(5′)G cap analog was from Ambion. [35S]methionine-cysteine (Tran35S-label) and [α-32P]-CTP were purchased from ICN Biomedicals, Inc. All restriction endonucleases and pSV-β-Gal, pGL3-control, and pGL3-basic plasmid vectors were from Promega. Oligonucleotide DNA primers were synthesized by Integrated DNA Technologies. Unless stated otherwise, all other compounds were reagent grade and obtained from Sigma-Aldrich.
Construction of dicistronic reporter DNAs
The pRL-HL dicistronic vector was generously provided by S.M. Lemon (Univ. of Texas, Medical Branch), and its design and construction have already been described (Honda et al. 2000). RNA transcribed from this vector contained an upstream cistron coding for Renilla luciferase and a downstream cistron coding for firefly luciferase with intervening sequence containing the HCV IRES. Renilla luciferase is translated from this template by canonical cap-dependent mechanisms, whereas firefly luciferase is translated by internal ribosome entry.
The pRL-FL construct, in which the intercistronic region was removed, was previously described (Van Eden et al. 2004). A ~460-bp DNA fragment corresponding to the region within the 5′ UTR of human XIAP (Genebank EST database: BX119811), just upstream of the reported XIAP ORF, was amplified from total cytoplasmic RNA (HeLa) by RT-PCR by using primers 5′-ctagcggccgc cttgtaaaaacaactttgatgccttga-3′ (sense) and 5′-cagccgcggtctcttgaaaat aggacttgtccacct-3′ (antisense; NotI and SacII sites underlined, respectively). The product was ligated into pGEM-T (Promega), sequenced, and then cloned into pRL-HL at NotI and SacII sites that flanked the HCV IRES to generate the pRL-XIAP-FL construct. DNAs amplified by long-range PCR from pRL-XIAP-FL, so as to exclude the CMV promoter, were blunt-end ligated to create the promoter-less p(ΔCMV)RL-XIAP-FL construct. Mutagenesis of pRL-XIAP-FL dicistronic constructs was performed according to the QuikChange site-directed mutagenesis protocol (Stratagene), and these constructs are described in the figure legends. In addition, constructs used for transient RNA transfection of cells contained a 35-nt poly(A) inserted 12 nt downstream of the firefly luciferase ORF. All constructs were verified by restriction enzyme analyses and automated DNA sequencing, the latter as a service provided by Lone Star Laboratories.
RNAi constructs
The pBS/U6ApaI vector was the generous gift of Y. Shi (Harvard Medical School), and its construction has been previously described (Sui et al. 2002). The pBS/U6-RLi construct was generated by using a two-step subcloning procedure. Briefly, oligonucleotides (5′-ggtgcctgtagtgtgatgtga3′ and 5′-agcttcacatcacactacaggcacc-3′) were annealed then ligated into pBS/U6, which had previously been digested with ApaI, treated with T4 DNA polymerase to blunt the ends, and finally digested with HindIII. The resulting construct was isolated, and a second set of oligonucleotides (5′-agc ttcacatcacactacaggcaccctttttg-3′ and 5′-aattcaaaaagggtgcctgtagtgtg atgtga-3′) were annealed and then ligated into the plasmid, which had previously been digested with HindIII and EcoRI. The resulting pBS/U6-RLi construct contained a 50-nt RNA hairpin downstream of the murine U6 promoter designed to elicit an siRNA response targeting the RL ORF.
Cell culture and DNA/RNA transfection
HeLa S3 cervical adenocarcinoma cells, 293T embryonic kidney cells, and HepG2 hepatocellular carcinoma cells were obtained from the American Type Culture Collection. All cultures were passaged as monolayer cultures in DMEM (Invitrogen) supplemented with 10% (v/v) FBS, 100 U penicillin, and 100 μg/mL streptomycin or in EAGLES medium supplemented as described above (HepG2). All cultures were grown in a humidified incubator at 37°C in an atmosphere of 5% CO2.
Transient DNA transfections were performed by using LipofectAMINE and Plus reagents (Invitrogen) according to the specifications of the manufacturer. Briefly, 3 × 105 cells were seeded per well of a 12-well plate the day prior to transfection. Cells were transfected with DNA (1 μg/well) and cultured for 20 h at 37°C prior to harvesting by removing the medium and lysing the cells in passive lysis buffer (PLB; Promega).
Alternatively, cells were transfected with RNA. Plasmid constructs were linearized with AgeI (XhoI for pRL-HL) so all runoff RNA transcripts contained a 35-nt poly(A) tail. Capped, poly(A) runoff RNA transcripts (+ or − m7GpppG) were generated in vitro by using recombinant T7 RNA Polymerase in the presence of [α32P]-CTP. Transient RNA transfections were performed by using DMRIE-C (Invitrogen) according to the specifications of the manufacturer. Approximately 3 × 105 cells were seeded per well of a 12-well plate the day prior to transfection. Cells were transfected with RNA (3 μg/well), cultured for ~8 h at 37°C, and then harvested by removing the medium and lysing the cells in PLB. Quantitation of transfected RNAs was via scintillation counting or glyoxol/DMSO-agarose gel electrophoresis and autoradiography of TriZol (Sigma) extracted RNA (results were similar in both cases).
In vitro transcription/translation
Plasmid DNA was linearized prior to transcription by using XhoI (pRL-HL and pRL-XIAP-FL) or BamHI (pRL-FL) to generate RNA for in vitro translation. Capped, runoff RNA transcripts were synthesized by using recombinant T7 RNA polymerase as previously described (Byrd et al. 2002). Following transcription in vitro, NucAway spin columns (Ambion) were used to separate nonincorporated nucleotides. RNA for transfection was prepared as above, but plasmids were linearized with AgeI (pRL-FL and pRL-XIAP-FL) or XhoI (pRL-HL). Transcription was via recombinant T7 polymerase. Following transcription, reactions were treated with DNase I, phenol-chloroform extracted, and purified over NucAway spin columns. In both cases RNA integrity was assessed by glyoxol/DMSO-agarose gel electrophoresis, and the concentration of RNA was estimated by UV spectroscopy. Nuclease-treated rabbit reticulocyte lysates (RRLs; Promega) were programmed with RNA transcripts (~80 pmole/ml) in the presence of [35S]methionine-cysteine for 90 min at 30°C in accordance with the instructions supplied by the manufacturer. Translation reactions were analyzed by autoradiography of SDS-PAGE gels (12.5% [w/v] acrylamide) using Biomax MR film (Eastman Kodak).
Assays for luciferase
RL and FL activities were quantitated by using the Dual-Luciferase Reporter System (Promega) and a Sirius luminometer (Berthold Detection Systems). Typically, 5–20 μL cell lysate (post-transfection) was combined sequentially with FL and then RL luciferase-specific substrates (i.e., beetle luciferin and coelenterazine, respectively) according to the protocol supplied by the manufacturer. Light emission was measured 3 sec after addition of each of the substrates and integrated over a 10-sec interval.
RNA analyses
RT-PCR was performed as a test for mRNA splicing in cells following DNA transfection. Cytoplasmic RNA was isolated from cells by using TriZol reagent according to the instructions of the manufacturer. After DNase I treatment, 1 μg purified cellular RNA was reverse-transcribed using the ImProm-II Reverse Transcription System (Promega) and an oligo-d(T) primer (Invitrogen). PCR was used to amplify the intercistronic region of cDNA by using the primers P1 5′-cagatcactagaagctttattgcg-3′ (sense) and P2 5′-tctcttcatagccttatgcagttgc-3′ (antisense). RT-PCR was performed similarly on RNA isolated from RNA transfected cells, except DNase I pretreatment was excluded. PCR in this case was with primers P3 5′-gctagccaccatgacttcgaaag-3′ (sense) just downstream of the T7 promoter and P2.
Northern blotting was performed with RNA purified from 293T cells following transfection with dicistronic DNAs as indicated above. Following extraction with TriZol, 2.5 μg total RNA was loaded per well of a 1% (w/v) glyoxol/DMSO-agarose gel, separated by electrophoresis, and transferred to a nylon membrane. Hybridization was in ULTRAhyb hybridization buffer (Ambion) at 68°C to a ~500-nt 32P-labeled riboprobe that annealed to the 3′-end of the FL ORF. Autoradiography of the membrane was performed overnight at −80°C with Kodak X-OMAT film to visualize the bands.
Acknowledgments
This work was supported by National Institutes of Health grants GM 59803 and AI 50237. M.E.V.E. and M.P.B. were supported by a National Institute of Allergy and Infectious Diseases (NIAID) training grant (T32 AI07471). We thank P.M. Younan for his technical assistance and R.F. Ramig for critical review of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5225204.
REFERENCES
- Blyn, L.B., Towner, J.S., Semler, B.L., and Ehrenfeld, E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71: 6243–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, C.B., Shen, X., Egan, M.A., Pierson, T.C., Walker, C.M., and Siliciano, R.F. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75: 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, M.P., Zamora, M., and Lloyd, R.E. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell Biol. 22: 4499–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y. and Sarnow, P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. [DOI] [PubMed] [Google Scholar]
- Choi, Y.D., Grabowski, P.J., Sharp. P.A., and Dreyfuss, G. 1986. Heterogenous nuclear ribonucleoproteins: Role in RNA splicing. Science 231: 1534–1539. [DOI] [PubMed] [Google Scholar]
- Coldwell, M.J., Mitchell, S.A., Stonely, M., MacFarlane, M., and Willis, A.E. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19: 899–905. [DOI] [PubMed] [Google Scholar]
- Ehrenfeld, E. 1996. Initiation of translation by picornavirus RNAs. In Translational control (eds. J.W.B. Hershey et al.), pp. 549–573. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Eu, J. and Andrade, J. 2001. Properties of firefly luciferase immobilized through a biotin carboxyl carrier protein domain. Luminescence 16: 57–63. [DOI] [PubMed] [Google Scholar]
- Falcone, D. and Andrews, D.W. 1991. Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell Biol. 11: 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino, N.A. and Cooper, T.A. 2003. Pre-mRNA splicing and human disease. Genes & Dev. 17: 419–437. [DOI] [PubMed] [Google Scholar]
- Fouraux, M.A., Kolkman, M.J., Van der Heijden, A., De Jong, A.S., Van Venrooij, W.J., and Pruijn, G.J. 2002. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA 8: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M., Tan, S., and Katze, M.G. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64: 239–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe, A.P. and Morris, D.R. 1994. Initiation codons within 5′-leaders of mRNAs as regulators of translation. Trends Biochem. Sci. 19: 159–164. [DOI] [PubMed] [Google Scholar]
- Hambidge, S.J. and Sarnow, P. 1992. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc. Natl. Acad. Sci. 89: 10272–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, B. and Zhang, J.T. 2002. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: The eIF4G story. Mol. Cell. Biol. 21: 7372–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U.T., Pestova, T.V., Litterst, M., and Wimmer, E. 1994. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J. Virol. 68: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit, S., Shani, G., Sines, T., Marash, L., Shohat, G., and Kimchi, A. 2002. The caspase-cleaved DAP5 protein supports internal ribosome entry site–mediated translation of death proteins. Proc. Natl. Acad. Sci. 99: 5400–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik, M. and Korneluk, R.G. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: Role of La autoantigen in XIAP translation. Mol. Cell Biol. 20: 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik, M., Lefebvre, C., Yeh, C., Chow, T., and Korneluk, R.G. 1999. A new internal-ribosome–entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell. Biol. 1: 190–192. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Gordon, B.W., and Korneluk, R.G. 2003. The internal ribosome entry site–mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell Biol. 23: 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, S., Kaneko, S., Matsushita, E., Kobayashi, K., Abell, G.A., and Lemon, S.M. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site–directed translation. Gastroenterology 118: 152–162. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J. and Kaminski, A. 1995. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA 1: 985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jang, S.K., Krausslich, H.G., Nicklin, M.J.H., Duke, A.C., Palmenberg, A.C., and Wimmer, E. 1988. A segment of the 5′ nontranslated region of the encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, G. and Sarnow, P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 12: 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev, G., Duncan, R.F., and Lai, M.M.C. 2002. Hepatitis C virus IRES-dependent translation is insensitive to an eIF2α-independent mechanism of inhibition by interferon in hepatocyte cell lines. Virology 297: 195–202. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- ———. 2001. New ways of initiating translation in eukaryotes? Mol. Cell Biol. 21: 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene 318: 1–23. [DOI] [PubMed] [Google Scholar]
- Macejak, D.G. and Sarnow, P. 1991. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature 353: 90–94. [DOI] [PubMed] [Google Scholar]
- Meerovitch, K., Svitkin, Y.V., Lee, H.S., Lejbkowicz, F., Kenan, D.J., Chan, E.K.L., Agol, V.I., Keene, J.D., and Sonenberg, N. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67: 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Brown, E.C., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2001. Protein factor requirements of the Apaf-1 internal ribosome entry segment: Roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell Biol. 21: 3364–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins, T.A., Harder, Z.M., Korneluk, R.G., and Holcik, M. 2003. Distinct regulation of internal ribosome entry site–mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 278: 3572–3579. [DOI] [PubMed] [Google Scholar]
- Olsson, O., Escher, A., Sandberg, G., Schell, J., Koncz, C., and Szalay, A.A. 1989. Engineering of monomeric bacterial luciferases by fusion of luxA and luxB genes in Vibrio harveyi. Gene 81: 335–347. [DOI] [PubMed] [Google Scholar]
- Pelletier, J. and Sonenberg, N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., and Hellen, C.U.T. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 98: 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, I., Itin, A., Einat, P., Skaliter, R., Grossman, Z., and Keshet, E. 1998. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: Implications for translation under hypoxia. Mol. Cell Biol. 18: 3112–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley, M., Paulin, F.E., Le Quesne, J.P., Chappell, S.A., and Willis, A.E. 1998. C-myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 22: 423–428. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Subkhankulova, T., Le Quesne, J.P.C., Coldwell, M.J., Jopling, C.L., Belsham, G.J., and Willis, A.E. 2000. Analysis of the c-myc IRES: A potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y., Forrester, W.C., and Shi, Y. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. 99: 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm, I., Kornblau, S.M., Segall, H., Krajewski, S., Welsh, K., Kitada, S., Scudiero, D.A., Tudor, G., Qui, Y.H., Monks, A., et al. 2000. Expression and prognostic significance of IAP-family genes in human cancers and myeloid luekemias. Clin. Cancer Res. 6: 1796–1803. [PubMed] [Google Scholar]
- Thompson, S.R. and Sarnow, P. 2003. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology 315: 259–266. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara, K., Iizuka, N., Kohara, M., and Nomoto, A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66: 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Gensac, M.C., Maret, A., Bayard, F., Amalric, F., Prats, H., and Prats, A.C. 1995. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell Biol. 15: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eden, M.E., Byrd, M.P., Sherrill, K.W., and Lloyd, R.E. 2004. Translation of cellular inhibitor of apoptosis protein 1 (c-IAP1) mRNA is IRES mediated and regulated during cell stress. RNA 10: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polyprimidine tract binding protein antagonized exon definition. Mol. Cell Biol. 21: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnakulasuriyarachchi, D., Ungureanu, N.H., and Holcik, M. 2003. The translation of an antiapoptotic protein HIAP2 is regulated by an upstream open reading frame. Cell Death Differ. 10: 899– 904. [DOI] [PubMed] [Google Scholar]