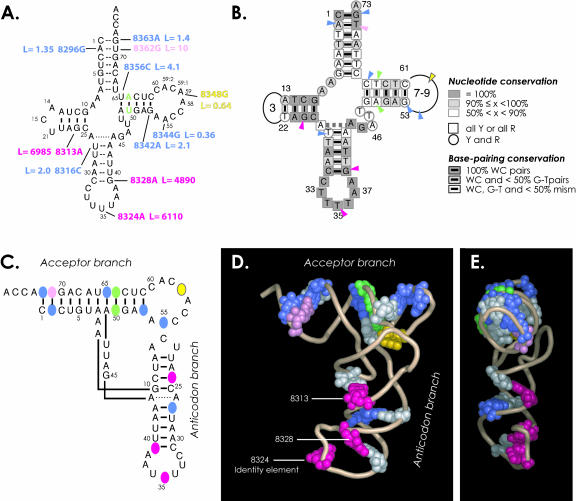
FIGURE 6.
Structural insight on pathology-related mutations. (A) Summary of the effects of mutations on tRNALys aminoacylation efficiencies. Values are from Table 1 ▶ and the color code is as follows: structural mutations introduced at positions 50 and 64, which prevent alternate folding into extended hairpin (Helm et al. 1998), are indicated in green. Polymorphic mutation A8348G is in yellow. Neutral, mild, and highly deleterious mutations are, respectively, in blue, pink, and magenta. (B) Sequence conservation of mammalian mt tRNALys genes (Helm et al. 2000) and location of pathology-related mutations studied in the present work (arrowheads). The color code for arrowheads is as in A. (C–E) Location of mutations in three-dimensional representations of the tRNA. The color code is the same as in parts A and B. Nucleotides complementary to mutated positions and thus involved in base-pairing are in white. The model tRNA is a ribbon representation of the crystal structure of yeast tRNAAsp (in its complexed form with AspRS; Ruff et al. 1991; Cavarelli et al. 1993). (E) Profile representation emphasizing the location of harmful mutations within a same face of the tRNA. The CCA end is pointed toward the reader.