Abstract
We investigated the self-splicing properties of two introns from the bacterium Bacillus anthracis. One intron (B.a.I1) splices poorly in vitro despite having typical structural motifs, while the second (B.a.I2) splices well while having apparently degenerated features. The spliced exons of B.a.I2 were sequenced, and splicing was found to occur at a 3′ site shifted one nucleotide from the expected position, thus restoring missing γ-γ′ and IBS3–EBS3 pairings, but leaving the two conserved exonic ORFs out of frame. Because of the unexpected splice site, the principles for 3′ intron definition were examined, which showed that the 3′ splice site is flexible but contingent on γ-γ′ and IBS3–EBS3 pairings, and can be as far away as four nucleotides from the wild-type site. Surprisingly, alternative splicing occurs at position +4 for wild-type B.a.I2 intron, both in vitro and in vivo, and the alternative event fuses the two conserved exon ORFs, presumably leading to translation of the downstream ORF. The finding suggests that the structural irregularities of B.a.I2 may be an adaptation to facilitate gene expression in vivo.
Keywords: group II intron, alternative splicing, retroelement, ribozyme, bacteria
INTRODUCTION
Group II introns are retroelements found in bacteria and organelles of lower eukaryotes. They consist of a highly structured RNA and an internally encoded reverse transcriptase (RT) open reading frame (ORF). The intron RNA is a ribozyme that provides the catalytic activity for splicing, while the RT, in combination with the RNA, lends mobility to the intron. The ORF has four functionally defined domains, all of which are important for mobility: RT (reverse transcriptase), X (maturase, or splicing activity), D (DNA-binding), and En (endonuclease; San Filippo and Lambowitz 2002). Mobility occurs by a mechanism termed target-primed reverse transcription (TPRT), in which the intron reverse splices into the DNA target and is then reverse-transcribed by the RT using cleaved target as a primer (Lambowitz et al. 1999; Bonen and Vogel 2001; Belfort et al. 2002; Toro 2003).
Group II introns in bacteria appear to behave mainly as retroelements (Dai and Zimmerly 2002), but splicing still occurs in vivo and is required for the mobility reaction (Mills et al. 1996; Martínez-Abarca et al. 1998; Belfort et al. 2002). Splicing is intrinsically RNA-catalyzed, although the intron-encoded protein facilitates the reaction by binding to the RNA structure and inducing conformational changes (Saldanha et al. 1999; Wank et al. 1999; Matsuura et al. 2001; Noah 2003). The conserved RNA structure consists of six domains (Fig. 1A ▶; Michel and Ferat 1995; Qin and Pyle 1998). Domains I and V are most important for catalysis, with domain V considered the catalytic center of the ribozyme. Domain VI contains a bulged A that forms the 2′–5′ lariat connection during splicing. Domains II, III, and IV are less important for catalysis, but domain IV encodes the RT ORF in its loop.
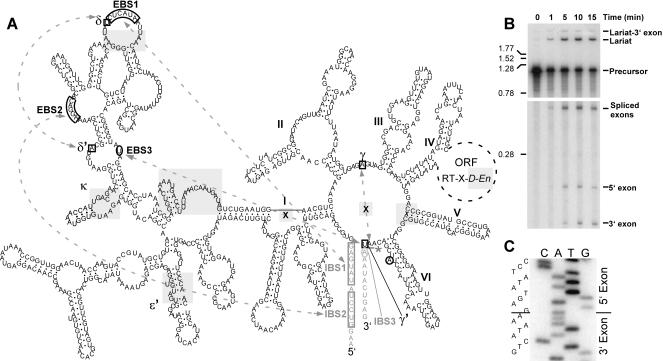
FIGURE 1.
Secondary structure and in vitro splicing of B.a.I2. (A) The B.a.I2 secondary structure is a IIB structure with the ORF encoded in domain IV. Irregularities are shaded in gray and include a mispair at the base of domain V (marked by an “X”), the absence of γ-γ′ and IBS3–EBS3 pairings at the predicted positions (marked by an “X”), unpaired nucleotides in the κ and EBS1 loops, bulged nucleotides in the ɛ′ region, an insertion of a stem–loop and single-stranded region in domain I, a frame shift in the ORF truncating the D and En domains, and a predicted lack of register between the conserved exonic ORFs in the spliced product. The intron is represented by black lettering, exons by gray lettering, and the ORF by a dotted circle. Arrows with dotted lines show long-range pairings critical for recognition of exons. There are no γ-γ′ and IBS3–EBS3 pairings at the expected positions (marked by “X”s); however, the actual γ′ and IBS3 positions are shifted one position, and are indicated by a square and circle, respectively. An asterisk indicates the observed 3′ splice site between γ′ and IBS3. (B) In vitro self-splicing of B.a.I2. Internally radiolabeled precursor transcript was self-spliced in vitro and analyzed on a polyacrylamide gel (see Materials and Methods). The bottom portion of the gel is a longer autoradiographic exposure to allow visualization of all splicing products. Size markers to the left are in kb. (C) Spliced exons from a self-splicing reaction were amplified by RT-PCR and sequenced, revealing a shifted 3′ splice site.
Accuracy in splice site selection, the topic of this study, is accomplished by base-pairing interactions between group II introns and their exons. The 5′ splice site is determined by two interactions immediately upstream of the splice site, IBS1 and IBS2 (intron binding sites 1 and 2), which pair with EBS1 and EBS2 (exon binding sites 1 and 2) in the intron to form roughly a dozen base pairs (Fig. 1A ▶). 3′ exon recognition involves two single-base-pair interactions. The γ-γ′ interaction is a base pair between the last position of the intron and an intron nucleotide between domains II and III (Fig. 1A ▶; Jacquier and Michel 1990). The second interaction differs for IIA and IIB introns. For IIB introns, it consists of a base pair between the first nucleotide of the 3′ exon (IBS3) and EBS3 in domain I (Michel and Jacquier 1987; Costa et al. 2000). An additional factor in determining the 3′ splice site is its location relative to the base of domain VI, which is conserved. For IIA introns, the 3′ splice site occurs 2 nt past the end of DVI and has the consensus sequence AY, whereas for IIB introns it occurs 3 nt past DVI and has the sequence GAY (Michel et al. 1989).
Comparatively few bacterial group II introns have been characterized for ribozyme properties. Characterized introns include Ll.ltrB of Lactococcus lactis, RmInt1 of Sinorhizobium, IntB of E. coli, Cal.X1 of Calothrix, GBSi1 of Streptococcus, and Avi.groEL of Azotobacter (Saldanha et al. 1999; Granlund et al. 2001; Adamidi et al. 2003; Ferat et al. 2003; Muñoz-Adelantado et al. 2003). In most cases, characterization does not proceed beyond a simple test for splicing competence. Interestingly, bacterial group II ribozymes are very diverse in RNA structure, with few introns conforming closely to the classical IIA and IIB structures that were defined based on organellar introns (Toor et al. 2001). Some bacterial introns are sufficiently different as to be designated a new structural class, IIC (Ferat et al. 2003; Toro 2003). Because group II introns likely evolved in bacteria and then migrated to organelles, it is possible that some ribozymes of bacterial introns correspond to more ancestral and less derived introns than those in organelles (Ferat and Michel 1993; Dai and Zimmerly 2002). Therefore, they might have novel properties compared to characterized organellar introns. One unusual property uncovered so far for a bacterial intron, Avi.groEL of Azotobacter vinelandii, is that it has a high temperature optimum (65°C) and is the most heat-stable of characterized group II introns (Adamidi et al. 2003).
Here we investigated two introns found in the pX01 virulence plasmid of Bacillus anthracis, the causative agent of anthrax. Unexpectedly, B.a.I2 was found to use a novel 3′ splice site and an alternative splice site both in vitro and in vivo. The unusual properties allowed the principles for 3′ splice site selection to be examined for this intron, which uncovered general properties for the flexibility of 3′ splice sites. This is the first example of alternative splicing for a group II intron, apart from the unusual introns of Euglena chloroplasts. The findings also have implications for predicting the properties of other introns identified in sequence databases that have not yet been characterized.
RESULTS
Self-splicing of B.a.I1 and B.a.I2
In an effort to characterize ribozyme activities of group II introns in bacteria, we investigated the splicing of two introns in B. anthracis, here called B.a.I1 and B.a.I2, which correspond to ORFs pX01-07 and pX01-23 in the genome annotation (Okinaka et al. 1999). B.a.I1 has a typical RNA secondary structure of the IIB1 subclass and standard ORF motifs, suggesting that the intron copy is functional for splicing and mobility (data not shown). In contrast, the B.a.I2 sequence appears defective. Its RNA structure lacks apparent γ-γ′ and IBS3–EBS3 pairings, and there is a mispair at the base of domain V, as well as several mispaired or unpaired regions and one insertion (shaded regions in Fig. 1A ▶). Moreover, the ORF is frameshifted near the end of the X domain, truncating the D and En domains, and the conserved ORFs in the 5′ and 3′ exons are predicted to be out of register in the spliced mRNA. Together these irregularities suggest that B.a.I2 is not fully functional for splicing and/or mobility in vivo.
Both B.a.I1 and B.a.I2 were cloned into pBluescript with the deletion of their ORFs (see Materials and Methods), and were subjected to in vitro self-splicing under a variety of reaction conditions. Unexpectedly, B.a.I1 spliced poorly under all conditions tested. RT-PCR amplification and sequencing of spliced exons confirmed correct splicing events, but most intron was unreacted based on data from polyacrylamide gels (data not shown). Splicing in vivo was investigated by RT-PCR of RNA isolated from cells. Spliced exons were readily detected, but the two unspliced intron–exon junctions were amplified more weakly, suggesting that splicing occurs adequately in vivo, if not in vitro.
Surprisingly, B.a.I2 self-spliced well under a variety of reaction conditions. A full description of reaction conditions and products formed will be reported elsewhere, but here the standard splicing conditions were 40 mM Tris-HCl (pH 7.6), 1 M NH4Cl, and 100 mM MgCl2 at 45°C. As expected, the major splicing products are lariat and spliced exons (Fig. 1B ▶), although there are also substantial amounts of 5′ and 3′ exons, formed by either spliced exon reopening (SER; Jarrell et al. 1988), or from failure to ligate the exons. Spliced exons were RT-PCR–amplified and sequenced, and the splice site was found to be shifted one nucleotide from the expected site, to a position having γ-γ′ and IBS3–EBS3 pairings (Fig. 1C ▶). The location of the 3′ splice site for diverse group II introns can be 2 and 3 nt past the base of domain VI, but the distances are conserved among either IIA or IIB structures (Michel et al. 1989; Toor et al. 2001), and we are not aware of any group II introns with splice sites farther than 3 nt after domain VI. Therefore, we were led to investigate the rules governing splice site position for this intron, with regard to γ-γ′ and IBS3–EBS3 pairings and their positions relative to domain VI.
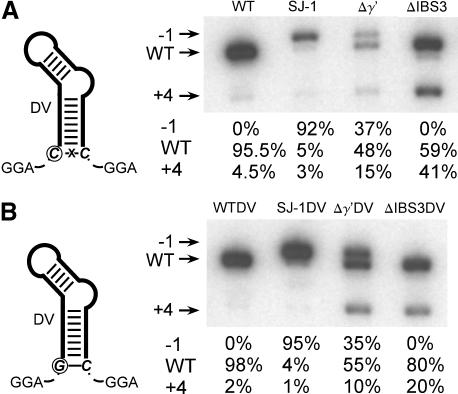
Figure 2A ▶ diagrams the observed wild-type 3′ intron–exon junction with γ-γ′ and IBS3–EBS3 pairings. To test for the effects of the positions of γ′ and IBS3, we created a series of mutants that shift the position of γ′ (U) by intervals of one nucleotide from +2 to −2 relative to the wild-type splice site (Fig. 2B ▶). Conveniently, a run of A residues surrounds the wild-type splice site, providing a uniform background of potential IBS3 pairings, thus simplifying experimental interpretation. At positions +4 and +5 are a U and A, which together provide competing γ-γ′ and IBS3–EBS3 pairings. In addition to these four mutants, we made single mutations blocking the γ-γ′ pairing (Δγ′) or IBS3–EBS3 pairing (ΔIBS3).
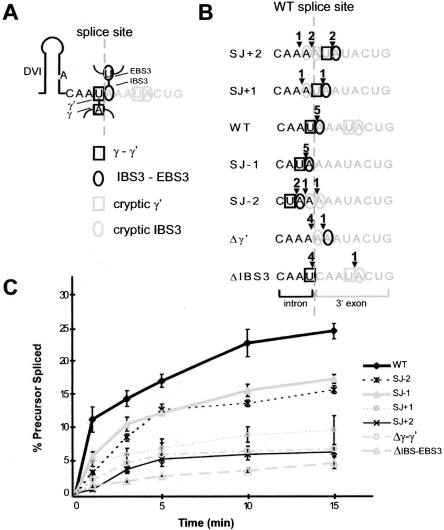
FIGURE 2.
Self-splicing of intron mutants. (A) Diagram of γ-γ′ and IBS3–EBS3 pairings at the 3′ junction of B.a.I2. The intron is in black; exons are gray, the splice site is a dotted gray line, and squares and circles indicate γ-γ′ and IBS3–EBS3 pairings, respectively. (B) Diagram of 3′ junction mutations. Six mutations were constructed that shift the positions of γ′ and IBS3 pairings or deleted them. The position of the wild-type splice junction is indicated as a dotted line, and squares and circles indicate the potential γ′ and IBS3 pairings. Arrowheads and numbers above the sequence indicate the number of splicing events observed at these positions in cloned and sequenced spliced exon junctions. (C) Time course of self-splicing reactions for B.a.I2 mutants. Mutants were self-spliced in vitro, resolved on a polyacrylamide gel, and quantitated by phosphorimaging (see Materials and Methods). Reactions were done in triplicate with at least two different RNA preparations for each mutation, and are expressed as averages with standard deviations.
The mutants were transcribed with T7 RNA polymerase and spliced in vitro (see Materials and Methods), and time-course reactions determined relative splicing efficiencies (Fig. 2C ▶). The wild-type intron had the greatest splicing efficiency, whereas the position −1 and −2 mutants spliced moderately well, and other mutants spliced poorly. Interestingly, the −1 mutant, which conforms most closely to the structural consensus of other IIB introns, in fact, spliced less well than the wild-type sequence, demonstrating contextual effects that have shifted the preferred position of splicing.
To determine the splice sites utilized in the mutants, spliced exons from parallel reactions were RT-PCR–amplified, cloned, and sequenced (Fig. 2B ▶). Wild-type intron was found to splice at the expected position in five independent clones, as did the −1 mutant. However, shifting the γ-γ′ and IBS3–EBS3 pairings to other positions resulted in mixed splice site usage, revealing competition between the position of splicing and the requirement for γ-γ′ and IBS3–EBS3 pairings.
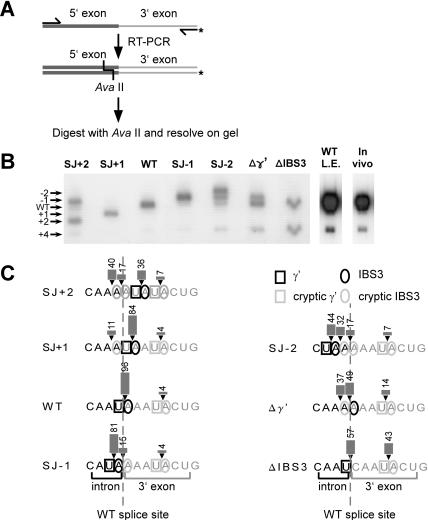
To obtain more quantitative data, a radioactive RT-PCR assay was developed (Fig. 3A ▶). In this assay, spliced exons were RT-PCR–amplified using one 5′ end-labeled primer, and the products were then cut with AvaII and resolved on a sequencing gel. Wild-type spliced exons would produce a 112-nt band, whereas bands of other sizes would indicate shifted splice sites. Quantitation of the bands provides an accurate measure of 3′ splice site usage. This assay is based on the assumption that the 5′ splice site is constant, which is predicted based on the sequences obtained in Figure 2B ▶.
FIGURE 3.
Quantitation of alternative splice sites using a radioactive RT-PCR assay. (A) Schematic of the assay. Spliced exons were amplified by RT-PCR using one 5′ end-labeled primer. Products were digested with AvaII and resolved on a sequencing gel (see Materials and Methods). Alternatively spliced exons are detected as bands shifted in position from the wild-type product. (B) Autoradiogram of data for wild-type and mutant introns. Numbers to the left indicate the positions of the 3′ splice site corresponding to the bands in the gel. “WT L.E.” is a long exposure of the wild-type intron lane in the left panel to show the band at +4 clearly. The “In vivo” lane shows splice sites detected for RNA isolated from logarithmically growing B. anthracis. (C) Summary of splicing products for wild-type and mutant introns. Quantitation of splice sites is shown by arrowheads, bars, and numbers (in percent). The wild-type splice site is indicated by a dotted line.
The data agree with Figure 2B ▶ but give added resolution (Fig. 3B ▶). Wild-type sequence has uniform splice site usage, except that 4% of splicing occurred at the competing site at +4 (see long exposure in “WT L.E.” lane). The apparent +4 splice site was confirmed by reamplifying the band from the gel, cloning, and sequencing. For the −1 mutant, only 81% of splicing occurred at the predicted −1 site; 15% occurred at position 0, and 4% occurred at +4. For the remaining mutants from −2 to +2, the general trend again is that the major splice site occurs at the position having both γ-γ′ and IBS3–EBS3 pairings, whereas there is a constant competition for the preferred spatial positions at −1 and 0, as well as a constant competition with the +4 site. Consistent with this trend, the Δγ′ mutant splices mainly at the −1 and 0 positions, and ΔIBS3 at 0. As perhaps expected, the mutants with poorest splicing efficiency in Figure 2C ▶ have the greatest competition for the alternative splicing event at position +4. So in summary, the 3′ splice site is flexible for this intron, occurring at positions as far away as +4, yet there is a position preference for the 0 position, and to a lesser extent the −1 position.
Are other group II introns variable in their 3′ splice sites?
We next asked whether this 3′ splice site flexibility is general to other group IIB introns. If so, then alternative splicing events might be a widespread occurrence for introns with cryptic γ′ and IBS3 pairings, and might affect the products of many genes in vivo. Other IIB introns were examined for potential γ-γ′ and IBS3–EBS3 pairings downstream of the wild-type splice site. aI5γ of yeast mitochondria was found to have a cryptic site at position +3; B.a.I1 has a potential site at +3; E.c.I2 of E. coli has a potential site at +20. The three introns were self-spliced in vitro, and tested for alternative splicing by the hot RT-PCR assay. No detectable alternative splicing was detected for any of the three examples within a level of 0.2% (data not shown).
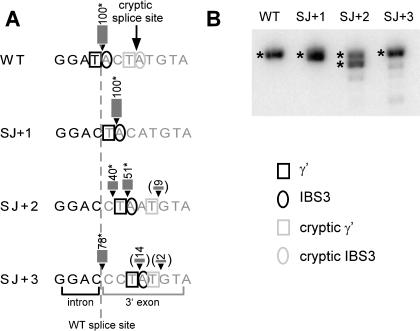
As a further test of splice site flexibility for aI5γ, we varied the γ′ and IBS3 residues from positions 0 to +3, and then tested for 3′ splice sites by the hot RT-PCR assay (Fig. 4 ▶). Similar to B.a.I2 splicing, the splice site was flexible by one position, contingent on γ-γ′ and IBS3–EBS3 pairings, but shifting the position more than one nucleotide resulted in heterogeneous splice sites. We conclude that the same principles of flexibility and dependence on γ-γ′ and IBS3–EBS3 pairings apply to aI5γ, and likely other IIB introns. However, because no alternatively spliced products were detected at position +3 for wild-type aI5γ intron, the flexibility of the 3′ splice site appears much reduced compared to B.a.I2. We predict that the alternative splicing event of B.a.I2 is an exception among group II introns, likely due to its structural variations.
FIGURE 4.
Assays of 3′ splice site mutations for aI5γ. (A) Mutations for the aI5γ intron in which γ′ and IBS3 positions are shifted. Wild-type intron has a cryptic splice site at +3. The wild-type splice site is indicated by a dotted line. Because the DNAs produced in the aI5γ assay are large and did not resolve unambiguously in the gel (B), the locations of the splice sites were determined by sequencing (marked by *), except for the figures in parentheses, which were estimated based on gel migration. (B) Radioactive RT-PCR assay of aI5γ mutants.
The mismatch at the base of domain V is partially responsible for 3′ splice site flexibility
To explain the alternative splicing properties of wild-type B.a.I2, we examined its structure for features that might influence 3′ splice site selection. The most obvious candidate was the mismatch at the base of domain V (Figs. 1A, 5 ▶ ▶), because the mispairing might increase the distance or flexibility between the 3′ splice site and the presumed active site in domain V. In fact, a precedent exists for this explanation. In one study that examined the size of the spacer between domains V and VI, it was found that increasing the spacer size in aI5γ resulted in an incorrect 3′ splice site, at a downstream exon position having both the γ′ and the not yet discovered IBS3 interaction (corresponding to position +3 in Fig. 4A ▶; Boulanger et al. 1996). To determine whether the domain V mispair contributes to 3′ flexibility of B.a.I2, the base pair was restored by a C-to-G change. The mutation was made in the context of otherwise wild-type intron, as well as three mutations (Fig. 2B ▶). Quantitation of splice sites by the hot RT-PCR assay showed that alternative splicing decreased by roughly a factor of two in the four cases. We conclude that the domain V mispair is partially responsible for splicing flexibility, but there are other contributing factors, because the alternative splice site at +4 persists.
FIGURE 5.
The mispair in domain V contributes to alternative splicing of B.a.I2. (A) Diagram of the mispaired domain V, and a radioactive RT-PCR assay showing alternative splice sites for wild-type intron and three mutants. Quantitation of splice site usage was by phosphorimaging of bands in the gel. (B) Diagram of domain V with restored pairing, and a radioactive RT-PCR assay showing the effect of restored pairing on alternative splicing, either in the context of otherwise wild-type intron or three mutants. Alternative splicing at +4 is reduced roughly 50% compared to panel A.
The alternative splicing event of B.a.I2 alters the predicted protein products in vivo
Curiously, splicing at the wild-type position does not join the two conserved ORFs in exons 1 and 2 (here called ORF1 and ORF2), but instead fuses two amino acids in exon 2 to ORF1 before a stop codon is encountered (Fig. 6A ▶). In contrast, the +4 splice site connects ORF1 with ORF2. This suggests that alternative splicing may be a mechanism for expressing ORF2. To determine whether alternative splicing at position +4 occurs in vivo, and thus might be responsible for translation of ORF2, we undertook the hot RT-PCR assay with RNA isolated from logarithmically growing B. anthracis (Fig. 3B ▶). The assay showed 4% alternative splicing in vivo, the same level as in vitro, and therefore it is predicted that splicing in vivo produces a ratio of ORF1 protein and ORF1-ORF2 fusion proteins, and it is possible that ORF2 translation in vivo is dependent on the alternative splicing event.
FIGURE 6.
The predicted consequences of B.a.I2 alternative splicing on the protein product of the host gene. (A) Predicted protein products for spliced and alternatively spliced exons. In the major splicing event, two amino acids are encoded in exon 2 before a stop codon is encountered. For the alternative splicing event at position +4, ORF2 is fused to ORF1, and is expected to be translated. (B) Organization of ORF1 and ORF2 in different bacterial genomes, as determined from BLAST searches of GenBank. In four cases, ORF1 and ORF2 are independent genes (bottom); in five cases the ORFs are fused into one reading frame, in either order; in four cases the ORFs are overlapping and frame-shifted, again in either order; in B. anthracis the ORFs are separated by an intron, but are fused by an alternative splicing event. ORF annotations and accession numbers are: A. pernix (APE0761/ NP_147473, APE0760/NP_147472), A. fulgidus (AF0948/NP_069781, AF0593/ NP_069427), B. anthracis (pXO1-24/ NP_052720, BXA0034/ NP_652800), G. violaceus (gll2293/BAC90234, gll2292/BAC90233), L. interrogans (LA2879/AAN50078, LA2880/AAN50079), M. jannaschii (MJ1305/NP_248305, MJ1304/NP_248304), P. aerophilum (PAE0273/ NP_558492; PAE1153/NP_559105, PAE1155/NP_559106; PAE1903/ NP_559624, PAE1902/NP_559623; PAE1320/NP_559219, PAE0044/ NP_558340), P. abyssi (PAB0900/CAB50264, PAB0899/CAB50263), P. horikoshii (PH0403/NP_877753, PH0690/NP_142637), S. solfataricus (SSO1251/NP_342700, SSO1161/NP_342621), S. tokodaii (ST0690/ NP_376587, ST0689/NP_376586; ST1573/NP_377540, ST1574/ NP_377541).
DISCUSSION
We investigated the self-splicing properties of two bacterial group II introns, and uncovered an alternative 3′ splice site usage that occurs both in vitro and in vivo. This is the first example of alternative splicing for a group II intron apart from the unusual introns of Euglena chloroplasts (below). The unexpected 3′ splice sites led to an investigation of the rules for 3′ definition and flexibility for this intron, and the inferred principles appear to be relevant to other IIB introns. Characterization of B.a.I2 also has implications for understanding other group II introns in genome sequences, as 3′ splice sites can now be more accurately predicted, and furthermore, introns having apparent defects, which we have considered to be inactive, dying copies (Dai and Zimmerly 2002) cannot automatically be assumed to be nonfunctional.
At the start of this study, the 3′ splice site of B.a.I2 was expected to be dependent on γ-γ′ and IBS3–EBS3 pairings and their relative positions from the base of domain VI. The mutational analysis presented here showed that the 3′ splice site is flexible by several positions, as directed by γ-γ′ and IBS3–EBS3 pairings, but there is a preferred spatial position located 4 nt after domain VI. This preferred position is different from other group IIB introns whose splice site is 3 nt after domain VI, and we rationalize the difference as being due to contextual effects from the structural variations of this intron.
Interestingly, the 3′ splice site of aI5γ also was readily shifted by one position, and shifting γ′ and EBS3 more than one nucleotide similarly resulted in heterogeneous 3′ splice sites. Thus, the principles of flexibility illustrated by B.a.I2 appear to apply to some extent to other group IIB introns. However, because no alternative splicing was observed for wild-type aI5γ or other introns tested, we do not anticipate that alternative 3′ splice sites will be common among other group II introns with potential γ-γ′ and IBS3–EBS3 pairings downstream of the true splice site. Instead, we rationalize the alternative splicing of B.a.I2 as being due to increased flexibility due to the mispair at the base of domain V and other contextual effects.
Alternative splicing is common for spliceosomal introns in higher eukaryotes, but is rare for other types of introns. The only other example of alternative splicing for group II introns comes from the unusual group II and group III introns and their twintron organizations in Euglena chloroplasts. In twintrons, one intron is nested inside another, and the two introns must be spliced sequentially in order to ligate together the exons of their host gene. For a number of Euglena introns and twintrons, including those in rpl16, rpoC1, ycf8, and roaA genes, a small proportion of splicing occurs at alternative 5′ or 3′ junctions, usually producing a truncated protein due to introduced frame shifts (Copertino et al. 1992; Hong and Hallick 1994; Jenkins et al. 1995). Alternative splicing has also been reported for some group I introns (Wallweber et al. 1997; Landthaler and Shub 1999; Vader et al. 2002), and in one case, alternative splicing may be responsible for translation of the maturase ORF encoded within the group I intron (Sellem and Belcour 1994).
In the case of B.a.I2, it is plausible that alternative splicing might contribute to regulation of ORF2, because ORF2 is in frame with its upstream exon ORF only for the minor splicing event but not for the major product. The functions of ORF1 and ORF2 are unknown, but both ORFs are conserved among a number of archaebacteria and eubacteria, as revealed by BLAST searches of GenBank. Interestingly, the organization of the ORFs with respect to each other is quite variable. In four cases, the two ORFs represent independent genes located far apart, whereas in nine cases the ORFs are either fused into one ORF, or are overlapping and frame-shifted (Fig. 6B ▶). In the fused or overlapping arrangements, the order can be either ORF1-ORF2 or ORF2-ORF1 (Fig. 6B ▶). B.a.I2 provides a sixth organization, in which the two ORFs are separated by an intron, and the splicing event produces mRNAs with the ORFs either out of frame or fused, depending on the splicing reaction (Fig. 6A ▶, B). Together, the different ORF1/ORF2 arrangements imply that the two genes interact functionally, possibly antagonistically. However, without knowledge of their exact cellular functions and assays to monitor them, one cannot be certain of the significance of alternative splicing events to the processes that these genes perform. For example, it is possible that each ORF is translated separately, although that would not explain the unusual combinations of overlapping organizations. In any case, given that alternative splicing occurs in vivo, it is plausible that alternative splicing might contribute to regulation of ORF2 expression, and that the structural irregularities of B.a.I2 are an adaptation to a biological role in the context of ORF1 and ORF2.
Finally, because group II introns are considered the evolutionary predecessors of spliceosomal introns (Sharp 1991), it is satisfying to find alternative splicing for a group II intron in nature. Although the mechanisms that underlie alternative splicing are not the same for group II and splice-osomal introns, the parallel nevertheless suggests that alternative splicing is encoded within the presumed ribozyme ancestors of spliceosomal introns, and that alternative splicing may be an ancient capability.
MATERIALS AND METHODS
DNA and RNA preparations from Bacillus anthracis
Bacillus anthracis Sterne (A0517) was grown in Brain Heart Infusion Broth at 37°C (Difco; Murray 1999). Genomic DNA was prepared using a MasterPure DNA Purification Kit (Epicentre) according to the manufacturer’s protocol. For RNA preparations, 50 mL of cells was harvested at an O.D600 of 0.3, washed, and resuspended in 0.5 mL of RNA extraction buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA). Then 0.5 mL of TE-saturated phenol was added (TE is 10 mM Tris-HCl, pH 7.6, 0.1 mM EDTA), and the suspension was incubated at 37°C for 30 min with mixing, followed by centrifugation and ethanol precipitation of the aqueous phase, treatment with DNase I (Amersham Pharmacia Biosciences), extraction with phenol-CIA (25:24:1 phenol:chloroform: isoamyl alcohol), and ethanol precipitation.
Plasmids
Plasmid constructs for self-splicing experiments were made by cloning PCR-amplified DNA segments of introns into pBluescript II KS+ (pKS+, Stratagene), in the BamHI and ClaI sites, in the orientation for T7 transcription. The introns were amplified in two fragments in order to delete the ORF. pKS-07ΔDIV is the construct made for B.a.I1, and contains 186 bp of the 5′ exon, 1095 bp of intron (741 bp 5′ sequence and 354 bp 3′ sequence), and 127 bp of the 3′ exon. pKS-23ΔDIV is the construct for B.a.I2, and contains 190 bp of the 5′ exon, 786 bp of intron (692 bp 5′ and 94 bp 3′ sequence), and 93 bp of the 3′ exon. pKS-aI5γ is the construct for aI5γ, and contains 21 bp of the 5′ exon, the entire 886-bp intron sequence, and 191 bp of the 3′ exon.
Site-directed mutagenesis was by PCR using the overlap extension method (Sambrook and Russell 2001). All PCR reactions for generation of constructs were made using Pfu DNA polymerase (Stratagene) to minimize mutations, and site-directed mutations were verified by sequencing.
In vitro transcription reactions
Plasmids were linearized by SalI (B.a.I1; B.a.I2) or PvuII (aI5γ). Transcription reactions were with 0.5 μg plasmid template and 50 U T7 RNA polymerase (Invitrogen) according to the manufacturer’s instructions. Transcripts were extracted with phenol-CIA, and ethanol precipitated in 2 M NH4OAc. The pellet was washed with 70% ethanol and resuspended immediately without drying in TE. For radiolabeled transcripts, transcription was performed using 10 μCi [α-32P] UTP (3000 Ci/mmole, Amersham Pharmacia Biosciences), 0.1 mM UTP, and 0.5 mM other NTPs. Transcripts were gel-purified on a 1% agarose TBE gel (90 mM Tris-borate, pH 8.3, 1 mM EDTA), and extracted by a MinElute gel extraction column (QIAGEN) according to the manufacturer’s protocol for dsDNA molecules.
In vitro self-splicing
Transcripts were denatured and refolded using a Perkin Elmer 2400 PCR machine, by incubating the transcript in TE at 90°C for 1 min, 75°C for 5 min, and then cooling to 45°C over 15 min. Intron transcripts were spliced with 20,000 cpm RNA (13 fmole) or 0.1 μg (~250 fmole) unlabeled transcript at 45°C in 40 mM Tris-HCl, pH 7.6, 100 mM MgCl2, and 1 M NH4Cl. Reactions were initiated by adding prewarmed splicing buffer to the transcript RNA, and stopped by placing in dry ice. Products were precipitated with 0.3 M NaOAc, pH 5.2 and 1 μg carrier tRNA (Sigma), resuspended in formamide loading buffer (Sambrook and Russell 2001), and heated to 85°C for 2 min before loading onto 4% polyacrylamide (19:1 acrylamide:bisacrylamide)/8 M urea gels. Dried gels were quantitated by BAS1000 phosphorimaging (Fuji Medical Systems USA). Unlabeled transcripts were spliced in the same way, but resuspended in ddH20 and used as templates for RT-PCR.
PCR and RT-PCR
cDNA synthesis of ligated exon products was performed using 100 U SuperScript II RNase H− reverse transcriptase (Invitrogen) at 42°C according to the manufacturer’s instructions, with 100 ng cold splicing products as templates (above) and the primers Ba-23-AS-C (5′-CCCATCGATGCAGTAGGGAAAGGGTTCATGTGAG) for B.a.I2, Ba-07-AS-C (5′-CCCATCGATCGGTATCCAA GATAGAAGGTCCTAC) for B.a.I1, and SK (5′-CGCTCTAGAACTAGTGGATC) for aI5γ. PCR conditions for all PCR and RT-PCR reactions were: 5 min at 94°C; 25 cycles of 30 sec at 94°C, 30 sec at 55°C, 1 min/kb at 72°C; and 7 min at 72°C using the manufacturer’s buffers and 5 mM MgCl2. For Pfu reactions, the extension time was 2 min/kb.
Radioactive RT-PCR assay for detection of alternative splice sites
Radioactive RT-PCR assays were performed using gel-purified primers (Ba-23-AS-C for B.a.I2; SK for aI5γ) end-labeled with γ-ATP (3000 Ci/mmole, Amersham Pharmacia Biosciences) and T4 polynucleotide kinase (Invitrogen) according to the manufacturer’s instructions to a specific activity of 106–107 cpm/μg. Cold cDNA synthesis reactions were as described above, and subsequent PCR reactions were with 5000 cpm (~4 pmole) of labeled primer, 1.25 pmole of corresponding cold primer, and 5 pmole of Ba-23-S-B (5′-CGCGGATCCCTGTATTGTTGTGGAAGGTGTAGGA) for B.a.I2 or T3 (5′-ATTAACCCTCACTAAAGGG) for aI5γ. PCR reactions were phenol-CIA extracted, ethanol-precipitated with 0.3M NaOAc, pH 5.2, and digested with AvaII for B.a.I2 (New England Biolabs) or EcoRI for aI5γ (Invitrogen) to ensure homogeneous ends. Products were ethanol-precipitated in 0.3M NaOAc, pH 5.2, resuspended in formamide dye, heated to 85°C for 2 min, and run on an 8% polyacrylamide gel containing 8 M urea and 40% formamide. The inclusion of formamide was found to be necessary to remove persistent secondary structure of the B.a.I2 exons under normal gel conditions.
Acknowledgments
This work is supported by the Natural Sciences and Engineering Research Council (NSERC) and the Alberta Heritage Foundation for Medical Research (AHFMR).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5246804.
REFERENCES
- Adamidi, C., Fedorova, O., and Pyle, A.M. 2003. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry 42: 3409–3418. [DOI] [PubMed] [Google Scholar]
- Belfort, M., Derbyshire, V., Parker, M.M., Cousineau, B., and Lambowitz, A.M. 2002. In Mobile DNA II (eds. N.L. Craig et al.), pp. 761–777. ASM Press, Washington, DC.
- Bonen, L. and Vogel, J. 2001. The ins and outs of group II introns. Trends Genet. 17: 322–331. [DOI] [PubMed] [Google Scholar]
- Boulanger, S.C., Faix, P.H., Yang, H., Zhuo, J., Franzen, J.S., Peebles, C.L., and Perlman, P.S. 1996. Length changes in the joining segment between domains 5 and 6 of a group II intron inhibit self-splicing and alter 3′ splice site selection. Mol. Cell Biol. 16: 5896–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copertino, D.W., Shigeoka, S., and Hallick, R.B. 1992. Chloroplast group III twintron excision utilizing multiple 5′ - and 3′ -splice sites. EMBO J. 11: 5041–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M., Michel, F., and Westhof, E. 2000. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 19: 5007–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L. and Zimmerly, S. 2002. Compilation and analysis of group II intron insertions in bacterial genomes: Evidence for retroelement behavior. Nucleic Acids Res. 30: 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferat, J.L. and Michel, F. 1993. Group II self-splicing introns in bacteria. Nature 364: 358–361. [DOI] [PubMed] [Google Scholar]
- Ferat, J.L., Le Gouar, M., and Michel, F. 2003. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol. Microbiol. 49: 1407–1423. [DOI] [PubMed] [Google Scholar]
- Granlund, M., Michel, F., and Norgren, M. 2001. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 183: 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L. and Hallick, R.B. 1994. A group III intron is formed from domains of two individual group II introns. Genes & Dev. 8: 1589–1599. [DOI] [PubMed] [Google Scholar]
- Jacquier, A. and Michel, F. 1990. Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 213: 437–447. [DOI] [PubMed] [Google Scholar]
- Jarrell, K.A., Peebles, C.L., Dietrich, R.C., Romiti, S.L., and Perlman, P.S. 1988. Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem. 263: 3432–3439. [PubMed] [Google Scholar]
- Jenkins, K.P., Hong, L., and Hallick, R.B. 1995. Alternative splicing of the Euglena gracilis chloroplast roaA transcript. RNA 1: 624–633. [PMC free article] [PubMed] [Google Scholar]
- Lambowitz, A.M., Caprara, M.G., Zimmerly, S., and Perlman, P.S. 1999. In The RNA world (eds. R.F. Gesteland et al.), pp. 451–484. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Landthaler, M. and Shub, D.A. 1999. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: Introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc. Natl. Acad. Sci. 96: 7005–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abarca, F., Zekri, S., and Toro, N. 1998. Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol. Microbiol. 28: 1295–1306. [DOI] [PubMed] [Google Scholar]
- Matsuura, M., Noah, J.W., and Lambowitz, A.M. 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J. 20: 7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F. and Ferat, J.L. 1995. Structure and activities of group II introns. Annu. Rev. Biochem. 64: 435–461. [DOI] [PubMed] [Google Scholar]
- Michel, F. and Jacquier, A. 1987. Long-range intron–exon and intron–intron pairings involved in self-splicing of class II catalytic introns. Cold Spring Harbor Symp. Quant. Biol. 52: 201–212. [DOI] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., and Ozeki, H. 1989. Comparative and functional anatomy of group II catalytic introns—A review. Gene 82: 5–30. [DOI] [PubMed] [Google Scholar]
- Mills, D.A., McKay, L.L., and Dunny, G.M. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lacto-cocci. J. Bacteriol. 178: 3531–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Adelantado, E., San Filippo, J., Martínez-Abarca, F., García-Rodríguez, F.M., Lambowitz, A.M., and Toro, N. 2003. Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J. Mol. Biol. 327: 931–943. [DOI] [PubMed] [Google Scholar]
- Murray, P.R. 1999. Manual of clinical microbiology, 7th ed., pp. 360. ASM Press, Washington, DC.
- Noah, J.W. and Lambowitz, A.M. 2003. Effects of maturase binding and Mg2+ concentration of group II intron RNA folding investigated by UV cross-linking. Biochemistry 42: 12466–12480. [DOI] [PubMed] [Google Scholar]
- Okinaka, R., Cloud, K., Hampton, O., Hoffmaster, A., Hill, K., Keim, P., Koehler, T., Lamke, G., Kumano, S., Manter, D., et al. 1999. Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 87: 261–262. [DOI] [PubMed] [Google Scholar]
- Qin, P.Z. and Pyle, A.M. 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 8: 301–308. [DOI] [PubMed] [Google Scholar]
- Saldanha, R., Chen, B., Wank, H., Matsuura, M., Edwards, J., and Lambowitz, A.M. 1999. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38: 9069–9083. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Mutagenesis. In Molecular cloning: A laboratory manual Vol. 2, pp. 13.36–13.39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- San Filippo, J. and Lambowitz, A.M. 2002. Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J. Mol. Biol. 324: 933–951. [DOI] [PubMed] [Google Scholar]
- Sellem, C.H. and Belcour, L. 1994. The in vivo use of alternate 3′-splice sites in group I introns. Nucleic Acids Res. 22: 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor, N., Hausner, G., and Zimmerly, S. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 17: 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, N. 2003. Bacteria and Archaea Group II introns: Additional mobile genetic elements in the environment. Environ. Microbiol. 5: 143–151. [DOI] [PubMed] [Google Scholar]
- Vader, A., Johansen, S., and Nielsen, H. 2002. The group I-like ribo-zyme DiGIR1 mediates alternative processing of pre-rRNA transcripts in Didymium iridis. Eur. J. Biochem. 269: 5804–5812. [DOI] [PubMed] [Google Scholar]
- Wallweber, G.J., Mohr, S., Rennard, R., Caprara, M.G., and Lambowitz, A.M. 1997. Characterization of Neurospora mitochondrial group I introns reveals different CYT-18 dependent and independent splicing strategies and an alternative 3′ splice site for an intron ORF. RNA 3: 114–131. [PMC free article] [PubMed] [Google Scholar]
- Wank, H., San Filippo, J., Singh, R.N., Matsuura, M., and Lambowitz, A.M. 1999. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell 4: 239–250. [DOI] [PubMed] [Google Scholar]