Abstract
In vitro transcription has proven to be a successful tool for preparation of functional RNAs, especially in the tRNA field, in which, despite the absence of post-transcriptional modifications, transcripts are correctly folded and functionally active. Human mitochondrial (mt) tRNALys deviates from this principle and folds into various inactive conformations, due to the absence of the post-transcriptional modification m1A9 which hinders base-pairing with U64 in the native tRNA. Unavailability of a functional transcript is a serious drawback for structure/function investigations as well as in deciphering the molecular mechanisms by which point mutations in the mt tRNALys gene cause severe human disorders. Here, we show that an engineered in vitro transcribed “pseudo-WT“ tRNALys variant is efficiently recognized by lysyl-tRNA synthetase and can substitute for the WT tRNA as a valuable reference molecule. This has been exploited in a systematic analysis of the effects on aminoacylation of nine pathology-related mutations described so far. The sole mutation located in a loop of the tRNA secondary structure, A8344G, does not affect aminoacylation efficiency. Out of eight mutations located in helical domains converting canonical Watson–Crick pairs into G–U pairs or C•A mismatches, six have no effect on aminoacylation (A8296G, U8316C, G8342A, U8356C, U8362G, G8363A), and two lead to drastic decreases (5000- to 7000-fold) in lysylation efficiencies (G8313A and G8328A). This screening, allowing for analysis of the primary impact level of all mutations affecting one tRNA under comparable conditions, indicates distinct molecular origins for different disorders.
Keywords: in vitro transcripts, tRNA, aminoacylation, human mitochondrial, MERRF, identity, post-transcriptional modification
INTRODUCTION
Specific aminoacylation of transfer RNAs (tRNA) by their cognate aminoacyl-tRNA synthetases (aaRS) is a key step in translation of genetic information. Rules governing the recognition process between the partner macromolecules have been well studied for a large number of prokaryotic and cytosolic eukaryotic systems (for review, see Giegé et al. 1998b; McClain 1993a,McClain 1995; Saks et al. 1994; Giegé and Frugier 2003). On the other hand, far less is known for mitochondrial (mt) tRNAs and aaRSs, especially from human and mammals in general (Florentz and Sissler 2003). This is a drawback not only in the frame of fundamental knowledge about these mt aminoacylation systems but also in regard to understanding molecular mechanisms underlying numerous human disorders. Over the past 15 years, an increasing number of point mutations within human mt tRNA genes have been correlated with severe neurodegenerative and/or muscular disorders (e.g., Larsson and Clayton 1995; Schon et al. 1997; Wallace 1999; DiMauro and Andreu 2000; Florentz and Sissler 2001). More than 90 point mutations have been reported so far, located in 21 of the 22 mt tRNA genes (e.g., Kogelnik et al. 1998; Servidei 2002). A large majority of these mutations lead to a decrease in the rate of mitochondrial protein synthesis. However, the detailed molecular level at which each mutation interferes with the tRNA lifecycle remains mostly unknown. tRNA aminoacylation being the key step of protein biosynthesis, a systematic investigation of the effects of point mutations in tRNAs on this process would allow for a straightforward comparison.
As opposed to studies on classical tRNAs, investigation of human mt tRNA aminoacylation systems suffers from a series of limitations linked to intrinsic properties of mitochondria. Their heteroplasmic status, that is, the presence of both WT and mutant DNA in the same cell, requires the creation of individual trans-mitochondrial cell-lines for in vivo investigation of each mutation (King and Attardi 1989). Although such cells allowed for evaluation of steady-state levels of aminoacylation (for review, see by Enriquez and Attardi 1996; Florentz and Sissler 2003), their creation is difficult so that only a handful of mutations could be analyzed in such a way so far. Moreover, the variety of cell types used as a basis for creation of cybrids (e.g., HeLa, osteosarcoma, lymphoblasts) does not allow for straightforward comparisons, due to variations in nuclear backgrounds (Jacobs and Holt 2000). Ex vivo analyses, based on extraction of a specific tRNA from cultured cells or from biopsies, are limited by the quantities of accessible material. This has only been reported in two cases so far (Yasukawa et al. 2001; Park et al. 2003). The alternative approach consists of producing tRNA through in vitro transcription of cloned WT and mutated human mt tRNA genes as has been the case for cohorts of canonical tRNAs (Giegé et al. 1998b; Giegé and Frugier 2003). Such an approach, based on tRNAs deprived of post-transcriptional modifications, has been validated over the past 20 years for a vast majority of aminoacylation systems from various organisms and allowed, for example, the deciphering of aminoacylation identity elements in specific tRNAs. Interestingly, the in vitro approach leads to similar results to those obtained by in vivo methods (for review, see McClain 1993a,b; Giegé et al. 1998b).
In regard of human mt tRNAs, the approach based on the in vitro synthesis of transcripts was found to be efficient and has already been explored in the case of isoleucine-specific (Degoul et al. 1998; Kelley et al. 2000 Kelley et al. 2001) and serine-specific (Shimada et al. 2001) tRNAs but lead to a number of difficulties for other mt tRNAs. Indeed, at least those transcripts specific for lysine (Helm et al. 1998; Tolkunova et al. 2000) and phenylalanine (Bullard et al. 1999) turned out to be unsatisfactory substrates for aaRSs due to unfavorable folding of the RNAs. In the case of tRNALeu(UUR), aminoacylation is possible despite structural flexibility of the transcript compared with native tRNA (Park et al. 2003; Sohm et al. 2003; Wittenhagen et al. 2003). Because in vitro transcription remains the sole conceivable straightforward approach to access human mt tRNAs (wild type and mutants) and to perform rigorous systematic comparative analysis of tRNA variants for a given function (e.g., maturation, aminoacylation), functional in vitro transcripts need to be engineered.
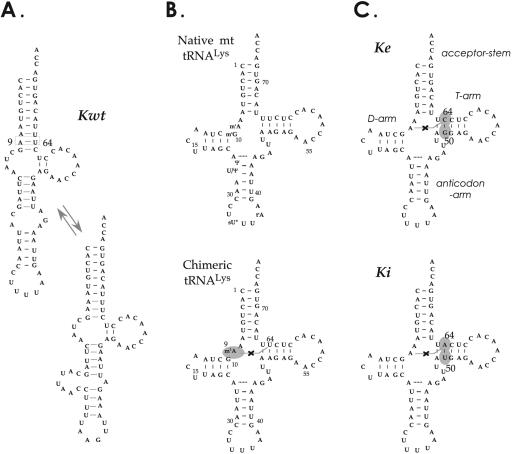
Here, we consider the specific case of human mt tRNALys. Previous work showed that an in vitro transcript of this tRNA does not fold into a functional cloverleaf but into alternative extended bulged hairpins (Fig. 1A ▶; Helm et al. 1998), and is poorly aminoacylated in the presence of a human mt crude enzymatic extract (Helm et al. 1998) or of an overexpressed and purified mt LysRS (Tolkunova et al. 2000). The requirement of a single post-transcriptional modification for cloverleaf folding of tRNALys was confirmed by synthesis and structural analysis of a chimeric tRNALys, which displays a unique modified base, a methylated adenosine at position 9 (Fig. 1B ▶; Helm et al. 1999b). This modification impairs the formation of a Watson–Crick base pair between nucleotides 9 and 64 responsible for the extension of the amino acid acceptor stem within the transcript. Interestingly, the structural chaperone effect of m1A9 modification can be simulated by mutating either nucleotide 9 or nucleotide 64 (provided base-pairing between residues 64–50 is conserved; Helm et al. 1998). Such variants, carrying an altered base pair at position 50–64 replacing A50–U64 by U50–A64 (Ki) or by G50–C64 (Ke), adopt the expected cloverleaf folding, as demonstrated by enzymatic and chemical probing (Fig. 1C ▶; Helm et al. 1998).
FIGURE 1.
Human mitochondrial wild-type and pseudo-WT tRNALys transcripts. (A) Determined secondary structures of human mt wild-type tRNALys transcript (Kwt; Helm et al. 1998). (B) Experimental secondary cloverleaf structures of native mt tRNALys (displaying the six post-transcriptional modifications, abbreviated in the conventional way; Sprinzl et al. 1998) and of a chimeric tRNALys methylated at position N1 of A9 as sole post-transcriptional modification (Helm et al. 1999b). (C) Pseudo-WT in vitro transcribed tRNALys derivatives Ke and Ki. tRNA domains are labeled for Ke. Structural mutations introduced at positions 50 and 64 (indicated by gray circles) prevent alternate folding into extended hairpin (black crosses; Helm et al. 1998).
Here, it is shown that these engineered variants are active substrates for LysRS and that they represent valuable reference molecules for further investigation of tRNALys nucleotides important for aminoacylation. The lysylation system has further been explored in a systematic investigation of the full set of mutations described so far for the human mt tRNALys gene, a hot spot for pathology-related mutations. Results are discussed both in the frame of molecular mechanisms involved in the expression of the diseases, and in the frame of understanding lysylation rules in human mitochondria.
RESULTS
Setting up an efficient lysylation system based on in vitro transcripts
The in vitro transcribed structural variants Ke and Ki of human mt tRNALys (Fig. 1C ▶) were previously shown to adopt the expected cloverleaf secondary structure (Helm et al. 1998). As the presence of a C-residue at the first position of tRNALys highly limits yields of in vitro transcription, a G being most favorable for T7 RNA polymerase efficiency (Sampson and Uhlenbeck 1988), new synthetic genes have been cloned. They consist of the T7 promoter followed by a hammerhead ribozyme (Rz; Fechter et al. 1998) and the tRNA gene, leading to wild-type (Rz-Kwt) and “pseudo-WT” (Rz-Ke and Rz-Ki) transcripts. These transcripts were tested for their lysylation properties compared with those of human mt native WT tRNALys (extracted from cybrid cell lines, see Experimental Procedures). Because it is laborious to obtain native tRNAs in the large amount required for determination of kinetic parameters, only charging levels have been established for this initial investigation (Fig. 2 ▶). The aminoacylation assays were performed in the presence of human cytosolic lysyl-tRNA synthetase. Both cytosolic and mt enzymes are encoded by the same nuclear gene and differ in their mature versions only by 21 to 49 (over 601) amino acids at their N terminus (Tolkunova et al. 2000).
FIGURE 2.
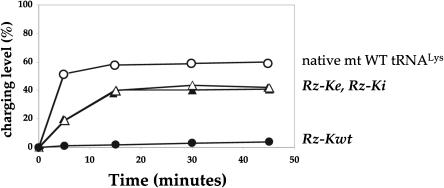
From inactive to active in vitro transcribed human mt tRNALys. Charging levels, determined with human LysRS at 37°C, are expressed as percentages of charged tRNA versus total amount of specific tRNA per experiment. Rz-Kwt, Rz-Ke, and Rz-Ki correspond to in vitro transcribed wild-type and structural variants of tRNALys, respectively, purified to single nucleotide resolution on polyacryl-amide/urea gels. Purified native wild-type mt tRNALys (by hybridization to a specific biotinylated oligonucleotide) form the positive control.
Although Rz-Kwt, the transcript which folds into extended hairpin structures, is poorly aminoacylated, Rz-Ke (with A50–U64 replaced by G50–C64) and Rz-Ki (with A50–U64 replaced by U50–A64) become active substrates for human LysRS with 42% ± 2% charging levels, an activity close to the 57 ± 2% measured for native mt WT tRNALys. Rz-Ke and Rz-Ki display identical functional properties, demonstrating that the residues paired at positions 50 and 64 do not influence recognition by LysRS.
Effect of MERRF mutation A8344G on aminoacylation levels and on tRNALys structure
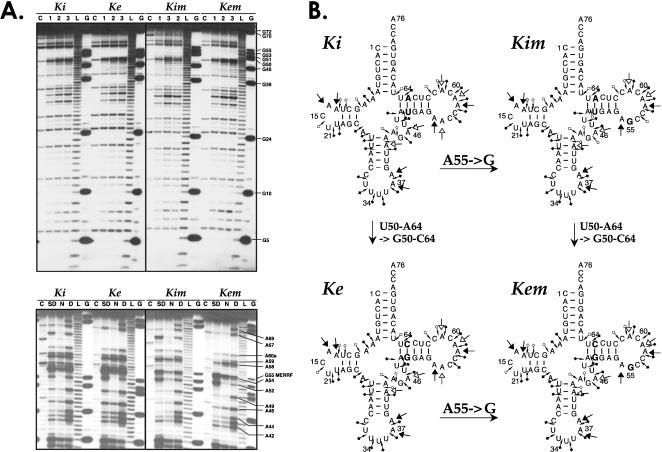
Among the 13 pathology-related mutations described within the mt tRNALys gene, transition A8344G,2 correlated with the MERRF syndrome (Shoffner et al. 1990; Servidei 2002), is the most prevalent mutation. This mutation, located in the T-loop of the cloverleaf-folded tRNA at position 55 (conventional tRNA numbering, see Fig. 5 ▶ below), has been introduced into Rz-Ke and Rz-Ki, leading to Rz-Kem and Rz-Kim. Aminoacylation properties as well as structural properties have been investigated for these variants. Interestingly, both Rz-Kem and Rz-Kim are aminoacylated to levels similar to the corresponding pseudo-WT transcripts (Fig. 3 ▶). Under optimal LysRS concentrations, all four transcripts are charged up to 42 ± 2% (Fig. 3 ▶).
FIGURE 5.
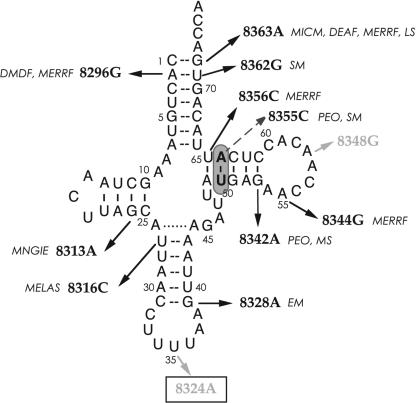
Investigated mutations in human mitochondrial tRNALys. Individual variants were prepared as derivatives of Rz-Ki, the pseudo-WT tRNALys with U50–A64 pair instead of A50–U64 pair. The 10 reported pathological mutations (http://www.mitomap.org/) are indicated in black; the dashed arrow represents the one not investigated because it overlaps the structural mutations. Control molecules, indicated in gray, are either the polymorphic A8348G mutation (positive control) or the U8324A sequence variation (which alters a major lysine identity element, negative control). Numbering is either according to the mt genome location (bold characters, for mutations; Anderson et al. 1981) or according to the conventional tRNA annotation (small numbers; Sprinzl et al. 1998). Related pathologies are abbreviated as follows: DEAF, maternally inherited deafness or aminoglycoside-induced deafness; DMDF, diabetes mellitus and deafness; EM, encephalomyopathy; LS, Leigh syndrome; MELAS, mitochondrial encephalomyopathy, lactic acidosis, stroke-like episodes; MERRF, myoclonic epilepsy and ragged red fibers; MICM, maternal inherited cardiomyopathy; MNGIE, mitochondrial neurogastrointestinal encephalomyopathy; MS, myoclonic seizures; PEO, progressive external ophthalmoplegia; and SM, skeletal myopathy.
FIGURE 3.
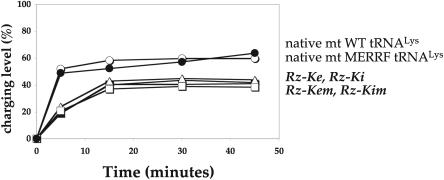
Effect of MERRF (A8344G) mutation on aminoacylation. Charging levels of wild type or pseudo-WT (Rz-Ke and Rz-Ki) versus MERRF or pseudo-MERRF (Rz-Kem and Rz-Kim) tRNAs. Native wild-type and MERRF-mutated mt tRNALys were extracted from mitochondria of R2–1A and R1–C3 cybrid cells (see Experimental Procedures) using biotinylated oligonucleotides complementary to the 3′ part of the tRNA. Rz- stands for self-cleaved in vitro transcribed tRNAs.
The aminoacylation properties of native human mt tRNALys, either wild type or with the MERRF mutation, have been established. tRNAs have been extracted from a specific couple of cybrid cell lines (R2–1A and R1–C3, respectively) by hybridization to a complementary biotinylated oligo-nucleotide. The yield of pure native tRNALys species is extremely low (0.3 μg of pure wild type or MERRF-mutated tRNAs were extracted from ~1 or ~1.3 L of corresponding adherent cell cultures, respectively), so that only charging levels could be established. Figure 3 ▶ shows that both native tRNALyss are lysylated to about the same level (57 ± 2%). Thus, as is the case for transcripts, no substantial effect of the MERRF mutation is observed on aminoacylation of native tRNAs.
Possible structural effects of MERRF mutation have been sought. End-labeled transcripts were submitted to chemical probing with DEPC and Pb(OAc)2, as previously performed for Kwt, Ke, and Ki (Helm et al. 1998). Autoradiographies of comparative structural probing experiments on Ke, Ki, Kem, and Kim are displayed in Figure 4A ▶. Cleavages were categorized weak (white), moderate (gray), or strong (black), and the results for individual nucleotides are summarized in Figure 4B ▶. The reactivities to probes of all four transcripts were virtually identical. The probes failed to detect any significant difference between the pseudo-WT analogs of mt tRNALys on one hand (Ki and Ke), and the “pseudo-MERRF” analogs on the other hand (Kim and Kem).
FIGURE 4.
Effect of MERRF mutation on the structure of in vitro transcribed tRNAs. (A) Autoradiographs of 15% denaturing polyacrylamide gels of chemical probing experiments. G indicates RNase T1 ladder; L, alkaline ladder; and C, control incubation without probe. (Top) Structural probing experiments with lead acetate (Pb(OAc)2). 5′-Labeled tRNAs are incubated at 25°C in 20 μL of 40 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 40 mM NaCl, and 0.1 μg/μL unlabeled tRNA without (C) or with 0.15 mM Pb(OAc)2 for 10 min (1), 20 min (2), or 30 min (3). (Bottom) Chemical probing with diethylpyrocarbonate (DEPC). N, SD, and D stand for native, semi-denaturating, and denaturating conditions, respectively. 5′-Labeled tRNAs are incubated for 40 min without (C) or with 5% (v/v) DEPC (N, SD, D) in 50 mM sodium cacodylate with 10 mM MgCl2 and 300 mM KCl (C, N) or with 1 mM EDTA (SD, D). Incubation was performed for 4 min at 60°C for denaturating condition (D). (B) Chemical probing patterns on cloverleaf structures of pseudo-WT (Ke and Ki) and pseudo-MERRF (Kem and Kim) transcripts. Structural mutations (at position 50–64) and MERRF mutation (at position 55) are highlighted in bold. Arrowheads correspond to positions cleaved by probes (Pb(OAc)2 and DEPC). Cleavages are categorized weak (white), modest (gray), or strong (black).
Systematic mutagenic analysis
So far, 10 mutations in the human mt tRNALys gene have been reported to be related to pathologies such as muscular disorders, encephalopathies, deafness, ophthalmoplegia, or diabetes (Kogelnik et al. 1998; Servidei 2002). A8348G, a polymorphism unrelated to disorders, was also described (Ingman et al. 2000). These 11 mutations have been individually introduced into the pseudo-wild-type Rz-Ki sequence (Fig. 5 ▶). Only the variant with transition T8355C could not be prepared because the mutation overlaps with the Rz-Ki structural mutation. An additional transcript has been constructed with a sequence modification in the anticodon central position (T8324A, or U35A in classical tRNA nomenclature). This mutation was hypothesized to have a strongly deleterious effect on the lysylation reaction because nucleotides U35 and U36 from the anticodon triplet are known as major lysine specific identity elements in canonical lysylation systems (e.g., in Escherichia coli [McClain et al. 1990; Normanly et al. 1990; Tamura et al. 1992] and human cytosolic tRNALys [Stello et al. 1999]). Three mutations are located in loops (MERRF mutation A8344G, polymorphism A8348G, and the rationally designed mutation in the anticodon triplet). All other mutations are located in stems.
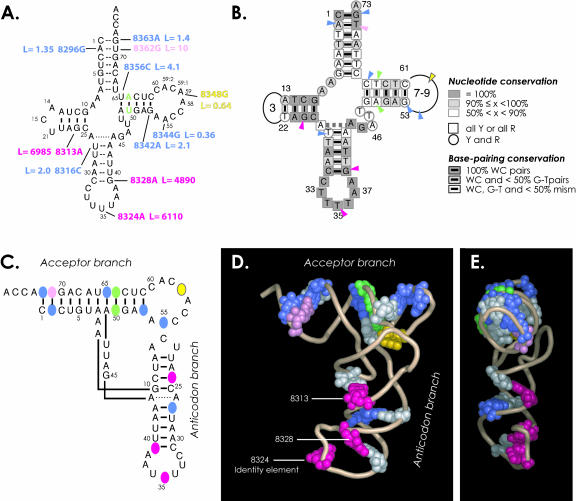
All transcripts have been tested for their ability to be lysylated. Kinetic parameters are presented in Table 1 ▶ (right part). As anticipated, the polymorphic mutation (A8348G) in the T-loop has no significant effect on aminoacylation, with a charging level of 60% (data not shown) and a negligible loss in aminoacylation efficiency. Mutation within the anticodon triplet, replacing U35 by A35, lead to a drastic decrease in lysylation efficiency (~6000-fold). These two variants can thus be considered as positive and negative controls in the lysylation reaction. The nine pathology-related mutants (including the MERRF-related A8344G mutation discussed above) have aminoacylation properties ranking between these substrates. For six of them (A8296G, T8316C, G8342A, A8344G, T8356C, and G8363A), amino-acylation is not significantly impaired by the mutation, with charging levels between 20% and 50% (data not shown) and, at most, a fourfold loss of catalytic efficiency. A small (2.8-fold) gain in efficiency is observed for the MERRF-related mutant A8344G (gain = 1/L). Variant T8362G shows a 10-fold decrease in efficiency. The two remaining variants, G8313A and G8328A, drastically impair lysylation. Charging levels are <1% (data not shown), and losses in catalytic efficiencies reach 4900- and 7000-fold. This poor charging did not allow for direct establishment of individual kinetic parameters kcat and KM for these two variants.
TABLE 1.
Aminoacylation properties of in vitro transcribed pseudo-WT and variant human mt tRNALys in the presence of human LysRS
| In vitro transcripts | tRNA position | Wild-type sequence | Mutated sequence | kcat (10−3 sec−1) | KM (μM) | kcat/KM (10−6 sec−1 • nM−1) | Loss |
| Reference molecule | |||||||
| Rz-Ki | 50–64 (TS) | A–U | U •A | 12 | 4.0 | 3.0 | 1 |
| Polymorphism | |||||||
| A8348G | 59 (AccS) | A | G | 11.8 | 2.4 | 4.9 | 0.6 |
| Anticodon triplet mutation | |||||||
| T8324A | 35 (ACL) | U | A | nd | nd | *0.49 10−3 | 6100 |
| Disease-related mutations | |||||||
| A8296G | 2 (AccS) | A–U | G •U | 10.2 | 4.4 | 2.3 | 1.3 |
| G8313A | 24 (DS) | C–G | C •A | nd | nd | *0.43 10−3 | 7000 |
| T8316C | 27 (ACS) | U–A | C • | 2.4 | 1.5 | 1.8 | 2.0 |
| G8328A | 39 (ACS) | C–G | C •A | nd | nd | *0.61 10−3 | 4900 |
| G8342A | 53 (TL) | G–C | A •C | 8.5 | 4.1 | 2.1 | 1.4 |
| A8344G | 55 (TS) | A | G | 14.3 | 1.7 | 8.4 | 0.3 |
| T8356C | 65 (TS) | A–U | A •C | 0.9 | 1.2 | 0.75 | 4.0 |
| T8362G | 71 (AccS) | A–U | A •G | nd | nd | *0.3 | 10 |
| G8363A | 72 (AccS) | C–G | C •A | 22.9 | 10.3 | 2.2 | 1.4 |
Names of the tRNA variants, affected tRNA positions (conventional tRNA numbering, according to Sprinzl et al. 1998), and sequence changes are given in the left part of the table. tRNA structural domains are abbreviated as follows: AccS, acceptor stem; TS, T-stem; DS, D-stem; ACS, anticodon stem; and TL, T-loop. Aminoacylation parameters measured at 25°C in the presence of human cytosolic LysRS are presented on the right. Loss-values (L) are defined as ratio (kcat/KM)Rz-Kl/(kcat/KM)variant. L-values varied by <10%.
*Ratios kcat/KM were determined directly as described (Schulman and Pelka 1988).
Mutations with low effects on aminoacylation are mainly located in the acceptor- and T-stems of the tRNA and at the upper part of the anticodon stem (Table 1 ▶, left). These mutations can be segregated into two categories from a mechanistic point of view because some have unaffected kinetic parameters kcat and KM (A8296G, G8342A, A8344G, G8363A), whereas others have both a decrease in kcat and in KM (fourfold to 10-fold), leading also to unaffected amino-acylation efficiencies (T8316C, T8356C). Interestingly, both mutations A8296G and T8362G concern the same base pair but lead to low but significant differences in effects on aminoacylation. Mutation A8296G, converting the A2–U71 pair into G2–U71 has no effect, whereas mutation T8362G, leading to an A2 •G71 mismatch, leads to a 10-fold loss in lysylation efficiency. This shows that LysRS is sensitive to some extent to the structural integrity of base pair 2–71 rather than to the nature of the nucleotides present at these positions. Mutations with large negative effects are located within the anticodon domain and within the D-stem (Table 1 ▶, left).
DISCUSSION
From inactive to active in vitro transcribed human mitochondrial tRNALys
In the present work, in vitro transcription was used to produce active molecules despite the absence of post-transcriptional modification required to produce the canonical secondary structure. In the specific case of tRNALys, the extremely low aminoacylation capacity of the wild-type in vitro transcript has been overcome by molecular engineering, leading to an efficient substrate for human LysRS. Exchange of a single Watson–Crick pair by an alternative Watson–Crick pair within the tRNA T-stem simulates the structural chaperone effect of post-transcriptional modification m1A9, favoring the cloverleaf structure (Helm et al. 1999b). Two pseudo-WT ribozyme-derived transcripts, Rz-Ki and Rz-Ke, with structural mutations A50–U64 →U50–A64 or A50–U64 →G50–C64, respectively, become lysylated with aminoacylation capacities significantly increased compared with WT tRNALys transcript Rz-Kwt, close to those of native tRNALys extracted from mitochondria. As the two base pair exchanges lead to identical functional effects, and the corresponding nucleotide positions have not been shown in tRNALys from other species to be involved in LysRS recognition and aminoacylation efficiency, both Rz-Ki and Rz-Ke can be used as reference molecules for further functional exploration of tRNALys.
This has been further confirmed in the case of Rz-Ki by the aminoacylation properties of four variants, namely, those with the polymorphism A8348G, the rationally designed mutation in the anticodon triplet, and the two pathology-related mutations A8344G and G8313A. Polymorphic mutations have been discovered by comparing mt DNA sequences among large human populations and have been explored to reconstruct human prehistory and population movements (e.g., Allen and Raven 1996; Ingman et al. 2000). Such mutations are assumed to be neutral in molecular and phenotype effects. We show here that this is indeed the case for mutation A8348G in regard to aminoacylation properties of the corresponding Rz-Ki derivative. Mutating the anticodon triplet was expected to have a strong negative effect on lysylation. Although aminoacylation identity elements for the human mt lysine system were not explored so far, it could be foreseen by analogy with known lysylation systems (McClain et al. 1990; Tamura et al. 1992; Stello et al. 1999) and on the basis of the general concept that major identity elements are conserved among species (Giegé et al. 1998b; Giegé and Frugier 2003), that mutating the central residue of the anticodon triplet would be detrimental to aminoacylation. Indeed, replacing U35 by A35 in the Rz-Ki framework leads to a ~6000-fold loss in aminoacylation efficiency, demonstrating that the initial replacement of A50–U64 by U50–A64 does not interfere with the expression of identity elements in the tRNA scaffold. Finally, two pathology-related mutations within tRNALys have already been investigated for their impact on lysylation by approaches not including in vitro transcribed tRNAs. The impact of MERRF mutation A8344G has been tested by an in vitro approach based on purification of low amounts of native WT tRNALys and tRNALys with mutation A8344G from large-scale cultures of HeLa-derived cybrid cells (Yasukawa et al. 2001). Kinetic parameters for lysylation of these natural, post-transcriptionally modified molecules revealed a threefold increase in aminoacylation efficiency for the mutant tRNA compared with wild type. This is in line with a 2.8-fold increase observed in the present work on the Rz-Ki derivative with mutation A8344G compared with Rz-Ki and with unchanged aminoacylation plateau levels for native tRNALys species extracted from osteosarcoma-derived cybrid cell lines. Further, in vivo measurements of aminoacylation levels of mutated tRNALys have been performed both for mutation A8344G (Enriquez et al. 1995; Börner et al. 2000) and G8313A (Bacman et al. 2003). Although in the first case, variable results were obtained according to the cell type used (decrease in aminoacylation in osteosarcomaderived cybrid cell-lines, no effect in biopsies), a strikingly negative effect of mutation G8313A has been reported in cybrid cell-lines. This result is in line with the 7000-fold loss in lysylation observed in the present work for Rz-Ki derivative bearing second site mutation G8313A. Based on these results, we conclude that the in vitro transcribed derivative of tRNALys Rz-Ki can indeed be considered as a pseudo-wild-type molecule, relevant for further mutagenic analyses.
Variable impacts of pathology-related mutations on lysylation
Ten mt tRNALys gene mutations have been reported to be associated with phenotypically wide-ranging human disorders. Nine could be comparatively analyzed in regard to their effect on aminoacylation of in vitro transcripts. They segregate into two categories, with seven leading to little effect on aminoacylation (<10-fold loss in aminoacylation efficiency) and two (mutations G8313A and G8328A) leading to severe decreases (4900- to 7000-fold losses in efficiency). These large differences suggest that a defect in aminoacylation efficiency would not be a common primary molecular impact of pathology-related mutations in tRNALys. However, it cannot be ruled out that even a weak effect on aminoacylation, amplified by additional molecular events, might be sufficient to contribute to the disease status. Similar conclusions have been drawn from aminoacylation studies performed on tRNAIle mutations (Kelley et al. 2000,Kelley et al. 2001) and tRNALeu(UUR) mutations (Sohm et al. 2003). As alternatives to aminoacylation, the impact of individual mutations in various tRNAs were found at other functional levels, including transcription, maturation, post-transcriptional modification, stability, binding to elongation factor EF-Tu, and codon reading (for review, see Florentz et al. 2003). In the specific case of tRNALys mutations, only MERRF mutation A8344G has been investigated at different levels. This mutation does not affect mitochondrial transcription (Hanna et al. 1995; Masucci et al. 1995). It does not affect the global post-transcriptional modification pattern of the tRNA (Helm et al. 1999a), except modification at nucleotide 34, which is completely abolished (Yasukawa et al. 2000). The detailed chemical probing analysis in solution performed herein shows that MERRF-mutation A8344G does not alter the tRNA structure. However, the stability of the mutated tRNA is reduced (Enriquez et al. 1995; Yasukawa et al. 2000). Further, its interaction with EF-Tu is decreased (Yasukawa et al. 2001), and most important, its capacity to read the lysine codons is strongly affected (Yasukawa et al. 2001). For most other mutations affecting the human mt tRNALys gene, no information is available as to primary molecular impact. Access to in vitro transcripts has allowed a systematic investigation of the effect of individual mutations on aminoacylation and should allow a number of other possibilities, including maturation, stability, structure, and recognition by elongation factor, to be investigated in a systematic way.
The comparative analysis of mutations within the same tRNA also shows only partial correlation between the primary impact of the mutation and the phenotype of the disorder. None of the four mutations correlated with MERRF syndrome (G8363A, T8356C, A8344G, A8296G) or either of the two mutations correlated with deafness (G8363A, A8296G) affect tRNALys aminoacylation. On the other hand, two mutations linked to encephalopathies do affect this reaction (G8313A and G8328A), whereas a third one (T8316C, linked to MELAS) does not. MELAS syndrome correlates with mutation A3243G in tRNALeu(UUR), in which it leads to a significant decrease in leucylation (see Börner et al. 2000; Chomyn et al. 2000; Park et al. 2003; Sohm et al. 2003). Thus, primary mechanisms leading to the same disorder can be different according to the tRNA in which the deleterious mutation occurs.
Rules governing lysylation of human mitochondrial tRNALys
Several observations from the present work allow one to distinguish features within human mt tRNALys regarding their importance for lysylation. Comparison of aminoacylation properties of native tRNA extracted from human mitochondria with in vitro transcripts defines the contribution of post-transcriptional base modifications to the aminoacylation process. The natural tRNA was shown to possess six modifications: m1A9, m2G10, ψ27, ψ28, sU*34, and t6A37 (Helm et al. 1998; Yasukawa et al. 2001). As already discussed, m1A9 plays an essential role as a structural chaperone, promoting cloverleaf folding. The other modifications, especially those in helical domains, probably also contribute in a limited way to structure stabilization, as was shown for other tRNAs (Agris 1996), and in consequence improve aminoacylation efficiency. Such considerations may explain the slight difference in aminoacylation plateaus observed for the tRNALys transcripts Rz-Ke and Rz-Ki compared with native tRNALys. Modifications of anticodon loop nucleotides of tRNAs can contribute directly to aminoacylation identity (Giegé et al. 1998b; Giegé and Frugier 2003). This is probably not the case for human mt tRNALys, as can be deduced from aminoacylation properties of native wild type and native MERRF mutated tRNALys (Yasukawa et al. 2001). Both tRNAs are lysylated to similar levels (this work) with very similar efficiencies (Yasukawa et al. 2001) despite the complete absence of modification sU*34 in the MERRF derivative (Yasukawa et al. 2000). In the crystal structures available of lysyl-tRNA synthetase from T. thermophilus complexed either to fully modified E. coli tRNALys or to in vitro transcribed T. thermophilus tRNALys (Cusack et al. 1996), the mode of binding of the anticodon loops is the same, and both mnm5s2U or a nonmodified C-residue can be accommodated in the anticodon binding site of the enzyme. Although access to comparative kinetic parameters of transcripts and native tRNALys are not available (due to the difficulty in purifying sufficient quantities of native tRNAs), the above data lead to the conclusion that, beyond the major structural role of m1A9, post-transcriptional modifications make no major contribution to the lysylation properties of human mt tRNALys. This is in line with the low decrease (10-fold) in kcat/KM observed for human cytosolic tRNALysIII transcribed in vitro compared with native tRNA (Stello et al. 1999). In E. coli, post-transcriptional modifications make a more pronounced contribution because they improve aminoacylation efficiency 140-fold (Tamura et al. 1992).
Mutagenesis of in vitro transcribed tRNAs has been explored intensively in a large number of aminoacylation systems with the goal of deciphering aminoacylation identity elements (Giegé et al. 1998b; Giegé and Frugier 2003). As an outcome of the investigation of the 11 tRNALys variants investigated here, it can be concluded that the central U-residue from the anticodon loop (U35) is a major lysylation identity nucleotide. This was anticipated based on results from all lysylation systems explored so far both in vitro (Tamura et al. 1992; Shiba et al. 1997) and in vivo (McClain et al. 1990; Normanly et al. 1990). It is also clear that nucleotides 24 and 39 (and/or the base pairs in which they are involved) in the D-stem and anticodonstem respectively, contribute in an essential way to lysylation. On the other hand, nucleotides or base pairs involving nucleotides 2, 71, and 72 in the acceptor stem; nucleotide 27 in the anticodon stem; nucleotides 50, 53, 64, 65 in the T-stem; and nucleotides 53, 55, and 59 in the T-loop are only of limited importance.
Interestingly, the mutations investigated here are distributed all over the secondary cloverleaf structure of the tRNA, with only three mutations in loops and all others in stems. Within stems, each mutation converts a classical Watson–Crick pair into a weaker base pair. Eight mutations lead to C•A, A•C, or A•G mismatches and one to a G•U weak pair. Some drastically reduce aminoacylation; others do not affect this process (Fig. 6A ▶). Occurrence of mismatches of the above type is not specific to tRNALys mutations, or to pathology-related mutations, but is a general observation for mt tRNA gene mutations (Florentz and Sissler 2001). These mismatches are expected to destabilize helical domains to some extent (Varani and McClain 2000). Because aminoacylation of tRNAs by their cognate aminoacyl-tRNA synthetase is sensitive to structural properties of the tRNA (Giegé and Frugier 2003), some mismatches can be expected to interfere with the lysylation properties of the mt tRNA. In an attempt to understand these distinct effects, mutated nucleotides have been located within a three-dimensional tRNA structure (Fig. 6C ▶–E). In absence of crystallographic data for human mt tRNALys, the structure of yeast tRNAAsp complexed to its cognate synthetase (Ruff et al. 1991; Cavarelli et al. 1993) has been chosen as a model. This structure is not expected to correspond exactly to that of tRNALys, especially in regard of the D/T-loops interactions, which obviously are different (the mt tRNA has a very short D-loop of three nucleotides and an enlarged T-loop of nine nucleotides; further, classical nucleotides G18, G19, U56, and C57 allowing for tertiary interactions between the two loops are missing). Both tRNALys and tRNAAsp are, however, recognized by aminoacyl-tRNA synthetases belonging to the same subclass IIb of synthetases (Cusack et al. 1990; Eriani et al. 1990). Figure 6 ▶, C through E, highlights that mutations affecting aminoacylation are all in the anticodon branch of the L-shaped three-dimensional structure, along the same side of the molecule. These mutations are in line with the single acceptor branch mutation leading to a weak loss in aminoacylation efficiency. All other mutations not affecting aminoacylation either are in the acceptor branch or, if present in the anticodon branch, are located on the opposite side.
FIGURE 6.
Structural insight on pathology-related mutations. (A) Summary of the effects of mutations on tRNALys aminoacylation efficiencies. Values are from Table 1 ▶ and the color code is as follows: structural mutations introduced at positions 50 and 64, which prevent alternate folding into extended hairpin (Helm et al. 1998), are indicated in green. Polymorphic mutation A8348G is in yellow. Neutral, mild, and highly deleterious mutations are, respectively, in blue, pink, and magenta. (B) Sequence conservation of mammalian mt tRNALys genes (Helm et al. 2000) and location of pathology-related mutations studied in the present work (arrowheads). The color code for arrowheads is as in A. (C–E) Location of mutations in three-dimensional representations of the tRNA. The color code is the same as in parts A and B. Nucleotides complementary to mutated positions and thus involved in base-pairing are in white. The model tRNA is a ribbon representation of the crystal structure of yeast tRNAAsp (in its complexed form with AspRS; Ruff et al. 1991; Cavarelli et al. 1993). (E) Profile representation emphasizing the location of harmful mutations within a same face of the tRNA. The CCA end is pointed toward the reader.
The mechanisms by which both deleterious mutations, affecting highly conserved base pairs in mammalian mt tRNALys sequences (Fig. 6B ▶; Helm et al. 2000), interfere with lysylation remain to be established. Mutations may either remove identity elements or introduce antideterminants, hindering recognition by the cognate synthetase. In both situations, direct contacts between the enzyme and its substrate would be hindered. Alternatively, if the affected nucleotides are not in direct contact with the synthetase, their sequence variations may lead to perturbations of the tRNA architectural frame, thus hindering signal transmission from the anticodon loop identity elements (in particular, the central anticodon triplet nucleotide) to the 3′-end of the tRNA, where aminoacylation takes place. Finally, mutations with negative effects on aminoacylation may simply remove essential permissive elements. These concepts, already described and illustrated for a number of canonical tRNA aminoacylation systems (see Frugier et al. 1998; Giegé et al. 1998a,b; Choi et al. 2003), now need to be tested experimentally for mt tRNALys.
Conclusion and outlook
The present work demonstrates that despite the crucial requirement of a post-transcriptional modification in human mt tRNALys folding, it is possible to create well-folded in vitro transcripts, efficient in aminoacylation, and thus to explore the contribution of individual nucleotides on tRNA structural and functional properties by second-site mutagenesis. Systematic exploration of aminoacylation properties of the full set of pathology-related and polymorphic mutations in tRNALys represents an initial application of such engineered substrates. It enlarges both our understanding of lysylation properties of a mt tRNA and of potential molecular mechanisms underlying various human disorders. It also offers new perspectives for exploring further structural and functional properties of this tRNA and of the related variants, including mischarging properties or, formulated differently, the gain of function hypothesis in mitochondrial disorders (Jacobs and Holt 2000). Experiments are underway to fully decipher the lysine identity elements and their mechanism of action.
From a theoretical point of view, most human mt tRNAs have peculiar structural features (Helm et al. 2000; Florentz et al. 2003), including weak base pairs; the presence of mismatches, unusual D-loop, and T-loop sizes; and the absence of classical tertiary interactions. Engineered structural stabilization of in vitro transcripts might represent a successful approach to enable a systematic investigation of structural and functional properties of any member of this tRNA family and to screen for the effect of pathology-related mutations on a simple basis, before undertaking indispensable but laborious in vivo analyses.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes were from New England Biolabs and T4 DNA ligase from Boehringer-Mannheim. PCR-purified oligonucleotides were from NAPS or Interactiva. GoldStar Taq DNA polymerase was purchased from Eurogentech. Phage T7 RNA polymerase was prepared from an overexpression clone according to the method of Becker et al. (1996). Plasmid pM368 Hs-LysRS-HisTag encoding the full-length human cytosolic LysRS (LysRS) was kindly provided by Karin Musier-Forsyth (University of Minnesota, Min-neapolis, MN). Initial cybrid cell lines R2–1A and R1–C3, displaying, respectively, wild-type and MERRF-mutated human mt DNA, were kindly provided by Guiseppe Attardi and Anne Chomyn (Caltech, Pasadena, CA).
Production of mitochondrial tRNAs by in vitro transcription
Synthetic genes, corresponding to the T7 RNA polymerase promoter followed by the human mt wild-type tRNALys gene (Kwt) or variants Ke (G50–C64) and Ki (U50–A64), and terminating at a BstN1 site, were previously constructed from overlapping and complementary oligonucleotides and cloned into pTFMA as described in (Helm et al. 1998). The synthetic gene corresponding to the T7 promoter connected to the downstream sequence of a hammerhead ribozyme (Fechter et al. 1998) followed by the human mt tRNALys sequence (Rz-Kwt) was also previously cloned (Helm et al. 1999b). Mutated tRNAs, bearing in all cases the A50U/U64A structural mutations and, respectively, mutation A8296G, G8313A, T8316C, G8328A, G8342A, T8356C, T8362G, G8363A (pathology-related mutations; numbering according to the location in the mitochondrial DNA), A8348G (polymorphic mutation), or T8324A (altering the anticodon triplet), were cloned following the same protocol (Helm et al. 1999b). Clones Rz-Ke, Rz-Ki, Rz-Kem, and Rz-Kim were derived from Rz-Kwt by PCR amplification, using in all cases oligonucleotide 659 (5′-CACCAAGCTTAATACGACTCACTATA-3′) and, respectively, oligonucleotide 660 (5′-CGGGATCCCCTGGTCACTGTAAGGAGGTGTTGGTTCTCCTAATCTTTAA-3′ for mutations A50G and U64C in Rz-Ke), oligonucleotide 662 (5′-CGGGATCCCCTGGTCACTGTAATGAGGTGTTGGTTCTCATAATCTTTAA-3′ for mutations A50U and U64A in Rz-Ki), oligonucleotide 663 (5′-CGGGATCCCCTGGTCACTGTAAGGAGGTGTTGGCTCTCCTAATCTTTAA-3′ for mutations A50G, U64C, and A55G in Rz-Kem), and oligonucleotide 661 (5′-CGGGATCCCCTGGTCACTGTAATGAGGTGTTGGCTCTCATAATCTTTAA-3′ for mutations A50U, U64A, and A55G in Rz-Kim). Oligonucleotide 659 introduces a HindIII restriction site, and oligonucleotides 660 to 663 introduce BstN1 and BamHI restriction sites. Prior to ligation into pTFMA, PCR products were purified on low-melting 1% agarose gel, extracted with the QIAquick Gel Extraction kit (Qiagen S.A.) and double-digested with HindIII and BamHI (according to manufacturer’s instructions). Plasmids were isolated from transformed E. coli TG1 cells. In vitro transcription of 0.1 mg/mL BstN1 linearized plasmids was performed for 4 h at 37°C in 40 mM Tris-HCl (pH 8.1 at 37°C), 30 mM MgCl2, 5 mM dithiothreitol, 0.01% Triton-X100, 1 mM spermidine, and 4 mM each nucleoside triphosphate and T7 RNA polymerase. The hammerhead ribozyme, which contains four nucleotides complementary to the first four nucleotides of the 5′-end of the tRNA, cleaves the phosphodiester linkage directly upstream of residue C1 of the tRNA. After phenol/chloroform extraction, transcripts were purified to single nucleotide resolution on denaturing 12% polyacrylamide/urea gels, electro-eluted, and ethanol precipitated.
Preparation of native tRNALys
Human cybrid cells (homoplasmic for either wild-type, R2–1A, or MERRF-mutated, R1-C3, human mt tRNALys gene) were grown, mitochondria were isolated, and specific tRNAs were extracted by hybridization to solid-phase oligonucleotide as previously described (Chomyn et al. 1991; Helm et al. 1999a). Briefly, mitochondria were isolated from cybrid cells by differential centrifugation and total mt tRNAs were obtained by subsequent extraction using guanidium isothiocyanate. Human mt tRNALys was further purified by hybridization to a 5′-biotinylated labeled 30-mer, complementary to its 3′-end as described (Helm et al. 1998). tRNA was recovered from the streptavidin-coated beads by heating for 5 min at 80°C in 2 mM EDTA and immediately chilling on ice. About 0.3 μg of native tRNALys were recovered from 3.3 g of pelleted cells (corresponding to ~1 L adherent cell-cultures).
Lysyl-tRNA synthetase
The human cytosolic histidinetagged LysRS was purified as previously described (Shiba et al. 1997), using a nickel-nitrilotriacetic acid resin (Qiagen) according to the manufacturer’s instructions. Enzymatic activity for LysRS was 51 UE/mg (nanomoles of lysine incorporated per mg of protein per min), in aminoacylation conditions as described below using E. coli total tRNAs as substrate.
Aminoacylation reactions
Aminoacylation reactions have been performed as described (Perret et al. 1990; Shiba et al. 1997) in a medium containing 50 mM HEPES-KOH (pH 7.5), 20 mM KCl, 10 mM MgCl2, 4 mM ATP, 0.1 μg/μL BSA, 20 μM [3H]-labeled lysine (specific activity of ~1400 cpm/pmole), and adequate amounts of tRNA (0.1 to 10 μM) and LysRS (1 to 600 nM) at 37°C or 25°C. Before aminoacylation, transcripts were renatured by heating for 2 min at 60°C and slow cooling to room temperature. Kinetic parameters were derived from Lineweaver-Burk plots. Because the concentration of amino acid was subsaturating, only apparent kinetic parameters are given. In all cases, two independent preparations of transcripts have been done and at least two sets of experiments performed to define kinetic parameters. If the variability in functional results varied >10%–20%, a third preparation of molecules was done and analyzed. Doing so, kinetic parameters differing by <10% were obtained for each tRNA. The charging levels (optimal lysylation plateaus) are expressed as percentages of charged tRNA versus total amount of specific tRNA per experiment. For poor tRNA substrates, direct determination of the ratio kcat/KM was established as the slope of the linear plot of initial rate versus high tRNA concentrations (Schulman and Pelka 1988).
Structural mapping in solution
5′-Labeling and subsequent purification of tRNA transcripts were carried out as previously described (Helm et al. 1998). Cleavage with lead acetate was performed in 40 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 40 mM NaCl, and 0.1 μg/μl carrier tRNA in the presence of 0.15 mM Pb(OAc)2 for 10 to 30 min at 25°C. Reactions were stopped by addition of 1 volume RNA loading buffer (8 M urea, 100 mM Tris-borate at pH 8.3, 10 mM EDTA). Modification with diethylpyrocarbonate (DEPC) was performed in 50 mM sodium cacodylate (pH 8.0), 10 mM MgCl2, 300 mM KCl(control, C, and native, N, conditions), or 50 mM sodium cacodylate (pH 8.0), 1 mM EDTA (semi-denaturing, SD, and denaturing, D, conditions), respectively. Aliquots of 50 000 cpm 5′-labeled RNA supplemented with 0.05 to 0.1 μg/μL carrier tRNA were incubated with 5% (v/v) DEPC for 40 min at 25°C (N, SD) or 4 min at 60°C (D), respectively. Reactions were stopped by addition of 1 volume 0.6 M NaOAc (pH 7.0) and 20 μg carrier tRNA and subsequent ethanol precipitation. Cleavage of RNA at DEPC-modified adenine residues was done according to the method of Peattie and Gilbert (1980). Cleavage products were analyzed by denaturing separation on 12% PAGE as described (Helm et al. 1998).
Acknowledgments
Karin Musier-Forsyth (Minneapolis, USA), Guillaume Bec (Strasbourg, France), and Guiseppe Attardi and Anne Chomyn (Pasadena, USA) are acknowledged for their generous gifts of materials. We are indebted to Caroline Paulus for excellent technical assistance and to Louis Levinger for stylistic improvements of the manuscript. This work was supported by CNRS (Centre National de la Recherche Scientifique), ULP (Université Louis Pasteur), European Commission (FP5 RTD QLG2-CT-1999-00660 grant), and AFM (Association Française contre les myopathies).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
tRNA, transfer RNA
aaRS, aminoacyl-tRNA synthetase
mt, mitochondrial
DEPC, diethylpyrocarbonate
WT, wild-type
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5267604.
Footnotes
Numbering is according to location in the mitochondrial genome; Anderson et al. 1981.
REFERENCES
- Agris, P.F. 1996. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 53: 79–129. [DOI] [PubMed] [Google Scholar]
- Allen, J.F. and Raven, J.A. 1996. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J. Mol. Evol. 42: 482–492. [DOI] [PubMed] [Google Scholar]
- Anderson, S., Bankier, A.T., Barrel, B.G., de Bruijn, M.H.L., Coulson, A.R., Drouin, J., Eperon, J.C., Nierlich, D.P., Roe, B.A., Sanger, F., et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. [DOI] [PubMed] [Google Scholar]
- Bacman, S.R., Atencio, D.P., and Moraes, C.T. 2003. Decreased mitochondrial tRNALys steady-state levels and aminoacylation are associated with the pathogenic G8313A mitochondrial DNA mutation. Biochem. J. 374: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, H., Giegé, R., and Kern, D. 1996. Identity of prokaryotic and eukaryotic tRNAAsp for aminoacylation by aspartyl-tRNA synthetase from Thermus thermophilus. Biochemistry 35: 7447–7458. [DOI] [PubMed] [Google Scholar]
- Börner, G.V., Zeviani, M., Tiranti, V., Carrara, F., Hoffmann, S., Gerbitz, K.D., Lochmuller, H., Pongratz, D., Klopstock, T., Melberg, A., et al. 2000. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 9: 467–475. [DOI] [PubMed] [Google Scholar]
- Bullard, J., Cai, Y.-C., Demeler, B., and Spremulli, L. 1999. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J. Mol. Biol. 288: 567–577. [DOI] [PubMed] [Google Scholar]
- Cavarelli, J., Rees, B., Ruff, M., Thierry, J.-C., and Moras, D. 1993. Yeast tRNAAsp recognition by its cognate class II aminoacyl-tRNA synthetase. Nature 362: 181–184. [DOI] [PubMed] [Google Scholar]
- Choi, H., Kay, G., Schneider, J.A., Otten, S., and McClain, W.H. 2003. Recognition of acceptor-stem structure of tRNAAsp by Escherichia coli aspartyl-tRNA synthetase. RNA 9: 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn, A., Meola, G., Bresolin, N., Lai, S.T., Scarlato, G., and Attardi, G. 1991. In vitro genetic transfer of protein synthesis and respiration defects to mitochondrial DNA-less cells with myopathy-patient mitochondria. Mol. Cell. Biol. 11: 2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn, A., Enriquez, J.A., Micol, V., Fernandez-Silva, P., and Attardi, G. 2000. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem. 275: 19198–19209. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Berthet-Colominas, C., Härtlein, M., Nassar, N., and Leberman, R. 1990. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase. Nature 347: 249–255. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Yaremchuk, A., and Tukalo, M. 1996. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNALys and a T. thermophilus tRNALys transcript: Anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 15: 6321–6334. [PMC free article] [PubMed] [Google Scholar]
- Degoul, F., Brulé, H., Cepanec, C., Helm, M., Marsac, C., Leroux, J.-P., Giegé, R., and Florentz, C. 1998. Isoleucylation properties of native human mitochondrial tRNAIle and tRNAIle transcripts. Implications for cardiomyopathy-related point mutations (4269, 4317) in the tRNAIle gene. Hum. Mol. Gen. 7: 347–354. [DOI] [PubMed] [Google Scholar]
- DiMauro, S. and Andreu, A. 2000. Mutations in mtDNA: Are we scraping the bottom of the barrel? Brain Pathol. 10: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez, J.A. and Attardi, G. 1996. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 264: 183–196. [DOI] [PubMed] [Google Scholar]
- Enriquez, J.A., Chomyn, A., and Attardi, G. 1995. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNALys and premature translation termination. Nat. Genet. 10: 47–55. [DOI] [PubMed] [Google Scholar]
- Eriani, G., Delarue, M., Poch, O., Gangloff, J., and Moras, D. 1990. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347: 203–206. [DOI] [PubMed] [Google Scholar]
- Fechter, P., Rudinger, J., Giegé, R., and Théobald-Dietrich, A. 1998. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: Production and activity. FEBS Lett. 436: 99–103. [DOI] [PubMed] [Google Scholar]
- Florentz, C. and Sissler, M. 2001. Disease-related versus polymorphic mutations in human mitochondrial tRNAs: Where is the difference? EMBO Rep. 2: 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Mitochondrial tRNA aminoacylation and human diseases. In Translation mechanisms (J. Lapointe and L. Brakier-Gingras, eds.), pp. 129–143. Landes Bioscience, Georgetown, TX.
- Florentz, C., Sohm, B., Tryoen-Tóth, P., Pütz, J., and Sissler, M. 2003. Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 60: 1356–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier, M., Helm, M., Felden, B., Giegé, R., and Florentz, C. 1998. Sequences outside recognition sets are not neutral for tRNA aminoacylation: Evidence for non-permissive combinations of nucleotides in the acceptor stem of yeast tRNAPhe. J. Biol. Chem. 273: 11605–11610. [DOI] [PubMed] [Google Scholar]
- Giegé, R. and Frugier, M. 2003. Transfer RNA structure and identity. In Translation mechanisms (J. Lapointe and L. Brakier-Gringas, eds.), pp. 1–24. Landes Bioscience, Georgetown, TX.
- Giegé, R., Frugier, M., and Rudinger, J. 1998a. tRNA mimics. Curr. Opin. Struct. Biol. 8: 286–293. [DOI] [PubMed] [Google Scholar]
- Giegé, R., Sissler, M., and Florentz, C. 1998b. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, M.G., Nelson, I.P., Morgan-Hughes, J.A., and Harding, A.E. 1995. Impaired mitochondrial translation in human myoblasts harbouring the mitochondrial DNA tRNA lysine 8344 A →G (MERRF) mutation: Relationship to proportion of mutant mitochondrial DNA. J. Neurol. Sci. 130: 154–160. [DOI] [PubMed] [Google Scholar]
- Helm, M., Brulé, H., Degoul, F., Cepanec, C., Leroux, J.-P., Giegé, R., and Florentz, C. 1998. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 26: 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Florentz, C., Chomyn, A., and Attardi, G. 1999a. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR). Nucleic Acids Res. 27: 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Giegé, R., and Florentz, C. 1999b. A Watson-Crick base-pair disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38: 13338–13346. [DOI] [PubMed] [Google Scholar]
- Helm, M., Brulé, H., Friede, D., Giegé, R., Pütz, J., and Florentz, C. 2000. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6: 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman, M., Kaessmann, H., Pääbo, S., and Gyllenstein, U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708–713. [DOI] [PubMed] [Google Scholar]
- Jacobs, H.T. and Holt, I.J. 2000. The np 3243 MELAS mutation: Damned if you aminoacylate, damned if you don’t. Hum. Mol. Genet. 9: 463–465. [DOI] [PubMed] [Google Scholar]
- Kelley, S., Steinberg, S., and Schimmel, P. 2000. Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nat. Struct. Biol. 7: 862–865. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J. Biol. Chem. 276: 10607–10611. [DOI] [PubMed] [Google Scholar]
- King, M.P. and Attardi, G. 1989. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 246: 500–503. [DOI] [PubMed] [Google Scholar]
- Kogelnik, A.M., Lott, M.T., Brown, M.D., Navathe, S.B., and Wallace, D.C. 1998. MITOMAP: A human mitochondrial genome database: 1998 update. Nucleic Acids Res. 26: 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, N.-G. and Clayton, D.A. 1995. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29: 151–178. [DOI] [PubMed] [Google Scholar]
- Masucci, J.P., Davidson, M., Koga, Y., Schon, E.A., and King, M.P. 1995. In vitro analysis of mutations causing myoclonus epilepsy with ragged-red fibers in the mitochondrial tRNALys gene : Two genotypes produce similar phenotypes. Mol. Cell. Biol. 15: 2872–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, W.H. 1993a. Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 234: 257–280. [DOI] [PubMed] [Google Scholar]
- ———. 1993b. Transfer RNA identity. FASEB J. 7: 72–78. [DOI] [PubMed] [Google Scholar]
- ———. 1995. The tRNA identity problem: past, present and future. In tRNA: Structure, biosynthesis, and function (D. Söll and U.L. RajBhandary, eds.), pp. 335–347. American Society of Microbiology Press, Washington, DC.
- McClain, W.H., Foss, K., Jenkins, R.A., and Schneider, J. 1990. Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc. Natl. Acad. Sci. 87: 9260–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly, J., Kleina, L.G., Masson, J.M., Abelson, J., and Miller, J.H. 1990. Construction of Escherichia coli amber suppressor tRNA genes, III: Determination of tRNA specificity. J. Mol. Biol. 213: 719–726. [DOI] [PubMed] [Google Scholar]
- Park, H., Davidson, E., and King, M.P. 2003. The pathogenic A3243G mutation in human mitochondrial tRNALeu(UUR) decreases the efficiency of aminoacylation. Biochemistry 4: 958–964. [DOI] [PubMed] [Google Scholar]
- Peattie, D.A. and Gilbert, W. 1980. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. 77: 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret, V., Florentz, C., and Giegé, R. 1990. Efficient aminoacylation of a yeast tRNAAsp transcript with a 5′-extension. FEBS Lett. 270: 4–8. [DOI] [PubMed] [Google Scholar]
- Ruff, M., Krishnaswamy, S., Boeglin, M., Poterszman, A., Mitschler, A., Podjarny, A., Rees, B., Thierry, J.-C., and Moras, D. 1991. Class II aminoacyl transfer RNA synthetases: Crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science 252: 1682–1689. [DOI] [PubMed] [Google Scholar]
- Saks, M.E., Sampson, J.R., and Abelson, J.N. 1994. The transfer RNA identity problem: A search for rules. Science 263: 191–197. [DOI] [PubMed] [Google Scholar]
- Sampson, J.R. and Uhlenbeck, O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon, E.A., Bonilla, E., and DiMauro, S. 1997. Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomemb. 29: 131–149. [DOI] [PubMed] [Google Scholar]
- Schulman, L.H. and Pelka, H. 1988. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science 242: 765–768. [DOI] [PubMed] [Google Scholar]
- Servidei, S. 2002. Mitochondrial encephalomyopathies: Gene mutation. Neuromuscul. Disord. 12: 524–529. [DOI] [PubMed] [Google Scholar]
- Shiba, K., Stello, T., Motegi, H., Noda, T., Musier-Forsyth, K., and Schimmel, P. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem. 272: 22809–22816. [DOI] [PubMed] [Google Scholar]
- Shimada, N., Suzuki, T., and Watanabe, K. 2001. Dual mode of recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J. Biol. Chem. 276: 46770–46778. [DOI] [PubMed] [Google Scholar]
- Shoffner, J., Lott, M., Lezza, A.M.S., Seibel, P., Ballinger, S.W., and Wallace, D.C. 1990. Myoclonic epilepsy and ragged red fiber disease (MERRF) is associated with mitochondrial DNA tRNALys mutation. Cell 61: 931–937. [DOI] [PubMed] [Google Scholar]
- Sohm, B., Frugier, M., Brulé, H., Olszak, K., Przykorska, A., and Florentz, C. 2003. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 328: 995–1010. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stello, T., Hong, M., and Musier-Forsyth, K. 1999. Efficient amino-acylation of tRNALys3 by human lysyl-tRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res. 27: 4823–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Himeno, H., Asahara, H., Hasegawa, T., and Shimizu, M. 1992. In vitro study of E. coli tRNAArg and tRNALys identity. Nucleic Acids Res. 20: 2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunova, E., Park, H., Xia, J., King, M.P., and Davidson, E. 2000. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual splicing of the primary transcript. J. Biol. Chem. 275: 35063–35069. [DOI] [PubMed] [Google Scholar]
- Varani, G. and McClain, W.H. 2000. The G–U wobble base pair: A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D.C. 1999. Mitochondrial diseases in man and mouse. Science 283: 1482–1488. [DOI] [PubMed] [Google Scholar]
- Wittenhagen, L.M., Roy, M.D., and Kelley, S.O. 2003. The pathogenic U3271C human mitochondrial tRNALeu(UUR) mutation disrupts a fragile anticodon stem. Nucleic Acids Res. 31: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa, T., Suzuki, T., Ishii, N., Ueda, T., and Ohta, S., Watanabe, K. 2000. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 467: 175–178. [DOI] [PubMed] [Google Scholar]
- Yasukawa, T., Suzuki, T., Ishii, N., Ohta, S., and Watanabe, K. 2001. Wobble modification defect in tRNA disturbs codon–anticodon interaction in a mitochondrial disease. EMBO J. 20: 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]