Abstract
The intergenic region (IGR) located upstream of the capsid protein gene in dicistroviruses contains an internal ribosome entry site (IRES). Translation initiation mediated by the IRES does not require initiator methionine tRNA. Comparison of the IGRs among dicistroviruses suggested that Taura syndrome virus (TSV) and acute bee paralysis virus have an extra side stem loop in the predicted IRES. We examined whether the side stem is responsible for translation activity mediated by the IGR using constructs with compensatory mutations. In vitro translation analysis showed that TSV has an IGR-IRES that is structurally distinct from those previously described. Because IGR-IRES elements determine the translation initiation site by virtue of their own tertiary structure formation, the discovery of this initiation mechanism suggests the possibility that eukaryotic mRNAs might have more extensive coding regions than previously predicted. To test this hypothesis, we searched full-length cDNA databases and whole genome sequences of eukaryotes using the pattern matching program, Scan For Matches, with parameters that can extract sequences containing secondary structure elements resembling those of IGR-IRES. Our search yielded several sequences, but their predicted secondary structures were suggested to be unstable in comparison to those of dicistroviruses. These results suggest that RNAs structurally similar to dicistroviruses are not common. If some eukaryotic mRNAs are translated independently of an initiator methionine tRNA, their structures are likely to be significantly distinct from those of dicistroviruses.
Keywords: IRES, Dicistroviridae, computational search, Scan For Matches
INTRODUCTION
The family Dicistroviridae is composed of positive-stranded RNA viruses from invertebrates (Mayo 2002). These viruses have an internal ribosome entry site (IRES) in an intergenic region (IGR) on their genome. IRES elements were first found in picornaviruses (Jang et al. 1988; Pelletier and Sonenberg 1988). Several viral and cellular mRNAs have also been shown to have an IRES within the 5′ UTR sequence (Hellen and Sarnow 2001; Vagner et al. 2001). Unlike other IRES-mediated initiation of translation, the IGR-IRES–mediated initiation of translation in dicistroviruses is shown to occur in the absence of base-pair interaction between the initiation codon and the anticodon triplet in initiator methionine tRNA (Met-tRNAi; Sasaki and Nakashima 1999 Sasaki and Nakashima 2000); thus, the coding region regulated by the IRES does not require a functional AUG initiation codon. Because we usually find a coding region in a cDNA sequence by identification of an AUG initiation codon, the discovery of IGR-IRES suggests the possibility that eukaryotic coding regions may be more extensive than previously believed (Wilson et al. 2000a).
We previously constructed a secondary structure model of the IGR-IRES of Plautia stali intestine virus (PSIV) using various mutational analysis methods (Kanamori and Nakashima 2001). This structural model was applicable to other dicistroviruses, suggesting a common secondary structure in the intergenic region. However, nucleotide sequence identity in this region was not high except for a few short nucleotide segments; thus, it is difficult to find counterparts using homology search tools based on primary nucleotide sequence similarity, such as FASTA and BLAST. “Scan For Matches” is a pattern-matching program written with C programming language. This program is designed to find a complex pattern, including potential secondary structure elements, in nucleic acid sequences by indicating parameters composed of a sequence of pattern units (Dsouza et al. 1997). Because this program extracts nucleotide sequences matched with the indicated parameter, we used this program to find counterparts of IGR-IRES from nucleotide sequence databases.
After construction of a secondary structure model of the IGR-IRES of PSIV, genome sequences of acute bee paralysis virus (ABPV) and Taura syndrome virus (TSV) were reported (Govan et al. 2000; Mari et al. 2002). Putative IRES regions in the two viruses were suggested in the 5′-half of the intergenic region, but the pseudo-knot (PK) I region responsible for initiation site determination in dicistroviral capsid protein translation was not identified. Instead of PK I, in-frame AUG triplets were found at 6- and 30-nt upstream of the 5′end of the capsid protein genes in TSV and ABPV, respectively (Govan et al. 2000; Mari et al. 2002). We recently found that ABPV and TSV may form an extra stem loop close to the 5′ end of the capsid protein coding region. If the stem loop forms, PK I would be folded and might serve as an initiator for translation (Nishiyama et al. 2003). Recently, the whole genome sequence of Kashmir bee virus (KBV) was released (accession number AY275710) and its IGR may also form the extra stem loop.
Here, we report that the upstream in-frame AUG triplet in TSV does not function as an initiator, and that the intergenic region in TSV functions as an IRES in vitro. Using a verified secondary structure of domain 3 of TSV, we searched for structures similar to IGR-IRES of dicistroviruses using “Scan For Matches.”
RESULTS AND DISCUSSION
The intergenic region of TSV functions as an IRES in vitro
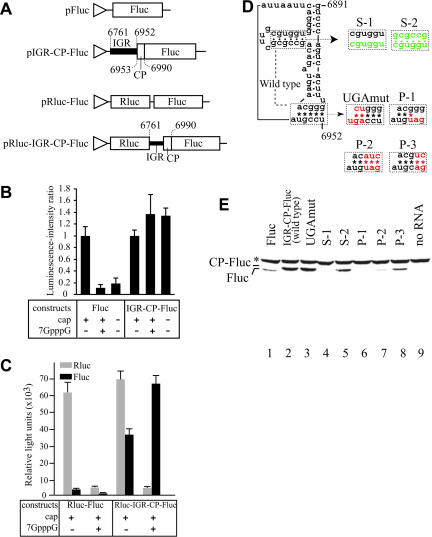
We previously predicted an extra side stem loop at domain 3 of the putative IGR-IRES elements of ABPV and TSV (Nishiyama et al. 2003). However, experimental data for IRES activity of the IGR in TSV and ABPV has not been provided. We first examined the effect of the cap on in vitro translation using transcripts from pFluc and pIGR-CP-Fluc (Fig. 1A ▶) because translation initiation mediated by the IGR-IRES of PSIV is cap independent (Sasaki and Nakashima 1999), while that mediated by the canonical ribosome scanning mechanism is cap dependent (Kozak 1999). When capped Fluc RNAs were translated in the presence of cap analog, the average luciferase activity decreased to less than 0.2 relative to control reactions lacking cap analogs (Fig. 1B ▶). These data suggest that Fluc RNA translation is cap dependent. In contrast, when capped IGR-CP-Fluc RNA was translated in the presence of the cap analog, the average luciferase activity increased to 1.3. In addition, uncapped transcripts of IGR-CP-Fluc also translated as well as the capped transcript, indicating that IGR-CP-Fluc RNA translation is cap independent (Fig. 1B ▶). To confirm internal initiation mediated by the IGR sequence of TSV, a dicistronic construct including a Rluc sequence as the first cistron (Fig. 1A ▶) was also examined. Capped Rluc-IGR-CP-Fluc construct showed highest Fluc activity in the presence of cap analog (Fig. 1C ▶), indicating that CP-Fluc is translated independently of the first cistron. These observations show that the IGR sequence of TSV has an IRES activity.
FIGURE 1.
Internal initiation of translation mediated by the intergenic region of Taura syndrome virus. (A) Structures of plasmids used for in vitro transcription. Triangles indicate T7 promoter sequences. Numbers mark nucleotide positions in the TSV genome sequence. CP indicates the 5′ part of the capsid coding sequence to produce a firefly luciferase (Fluc) fusion protein. (B) Effect of the 5′ cap and cap analogs on translation of transcripts from pIGR-CP-Fluc in vitro. (C) Effect of the 5′ cap and cap analogs on translation from the second cistron mediated by the IGR-sequence of TSV. (D) Compensatory mutations introduced into the predicted extra side stem and PK I. Dots and asterisks indicate base-pair formation for helical segments and PK I. Mutated nucleotides are colored. (E) Detection of translation products from various mutants. Asterisk marks position of endogenous biotinylated protein in wheat germ extracts.
In vitro translation of Fluc RNA produced a smaller protein than IGR-CP-Fluc RNA, showing that the introduced TSV capsid coding region of IGR-CP-Fluc was translated as a fusion protein with Fluc (Fig. 1E ▶, lanes 1,2). Next we tested the function of the in-frame AUG triplet (nt 6947–6949) in the IGR. The 5′ end of the capsid coding region of TSV has been mapped at nt 6953 by N-terminal sequence analysis of capsid proteins (Mari et al. 2002). When we mutated the in-frame AUG triplet located 6-nt upstream of the 5′ end of the capsid coding region to a UGA stop codon (Fig. 1D ▶), CP-Fluc was detected as well as the wild type (Fig. 1E ▶, lanes 2,3). This means that the AUG triplet does not function as an initiator in this construct, suggesting that the capsid protein message of TSV, as in dicistrovirus RNAs, is translated in an AUG-independent manner. Finally, compensatory mutations were introduced in the extra side stem loop and the putative PK I. When base-pair formation at the side stem was inhibited (Fig. 1D ▶), translation of CP-Fluc was also inhibited (Fig. 1E ▶, lane 4). Introduction of a compensatory mutation restored translation (Fig. 1E ▶, lane 5), indicating that base-pair formation in this side stem is essential for the translation. Also, the P-1 construct did not produce any product, but the P-2 construct did (Fig. 1E ▶, lanes 6,7). Although the amount of CP-Fluc was faint in the P-2 construct, we considered that this ineffective translation was caused by replacement of nucleotides comprising PK I. Because IRES-mediated translation depends on its tertiary structure, mutations introduced into the IRES sequence sometimes decrease translation efficiency. Indeed, inefficient translation of compensatory mutants in PK I was also observed in PSIV, cricket paralysis virus (CrPV), and Rhopalosiphum padi virus (RhPV; Domier et al. 2000; Sasaki and Nakashima 2000; Wilson et al. 2000b). In addition, the P-3 construct having the revertant GC base pair in the third pair in PK I showed improved translation efficiency (Fig. 1E ▶, lane 8). These observations indicate that the IGR structure of TSV is required for translation.
One of the reasons that the intergenic regions of TSV and ABPV have not been clearly identified as IGR-IRES is that the two viruses have in-frame AUG triplets at 6- and 30-nucleotides upstream of the 5′-end of their capsid coding regions (Govan et al. 2000; Mari et al. 2002). Here we have shown that the upstream in-frame AUG triplet in TSV does not function as an initiator and that the predicted extra side stem (Nishiyama et al. 2003) is essential for initiation. Structural similarity among IGRs in TSV, ABPV, and the recently sequenced KBV suggest that these three viruses have a structurally distinct domain 3 for IGR-IRES.
Computational search for IRES elements
Because Met-tRNAi is absolutely required for translation initiation in normal eukaryotic mRNAs (Drabkin et al. 1998), the finding of IGR-IRES-mediated initiation of translation raises a question of whether eukaryotic cellular mRNAs initiate translation at AUG-unrelated codons, suggesting the possibility that IGR-IRES–mediated initiation of translation may be a more general phenomena (McCarthy 2000; Pe’ery and Mathews 2000; Wilson et al. 2000a; Gerlitz et al. 2002). To identify sequences that might be able to form secondary structures similar to IGR-IRES elements, we searched nucleotide sequence databases listed in Table 1 ▶ using the pattern-matching program Scan For Matches with parameters representing the secondary structure of the IGR-IRES elements.
TABLE 1.
Databases used for computational search
| Database | Source | URL | Upload date |
| Viruses | DNA Data Bank of Japan | http://www.ddbj.nig.ac.jp/ | September 2003 |
| Drosophila melanogaster | Berkeley Drosophila Genome Project | http://www.fruitfly.org/ | January 2003 |
| Caenorhabditis elegans | WormBase | http://www.wormbase.org/ | December 2002 |
| Arabidopsis thaliana | The Arabidopsis Information Resource | http://www.arabidopsis.org/info/agi.jsp | March 2003 |
| Homo sapiens | Human Genome Resources | http://www.ncbi.nlm.nih.gov/genome/guide/human/ | April 2003 |
| Full-length human cDNA sequencing project | http://cdna.ims.u-tokyo.ac.jp/ | April 2000 | |
| Mus musculus | Mouse Genome Resources | http://www.ncbi.nlm.nih.gov/genome/guide/mouse/ | February 2003 |
| Functional Annotation of Mouse | http://www.gsc.riken.go.jp/e/FANTOM/ | December 2002 | |
| Oryza sativa | Knowledge-based Oryza Molecular biological Encyclopedia | http://cdna01.dna.affrc.go.jp/cDNA | July 2003 |
| Saccharomyces cerevisiae | Comprehensive Yeast Genome Database | http://mips.gsf.de/genre/proj/yeast/index.jsp | February 2002 |
| 5′ UTR | UTR Database | http://bighost.area.ba.cnr.it/BIG/UTRHome/ | February 2003 |
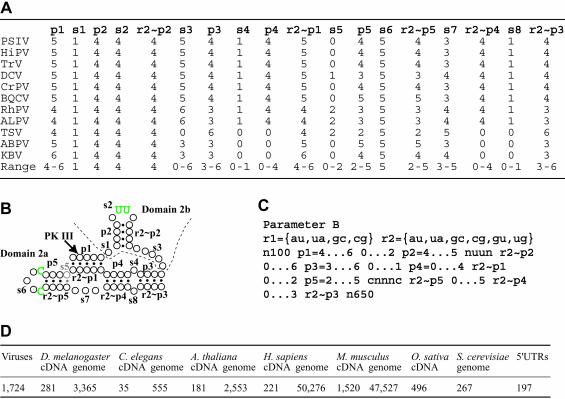
The initial search required a lot of CPU time because of the size of the target database and the need to search complementary sequences. This suggested that repeated searches with modified parameters to examine divergent structures would be difficult. To facilitate repeated searches, we first designed a screening parameter that extracts the most conserved region, domain 2 (Fig. 2B ▶). To date, whole genome sequences of 11 dicistroviruses have been reported. To consider upper and lower limits of length in each structural unit, nucleotide length of core domain 2 in 11 dicistroviruses were counted (Fig. 2A ▶) and parameter B was constructed (Fig. 2C ▶). The rule of parameters used in Scan for Matches is described in detail by Dsouza et al. (1997). In brief, r1 and r2 define base pairs permitted for helical segments. In our case, r1 permits AU and GC pairs and r2 permits AU, GC, and GU pairs. Paired segments are designated with the letter “p,” and each paired segment has been numbered. Upper and lower limits in the nucleotide length of each structural unit were defined by “n…n.” Although we used the letter “s” for single-stranded regions to facilitate description for Figure 2A and B ▶, the letter “s” is unnecessary for parameters. Complementary sequences for helical regions were defined with “~pn” according to permitted base pairs, r1 or r2. We defined nucleotides in domain 2b loop as nuun, because the two uracils interact with the 40S ribosome and these nucleotides are completely conserved in dicistroviruses (Nishiyama et al. 2003). Also, the loop in domain 2a was defined as cnnnc for similar reasons. In cases of Drosophila C virus (DCV), aphid lethal paralysis virus (ALPV), RhPV, and TSV, there is a single-stranded region between r2~p1 and p5. This region was described using s5 (Fig. 2A,B ▶). To avoid exclusion of putative domain 2-like sequences during the initial search, pattern units were loosened at s1, p2, s7, and s8 (Fig. 2A,C ▶). Parameter B was designed to retrieve 100 and 650 nucleotides upstream and downstream, respectively, of the putative domain 2 to allow a more detailed search later. For the UTR database search, the last pattern unit was shortened to 160 nt, because UTR sequences do not contain coding regions.
FIGURE 2.
Initial screening of domain 2-like sequences from databases. (A) List of number of nucleotides in each structural unit in domain 2 of IGR-IRES elements of dicistroviruses. The last line in each column represents the minimum and maximum number of nucleotides in the unit in dicistroviruses. Abbreviation of virus names: PSIV, Plautia stali intestine virus; HiPV, Himetobi P virus; TrV, Triatoma virus; DCV, Drosophila C virus; CrPV, Cricket paralysis virus; BQCV, Black queen-cell virus; RhPV, Rhopalosiphum padi virus; ALPV, Aphid lethal paralysis virus; TSV, Taura syndrome virus; ABPV, Acute bee paralysis virus; KBV, Kashmir bee virus. (B) A representative secondary structure model of domain 2 of IGR-IRES in dicistroviruses. Circles and dots indicate nucleotides and base-pair formations, respectively. Characters “p” and “s” denote sequentially designated structural units. Complementary sequences in the model are shown as r1~pn or r2~pn according to permission for nucleotide pairs comprising each helical segment. Green letters indicate nucleotides that are recognized by the 40S ribosome. s5, which is observed in DCV, RhPV, ALPV, and TSV, is shown in halftone. (C) Parameter B used for initial database search. (D) Result of the initial search against databases listed in Table 1 ▶ using parameter B. Extracted sequence files are available from http://cse.nias.affrc.go.jp/nakaji/RNA_F2DF3C.htm.
Parameter B extracted 1724 sequences from the viral database; in total, 2931 sequences from cDNA databases including 5′ UTRs, and 104,543 sequences from genomic databases (Fig. 2D ▶). Result files extracted by parameter B were concatenated to multifasta files for cDNAs and genomes, then used for detailed analysis.
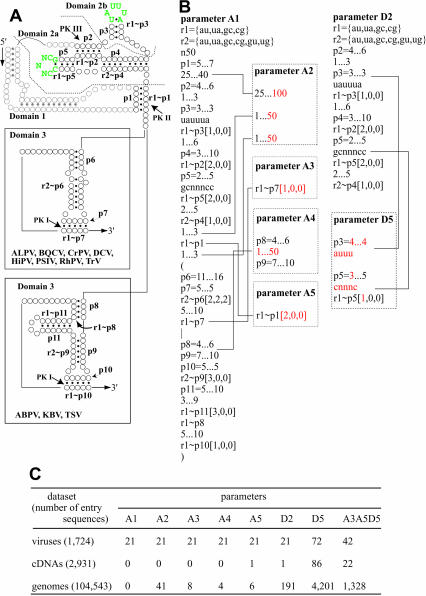
Figure 3A ▶ shows a representative secondary structure of IGR-IRES of dicistroviruses. The structural model has been experimentally verified for CrPV and PSIV (Jan and Sarnow 2002; Nishiyama et al. 2003). As shown in Figure 1 ▶, domain 3 of TSV and two related viruses has an additional side stem loop. To consider this divergent structure, pattern units for domain 3 were constructed using alternative modes A or B, indicated as (A|B) in Figure 3B ▶, parameter A1. To facilitate modification of parameters, short single-stranded regions located within or the end of helical regions were considered as mismatches, insertions, or deletions in the complementary sequences (Fig. 3B ▶). Because base pairs at the root of loop sequences in domains 2a and 2b are absolutely conserved in dicistroviruses, these regions are defined as GCNNNCC and UAUUUA.
FIGURE 3.
Detailed structure search against extracted data set in Figure 2D ▶. (A) A representative secondary structure model of IGR-IRES in dicistroviruses. Circles and dots indicate nucleotides and base-pair formations, respectively. Halftones in domain 1 means these regions are not included parameters. The two types of domain 3 are separately shown. Characters “p” denote sequentially designated structural units. Complementary sequences in the model are shown as rn~pn according to permission for nucleotide pairs comprising each helical segment. Green letters indicates conserved loop sequences in dicostroviruses. (B) Parameters used for detailed search. To extract dicistroviral IGR-IRES sequences, parameter A1 was constructed. Three numbers in square brackets represent the number of nucleotides permitted for mismatches, insertions, and deletions, respectively. Parameters A2, A3, A4, and A5 were modified from parameter A1 to loosen the search definitions. The loosened structural units were marked in red and linked to parameter A1 with lines. Parameter D2 extracts sequences similar to domain 2. Parameter D5 is a loosened version of parameter D2. (C) The result of a detailed search against data set extracted in Figure 2D ▶ using parameters shown in (B). Extracted sequence files are available from http://cse.nias.affrc.go.jp/nakaji/RNA_F2DF3C.htm.
When we searched the data set for viral sequences, parameter A1 extracted 21 sequences (Fig. 3C ▶). Among them, 11 sequences were IGR-IRES elements in distinct dicistroviruses and 10 sequences were those of geographic isolates of ABPV (Bakonyi et al. 2002). The parameter A1 identified the 3′ end of the IGR-IRESs of dicistroviruses exactly, except for that of TrV, which had C at the 3′ end of p6 and G at the 5′ end of the capsid coding region. This result indicated that structural units for PK II and domains 2 and 3 were sufficient to detect dicistroviral IGR-sequences from databases. Because parameter A1 did not extract sequences from cDNA and genome data sets (Fig. 3C ▶), we loosened the parameter at several regions. Parameter A2 extended the region between PK II and PK III up to 100 nt and also extended regions around PK II up to 50 nt (Fig. 3A,B ▶). Parameter A3 permitted one nucleotide mismatch at p7. Parameter A4 permitted up to a 50-nt insertion between P8 and P9. Parameter A5 permitted two nucleotide mismatches at PK II. These loosened parameters extracted several sequences (Fig. 3C ▶). Modeling of the secondary structures of the sequences extracted by parameters A2, A3, A4, and A5 suggested that the structures would be unstable in comparison to those of dicistroviruses. Because the instability was thought to be caused by mismatches at p4, p5, p6, and p9, we further examined tighter parameters in these regions but they did not extract sequences other than dicistroviruses.
It might be expected that invariant or closely related domain 2-like sequences may interact with ribosomes because synthesized domain 2 RNA of PSIV has been shown to bind with salt-washed 40S ribosomes (Nishiyama et al. 2003). When we searched the data sets using parameter D2 that extract invariant domain 2, one sequence was extracted from cDNAs, but this same sequence was also extracted with parameter A5. For the genomic data set, 191 sequences were extracted by parameter D2. Because a more relaxed parameter D5 extracted a lot of sequences, we again searched the data sets with parameter A3A5D5, including all modifications in parameter A3, parameter A5, and parameter D5. This extracted 42 sequences from the viral data set. When reading frames downstream of the PK I were analyzed in these sequences, all of candidates contained stop codons within 200 nt, except for sequences of dicistroviruses. Among 22 cDNA sequences, only one sequence could give a predicted translation product longer than 100 amino acids (BC015621, Homo sapiens chromosome 14, open reading frame 32); however, secondary structure modeling indicated that this sequence was also unstable because of an AU-rich PK III and disordered base pairs in the p6 stem (secondary structure models are located at http://cse.nias.affrc.go.jp/nakaji/RNA_F2DF3C.htm).
Is functional IGR-IRES specific for dicistroviruses?
Despite repeated search using loosened parameters, as shown in Figure 3B ▶, we could not identify sequences that form stable secondary structures resembling the IGR-IRES of dicistroviruses. These results suggested that IGR-IRES may be specific for dicistroviruses. Our parameters, however, might be too stringent to find divergent IGR-IRES-like sequences. Because allowing insertions among domains, as in parameter A2, increased extracted sequences, additional structures might be present in divergent IGR-IRES-like elements. In addition, our genomic database searches did not consider genes produced by splicing. Currently, 54 cellular mRNAs are proposed to have an IRES in the IRES database (Bonnal et al. 2003). To estimate the rate of sequences having introns in reported IRES elements, we compared reported IRES sequences and genomic sequences using the BLAST. As a result, proposed IRES elements in 18 of 54 mRNAs were suggested to be produced by splicing. This leaves the possibility that eukaryotic mRNAs that are not represented in cDNA databases may fold into divergent structures partly resembling IGR-IRES. To find such novel RNA structures, other approaches such as screening of mRNAs that can form functional complexes with ribosomes in the absence of eukaryotic initiation factors, would be required.
Recent analyses using the IGR-IRES of CrPV show that the elongation cycle occurs in the absence of eukaryotic initiation factors in vitro (Jan et al. 2003; Pestova and Hellen 2003). However, analyses using cell lines show that transfected IGR-IRES does not work effectively under normal cellular conditions (Thompson et al. 2001; Masoumi et al. 2003), indicating that the efficiency of IGR-IRES–mediated translation is correlated with viral regulation of host translation pathways. Taken together, these experimental observations and the results of our computer-based structure search suggest that, with the exception of the dicistroviruses, IGR-IRES–like sequences are not common. If eukaryotic mRNAs are translated independently of Met-tRNAi, their structures must be significantly distinct from those of dicistroviruses.
MATERIALS AND METHODS
Plasmid constructions
The cDNA sequence of TSV containing the IGR-IRES (nt 6741–6990, accession number, AF277675) was obtained by chemically synthesized oligo DNA fragments and polymer-ase chain reactions (PCRs). The synthesized IGR sequence, which had XbaI and BamHI recognition sequences on the 3′ and 5′ ends, respectively, were cloned into those sites of pBluescript SK- and sequences were confirmed by DNA sequencing (Takara Custom Services). To construct plasmids for translation assays, pSP-luc+ (Promega) was digested with KpnI, blunted with T4 DNA polymerase, and digested with EcoRI. The resultant 1.7-kb fragments were ligated into pT7Blue (Novagen), which had been digested with HincII and EcoRI, generating pFluc. The TSV sequences from nt 6741 to 6990 were amplified by PCR using a forward primer that contained a HindIII sequence at the 5′ end and reverse primer containing a NcoI sequence at the 5′ end. The amplified PCR products were digested with HindIII and NcoI, then ligated into those sites of pFluc, to construct pIGR-CP-Fluc. Mutations in domain 3 in the TSV sequence were introduced by PCR-based mutagenesis as described (Kanamori and Nakashima 2001). For dicistronic constructs, a fragment containing the Renilla luciferase (Rluc) gene, nt 309–1251 of pRL-null vector (Promega), was amplified with PCR. The fragment was phosphorylated, digested with XbaI, and then ligated into pT7Blue, which had been digested with HincII and XbaI, generating pRluc. The 1.7-kb fragment containing Fluc described above and a IGR-CP-Fluc fragment that had been obtained by HindIII digestion, blunting, and EcoRI digestion from pIGR-CP-Fluc, were used for second cistrons. These fragments were ligated into pRluc, which had been digested with XbaI, blunted with a Klenow fragment, and then digested with EcoRI, generating pRluc-Fluc and pRluc-IGR-CP-Fluc.
In vitro transcription and translation
Capped or uncapped RNAs were transcribed from linearized plasmids using a mMESSAGE mMACHINE (Ambion) or a RiboMAX Large Scale RNA Production System (Promega). In vitro translation using a wheat germ extract and biotinylated Lys-tRNA (Promega) was conducted as described (Shibuya et al. 2003). To examine the effect of cap analog (0.5 mM) on translation in vitro, Fluc and Rluc activities in the translation mixture were measured with the SteadyGlo Luciferase assay system (Promega) and Renilla Luciferase assay system (Promega).
Data set for computational search
Data sets containing full-length cDNA sequences or whole chromosome sequences were downloaded from the Web sites shown in Table 1 ▶. As for the 5′ UTR data set, we used data for fungi, human, invertebrates, mammals, vertebrates, plants, rodents, viruses, and mouse. The 5′ UTR sequences were converted to fasta format using the program ReadSeq (IUBIO Archive for Biology http://iubio.bio.indiana.edu/) and concatenated to a multifasta file. The downloaded cDNA or chromosome sequence files in each genome project and viral databases in DDBJ were also concatenated to single multifasta files.
Software and hardware
The Scan For Matches pattern match program (Dsouza et al. 1997) was downloaded from http://www-unix.mcs.anl.gov/compbio/PatScan/. The source file for Scan For Matches was compiled on the IRIX ver.6.5.9 in SGI Origin 3800 Scalar Operation Server (Silicon Graphics, Inc.) at the Ministry of Agriculture, Forestry, and Fisheries (MAFF) Scientific Computing System. The original program keeps 10 MB of memory prior to scanning the data set; however, this memory size was not enough for dealing with a large chromosome sequence data set. Therefore, we modified the source code of the program to read the largest size of entry sequences in the target multifasta file prior to memory storage. The modified program keeps memory at a maximum of 500 MB according to the largest entry sequence. Modification of the source code was done by consulting with MAFF Scientific Computing System Customer Support.
The sequence information of extracted sequences in output files was obtained by using Web-based sequence identifier, DBGET program on Genome Net (Japanese network of database and computational services for genome research and related research areas in molecular and cellular biology; http://www.genome.ad.jp/dbget/). The number of extracted sequences was counted by the program “Get Max Length,” originally provided by AFFRC Scientific Computing System Support.
For a brief search to check parameters, we used the Web site serving PatSearch, which is a sister program of Scan For Matches (Pesole et al. 2000; Grillo et al. 2003).
Acknowledgments
We thank M. Shimomura (Mitsubishi space software, Co., Ltd.) for the initial stage of this work, and members of MAFFIN for advice on the use of the Scientific Computing System. This work was supported by a grant from PROBRAIN, Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5208104.
REFERENCES
- Bakonyi, T., Grabensteiner, E., Kolodziejek, J., Rusvai, M., Topolska, G., Ritter, W., and Nowotny, N. 2002. Phylogenetic analysis of acute bee paralysis virus strains. Appl. Environ. Microbiol. 68: 6446–6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal, S., Boutonnet, C., Prado-Lourenco, L., and Vagner, S. 2003. IRESdb: The internal ribosome entry site database. Nucleic Acids Res. 31: 427–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier, L.L., McCoppin, N.K., and D’Arcy, C.J. 2000. Sequence requirements for translation initiation of Rhopalosiphum padi virus ORF 2. Virology 268: 264–271. [DOI] [PubMed] [Google Scholar]
- Drabkin, H.J., Estrella, M., and RajBhandary, U.L. 1998. Initiatorelongator discrimination in vertebrate tRNAs for protein synthesis. Mol. Cell. Biol. 18: 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza, M., Larsen, N., and Overbeek, R. 1997. Searching for patterns in genomic data. Trends Genet. 13: 497–498. [DOI] [PubMed] [Google Scholar]
- Gerlitz, G., Jagus, R., and Elroy-Stein, O. 2002. Phosphorylation of initiation factor-2α is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 269: 2810–2819. [DOI] [PubMed] [Google Scholar]
- Govan, V.A., Leat, N., Allsopp, M., and Davison, S. 2000. Analysis of the complete genome sequence of acute bee paralysis virus shows that it belongs to the novel group of insect-infecting RNA viruses. Virology 277: 457–463. [DOI] [PubMed] [Google Scholar]
- Grillo, G., Licciulli, F., Liuni, S., Sbisa, E., and Pesole, G. 2003. Pat-Search: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 31: 3608–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U.T. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Jan, E. and Sarnow, P. 2002. Factorless ribosome assembly on the internal ribosome entry site of Cricket paralysis virus. J. Mol. Biol. 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jan, E., Kinzy T.G., and Sarnow, P. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. 100: 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S.K., Krausslich, H.G., Nicklin, M.J.H., Duke, G.M., Palmenberg, A.C., and Wimmer, E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori, Y. and Nakashima, N. 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7: 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- Mari, J., Poulos, B.T., Lightner, D.V., and Bonami, J-R. 2002. Shrimp Taura syndrome virus: Genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol. 83: 915–926. [DOI] [PubMed] [Google Scholar]
- Masoumi, A., Hanzlik, T.N., and Christian, P.D. 2003. Functionality of the 5′- and intergenic IRES elements of cricket paralysis virus in a range of insect cell lines, and its relationship with viral activities. Virus Res. 94: 113–120. [DOI] [PubMed] [Google Scholar]
- Mayo, M.A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch Virol. 147: 1655–1656. [DOI] [PubMed] [Google Scholar]
- McCarthy, J.E.G. 2000. Translation initiation: Insect virus RNAs rewrite the rule book. Curr. Biol. 10: R715–R717. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Yamamoto, H., Shibuya, N., Hatakeyama, Y., Hachimori, A., Uchiumi, T., and Nakashima, N. 2003. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 31: 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’ery, T. and Mathews, M.B. 2000. Viral translational strategies and host defense mechanisms. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 371–424. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pelletier, J. and Sonenberg, N. 1988. Internal initiation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Pesole, G., Liuni, S., and D’Souza, M. 2000. PatSearch: A pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics 16: 439–450. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V. and Hellen, C.U.T. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, J. and Nakashima, N. 1999. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 73: 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl Acad. Sci. 97: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya, N., Nishiyama, T., Kanamori, Y., Saito, H., and Nakashima, N. 2003. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J. Virol. 77: 12002–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, S.R., Gulyas, K.D., and Sarnow P. 2001. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. 98: 12972–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Galy, B., and Pyronnet, S. 2001. Irresistible IRES, attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U.T., and Sarnow, P. 2000a. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Powell, M.J., Hoover, S.E., and Sarnow, P. 2000b. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20: 4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]