Abstract
It has been shown that the activity of the hammerhead ribozyme at μM magnesium ion concentrations is markedly increased by the inclusion of loops in helices I and II. We have studied the effect of such loops on the magnesium ion-induced folding of the ribozyme, using fluorescence resonance energy transfer. We find that with the loops in place, folding into the active conformation occurs in a single step, in the μM range of magnesium ion concentration. Disruption of the loop–loop interaction leads to a reversion to two-step folding, with the second stage requiring mM concentrations of magnesium ion. Sodium ions also promote the folding of the natural form of the ribozyme at high concentrations, but the folding occurs as a two-stage process. The loops clearly act as important auxiliary elements in the function of the ribozyme, permitting folding to occur efficiently under physiological conditions.
Keywords: RNA catalysis, RNA folding, metal ions, FRET
INTRODUCTION
Folding and activity are intimately related at a number of levels in catalytic RNA species. These molecules must adopt a structure that generates the local environment that accelerates a chemical reaction, while the RNA conformation may contribute directly to catalysis via orientation and proximity effects. In general, RNA folding requires metal ions to neutralize the electrostatic charge of the phosphodiester backbone. These ions can in principle be bound tightly to the RNA and may play a direct role in the chemistry of catalysis by acting as a general acid/base or through electrostatic effects.
The hammerhead ribozyme is the smallest member of the class of nucleolytic ribozymes (Takagi et al. 2001; Fedor 2002; Lilley 2003a); autocatalytic species that bring about a reversible site-specific cleavage of their backbone via a transesterification reaction in which the 2′-oxygen attacks the 3′-phosphorus, with departure of the 5′-oxygen to leave a cyclic 2′-3′-phosphate. The hammerhead ribozyme (Forster and Symons 1987; Uhlenbeck 1987; Hazeloff and Gerlach 1988) comprises three helical sections (I, II, and III) that are connected via an elaborated three-way helical junction (Fig. 1 ▶). Crystallography of the ribozyme has shown that the folded structure is built around two elements (Pley et al. 1994; Scott et al. 1995). Domain 1 is the probable active site of the ribozyme, in which the CUGA sequence of the single-stranded section connecting helices I and II forms a uridine turn adjacent to the scissile phosphate. Domain 2 mediates the coaxial alignment of helices II and III, formed by formation of consecutive G•A mismatches.
FIGURE 1.
The hammerhead ribozyme. (A) The sequence of the catalytic core of the ribozyme, with the position of cleavage arrowed. The critical nucleotides are indicated in bold, and the core and helices are numbered conventionally (Hertel et al. 1992). (B) Schematic showing the minimal ribozyme, comprising the core and open helices I, II, and III. (C) The natural form of the ribozyme found in many plant viroids, with terminal loops closing helices I and II. The lengths of the helices shown here are taken from the ribozyme of the tobacco ringspot virus satellite RNA (+) strand (Buzayan et al. 1986). (D) The form of the ribozyme found in a transcript of Schistosome satellite DNA (Ferbeyre et al. 1998), with an internal loop present in helix I.
All the folding studies on the hammerhead ribozyme to date have employed constructs that preserve the core sequences together with the helical arms that define the three-way junction (Bassi et al. 1995, 1997; Amiri and Hagerman 1996; Menger et al. 1996; Hammann et al. 2001b; Bondensgaard et al. 2002; Mikulecky and Feig 2002; Rueda et al. 2003). However, there has been no attempt to maintain the natural sequence within these arms, and they have been frequently extended or terminated with unnatural tetraloop sequences to suit particular experiments. A model for the ion-induced folding of the ribozyme has emerged (Hammann and Lilley 2002) based on electrophoretic (Bassi et al. 1995, 1996), fluorescence (Bassi et al. 1997, 1999; Menger et al. 2000), and nuclear magnetic resonance (Hammann et al. 2001b; Bondensgaard et al. 2002) studies. In the absence of added metal ions, the hammerhead ribozyme exists in an extended conformation with an open central core and no pairwise helical stacking. Folding occurs by a sequential, two-stage folding process, in which both stages are induced by the noncooperative binding of divalent metal ions. The first step involves the formation of the domain 2 structure and occurs in the μM range of magnesium ion concentration (half-magnesium ion concentration [Mg2+]1/2 ~ 100 μM). The uridine turn (domain 1) forms in the second stage of folding, requiring mM magnesium ion concentration ([Mg2+]1/2 ~ 2 mM). A two-stage, ion-induced folding process is also indicated by calorimetric measurements (Hammann et al. 2001a). The activity of the ribozyme as a function of magnesium ion concentration parallels the folding (Dahm and Uhlenbeck 1991), with half-maximal rates of cleavage at ~ 5 mM and virtually no detectable activity below 1 mM. This can be well understood in terms of the folding scheme, as the same (mM) concentrations of magnesium ions are required for formation of domain 1 and the acquisition of catalytic activity.
The magnesium ion concentration required for the folding and activity of this basic form of the ribozyme is higher than the free-ion concentration in the cell. However, it has recently been shown that forms of the ribozyme that preserve more of the natural sequence exhibit activity in μM concentrations of magnesium ions (De la Pena et al. 2003; Khvorova et al. 2003), a concentration that is two orders of magnitude lower than that required for previously studied forms of the ribozyme. This marked effect requires the presence of terminal loops found in helices II and III of some hammerhead ribozymes, and permutation experiments together with molecular modeling suggested that these would interact. Evidently, these features, which had been deleted from the species previously studied, were required for catalytic function under physiological conditions. It was consequently proposed that a tight interaction between the loops would change the folding pathway by stabilization of the final conformation in which helices II and III were physically close, perhaps allowing the ion-induced folding to proceed in a single stage at substantially lower magnesium ion concentration (Lilley 2003b).
In this work we have set out to examine whether or not the ion-induced folding of the natural form of the hammerhead ribozyme is altered from that of the minimal form. We have used fluorescence resonance energy transfer (FRET) to study the relative orientation of helical arms during the folding process. For these experiments we have used a slightly different form of the ribozyme, the one occurring in Schistosoma mansoni, where helix I contains an internal loop (Chartrand et al. 1995; Ferbeyre et al. 1998), as opposed to the terminal loop occurring in hammerhead species from other sources. We show that both folding and activity of this ribozyme occur in μM concentrations of magnesium ions, but that the properties revert to those of the minimal ribozyme if the loop–loop interaction is perturbed.
RESULTS
FRET analysis of the folding of the loop-carrying form of the hammerhead ribozyme
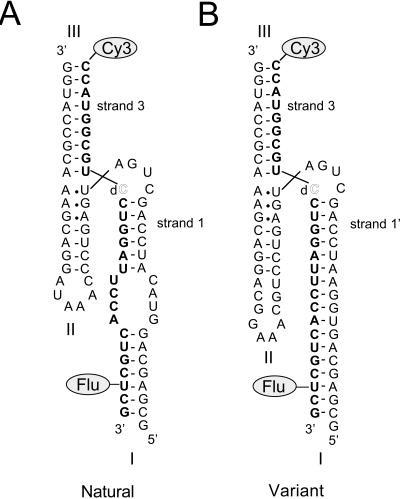
We have studied the ion-induced folding of the hammerhead ribozyme with the sequence of that found in Schistosoma mansoni (Chartrand et al. 1995; Ferbeyre et al. 1998) by means of FRET, using the construct shown in Figure 2 ▶. The ribozyme was constructed from two strands (named according to the helix that contains their 5′ termini), which is possible in this form of the ribozyme because of the absence of a terminal loop in helix I. Strand 1 is a 51-nt RNA molecule that was made by transcription. Strand 3 was prepared by chemical synthesis; it contains a Cy3 at the 5′ terminus and a 5-fluorescein-deoxyuracil substitution near the 3′ terminus. Ribozyme cleavage was prevented by substitution of 2′ -deoxyribocytidine at position 17, which removes the nucleophile in the transesterification reaction. Hybridization of these two strands generates the species shown in Figure 2A ▶, with the donor (fluorescein) and acceptor (Cy3) fluorophores attached at or near the ends of helices I and III, respectively. These should become further apart as the ribozyme folds into its active conformation, leading to a reduction in the efficiency of energy transfer as the concentration of divalent metal ions is increased.
FIGURE 2.
Nucleotide sequence of the hammerhead constructs used in the FRET analysis of folding. These are drawn in a way that approximates the final folded conformation. Both constructs were assembled from two strands. Strand 1 and 1′ (normal type) were generated by transcription, whereas strand 3 (bold type) was made by chemical synthesis. Strand 3 also incorporated C17 as a 2′-deoxyribonucleotide (open type), to prevent ribozyme action, Cy3 at the 5′ terminus, and 5-fluorescein deoxyribouridine (Flu) three nucleotides from the 3′terminus. This places the donor and acceptor fluorophores near the termini of helices I and III, respectively. (A) The natural sequence ribozyme, with terminal and internal loops. (B) Variant sequence generated by hybridization of an alternative strand 1′. This removes the internal loop of helix I by complementation, lengthens helix II by 3 bp, and replaces the terminal loop with a stable tetraloop sequence.
The loop-carrying form of the hammerhead ribozyme folds in μM concentrations of magnesium ions
We have analyzed the ion-induced folding of the ribozyme by measuring the efficiency of energy transfer (EFRET) between the fluorophores attached to helices I and III (Fig. 2A ▶) as a function of magnesium ion concentration (Fig. 3 ▶). On addition of magnesium ions, the value of EFRET reduced, corresponding to a conformational transition that resulted in helices I and III becoming further apart. The FRET efficiency reduced even for the smallest increases in ion concentration, and the process occurred principally in the μM range of magnesium ion concentration. The data are well fitted by a two-state model (equation 1), giving a Hill coefficient of n = 0.85 and a half-point of [Mg2+]1/2 = 160 μM (R = 0.998). No evidence for an intermediate state was detected; folding appeared to occur in a single step, induced by the noncooperative binding of magnesium ions. These data are in marked contrast to those obtained in analogous experiments using the minimal form of the ribozyme containing the core, but lacking the loop-carrying helices I and II (Bassi et al. 1997). The minimal ribozyme (i.e., with loops removed) folded in two stages, with an initial increase in FRET efficiency for the I–III vector, before reducing in a second, separate transition that occurred in the mM concentration range. No significant improvement in the fit to the data for the loop-carrying ribozyme could be obtained by allowing for a second transition. Thus, the natural form of the ribozyme undergoes magnesium ion-induced folding at a concentration that is substantially lower than that for the minimal form.
FIGURE 3.
FRET analysis of the folding of the natural ribozyme induced by addition of magnesium ions. Efficiency of fluorescein-Cy3 energy transfer (EFRET) for the construct shown in Figure 2A ▶ is plotted as a function of magnesium ion concentration. The data have been fitted to a two-state binding model, given by equation 1. The insert shows the same data and fit plotted on a logarithmic scale of magnesium ion concentration.
The natural form of the hammerhead ribozyme is active in μM concentrations of magnesium ions
Because the loop-carrying ribozyme folds in the presence of μM concentrations of magnesium ions, we examined the cleavage activity of the same form of the ribozyme under these conditions. Single-turnover cleavage experiments were performed using the ribozyme shown in Figure 4A ▶. The core and loop-carrying arms of the ribozyme were identical to those of the ribozyme used for the folding experiments, except that the deoxycytidine at position 17 was replaced by ribocytidine, and the fluorophores were not present. To ensure rapid dissociation of the 5′ product of cleavage, we used a version of the ribozyme in which helix III was only 5 bp in length, and it was consequently found necessary to add 500 mM NaCl to stabilize the ribozyme. Radioactively [5′-32P]-labeled strand 3 was incubated with a 1000-fold excess of strand 1 in the presence of 100 μM magnesium ions plus the indicated monovalent ion concentration, and conversion to product was analyzed by gel electrophoresis and phosphorimaging (Fig. 4B ▶, closed circles). Almost complete cleavage of the substrate strand occurred under these conditions, and the data fitted to a single exponential giving a rate of cleavage of kobs = 0.05 min−1. A modified form of the ribozyme in which the loop of helix I was removed by complementary base-pairing was incubated under identical conditions. This form gave virtually no cleavage during the 30-min incubation (Fig. 4B ▶, open circles).
FIGURE 4.
Ribozyme cleavage of the natural form of the hammerhead in the presence of low magnesium ion concentration. Cleavage activity of natural and variant ribozymes was studied under single-turnover conditions. (A) Sequences of the natural and variant ribozymes used in these experiments. The ribozyme is similar to that used in the FRET experiments (Fig. 2 ▶), except that helix III was reduced to 5 bp in length to facilitate product release. Strand 3 was synthesized with ribocytidine at position 17 to permit ribozyme activity. The variant was constructed using a modified strand 3 such that helix I is now fully base-paired, thereby removing the internal loop. Strand 3 was radioactively [5′-32P] labeled in these experiments. The site of ribozyme cleavage is arrowed. (B) Progress curve of ribozyme cleavage. The fraction of cleaved RNA is plotted as a function of time and is fitted to a single exponential (equation 3) for the natural (closed circles) and variant (open circles) ribozymes.
Disruption of loop—loop interaction abolishes folding at low magnesium ion concentration
Although it seemed very likely that the folding of the loop-carrying form of the ribozyme in μM magnesium ion concentrations was the result of an interaction between the loops, we tested this by disrupting the interaction and analyzing the folding of the modified species by FRET, as before. We therefore generated a modified strand 1′, resulting in a removal of the internal loop of helix I by complementation of base-pairing, and a lengthened stem-loop II terminated by a stable tetraloop (Fig. 2B ▶). This should leave no possibility of any residual interaction between the two loops. This strand was hybridized to the same fluorescein, Cy3-labeled strand 3 as before, and FRET efficiency was measured as a function of magnesium ion concentration (Fig. 5 ▶). The resulting changes in FRET are quite different from those exhibited by the natural form of the ribozyme. Folding occurs in two distinct steps, with an initial increase in EFRET at low magnesium ion concentration, followed by a reduction in the mM range. These results are very similar to those observed previously for the minimal form of the ribozyme (Bassi et al. 1997). A good fit of the data requires a model involving two separate transitions induced by non-cooperative ion binding (equation 2), with values of [Mg2+]1/2 = 55 μM and 2 mM (R = 0.997).
FIGURE 5.
FRET analysis of the folding of the variant ribozyme in which loop–loop interaction has been disrupted, induced by addition of magnesium ions. Efficiency of fluorescein-Cy3 energy transfer (EFRET) for the construct shown in Figure 2B ▶ is plotted as a function of magnesium ion concentration. The data have been fitted to a three-state binding model, given by equation 2. The inset shows the same data and fit plotted on a logarithmic scale of magnesium ion concentration.
Separate folding transitions observed in sodium ions
We have examined the folding of the loop-carrying form of the ribozyme induced by the addition of sodium ions. Using the form of the ribozyme shown in Figure 2A ▶, we studied the FRET efficiency as a function of sodium ion concentration (Fig. 6A ▶). We observed an overall reduction in FRET efficiency with addition of sodium ions, indicating that a similar conformational transition is induced. As expected, very much higher concentrations of sodium ions are required to induce folding. Interestingly, in contrast to the results using magnesium ions, we observed a small increase in EFRET at the lower sodium ion concentrations. It appears that the presence of monovalent ion results in a separation of two distinct transitions in the folding of the hammerhead ribozyme despite the presence of the loops. The main transition at higher sodium ion concentration has a [Na+]1/2 = 220 mM (R = 0.998). Very similar two-stage folding was observed by the addition of lithium ions, although the smaller ion was a little more efficient in inducing folding, with [Li+]1/2 = 110 mM (R = 0.999; data not shown). We also studied the sodium ion-induced folding of the loop-disrupted variant of Figure 2B ▶; the FRET data are plotted in Figure 6B ▶. In the absence of loop–loop interaction, both transitions require significantly higher ion concentrations, and the second stage is clearly incomplete at 700 mM sodium, with [Na+]1/2 = 540 mM (R = 0.995).
FIGURE 6.
FRET analysis of the folding of the hammerhead ribozyme induced by addition of sodium ions. Efficiency of fluorescein-Cy3 energy transfer (EFRET) for the construct shown in Figure 2A ▶ was studied as before. The data have been fitted to a three-state binding model, given by equation 2. (A) FRET efficiency for the natural ribozyme as a function of sodium ion concentration. (B) FRET efficiency for the variant (loop-disrupted, Fig. 2B ▶) ribozyme as a function of sodium ion concentration. (C) FRET efficiency for the natural ribozyme as a function of sodium ion concentration in the presence of a constant 100 μM magnesium ions.
In view of the different behavior of the natural form of the hammerhead in divalent and monovalent cations we examined the effect of addition of sodium ions to ribozyme that was already substantially folded in the presence of magnesium ions. Using the fluorescent vector shown in Figure 2A ▶, we studied FRET efficiency as a function of sodium ion concentration in the presence of a constant background concentration of 100 μM of magnesium ions. Despite beginning from a folded state before addition of the monovalent ions, addition of sodium ions up to 100 mM brought about an increase in EFRET, indicating an initial reversal of the folded state. At higher concentrations of sodium ions, the FRET efficiency reduced once again, consistent with a sodium ion-induced folding with [Na+]1/2 = 230 mM (R = 0.998). The ribozyme is substantially folded in the presence of 100 μM magnesium, 500 mM sodium; that is, the conditions under which the cleavage reaction was studied (Fig. 4 ▶).
DISCUSSION
Our results clearly demonstrate that the ion-induced folding of the hammerhead ribozyme is greatly enhanced by the presence of the loops in helices I and II. The observed activity of these natural forms of the ribozyme at μM magnesium ion concentrations may be explained by their ability to undergo folding at substantially lower ion concentrations compared with the minimal forms. Disruption of the proposed loop–loop interaction leads to a reversion to two-step folding at mM magnesium ion concentrations. These concentrations are significantly higher than those likely to exist inside living cells, and this explains why the loops are essential for the activity of transfected RNA in vivo (Khvorova et al. 2003).
The minimal form of the ribozyme (from which the loops have been removed) folds in two distinct steps, clearly shown by the FRET analysis of the variant species shown in Figure 5 ▶. We have shown previously that the two stages correspond to the formation of domain 2, occurring at μM magnesium ion concentration, followed by domain 1 at mM magnesium ion concentration (Hammann et al. 2001b). This is depicted in Figure 7 ▶. We were unable to fit the measured FRET efficiencies for the complete ribozyme (i.e., with the loops present) to a model that introduced an intermediate species in the folding process. It seems improbable that formation of domain 2 (i.e., interconversion of states U and I) would be affected by the presence of the loops, but a tight tertiary interaction should stabilize the final structure (state F), potentially increasing the rate k2, and almost certainly reducing k−2. Thus, the equilibrium between the intermediate (I) and final states will be shifted toward the fully folded ribozyme by the loop–loop interaction in the presence of magnesium ions. Sodium ions induce folding of the loop-carrying ribozyme at very high concentrations, but a two-stage transition is observed. Thus the monovalent ion is less effective at stabilizing the loop–loop interaction so that the final state is less favored in the conformational I <> F equilibrium. Interestingly, the folding of the natural ribozyme in magnesium ions is reversed by intermediate concentrations of sodium ions. This suggests that the magnesium ions are displaced by concentrations of monovalent ions that are unable to induce folding by themselves, consistent with two distinct modes of binding by the different metal ions.
FIGURE 7.
A folding scheme for the hammerhead ribozyme. Schematic to show the two-stage folding scheme previously proposed for the hammerhead ribozyme. State U exists in the absence of added metal ions, in which the three helical arms extend from an open central core. On addition of metal ions, the minimal ribozyme undergoes folding in two steps, corresponding to the two transitions observed in Figure 5 ▶. In the first stage, domain 2 is formed, resulting in a coaxial alignment of helices II and III (state I). Further increase in metal ion concentration leads to the folding of the catalytic core (domain 1), reorienting helix I into the same quadrant as helix II (state F). The presence of the loops in helices I and II leads to folding at μM magnesium ion concentration, with no form I detectable. The stabilization of form F by the loop–loop interaction drives the I <> F equilibrium such that form I is not observed. However, form I is detected at intermediate ionic concentrations in the same construct when sodium replaces magnesium ions.
A tight interaction between the loops might also affect the local structure within the core of the ribozyme. This may explain the different initial and final FRET efficiencies observed for the two forms of the ribozyme, but the interpretation of this state is complicated by the possible effects of the internal loop on the trajectory of helix I, altering the position of the fluorescein with respect to the Cy3. However, it has been observed that chemical cross-linking of helices I and II leads to significant enhancement in the efficiency of the ligation reaction by the hammerhead ribozyme (Stage-Zimmermann and Uhlenbeck 2001; Blount and Uhlenbeck 2002). Subtle changes in the ribozyme core might result in a better alignment of the substrates for ligation. It is also likely that some rearrangement within the core is required for the cleavage reaction. A series of conformational changes has been observed by crystallography of different forms of the ribozyme (Scott et al. 1996; Murray et al. 2000, 2002), including a form in which helices I and II have become tethered by strand exchange (Dunham et al. 2003). It has been proposed that a larger-scale rearrangement is required to achieve the activated complex (Wang et al. 1999). In principle, any such local changes in structure might be affected by the loop–loop interaction. This will require further analysis, focusing on more local conformational changes within the ribozyme core.
The role of the loops in the folding of the hammerhead ribozyme suggests a clear parallel with another nucleolytic ribozyme, the hairpin ribozyme (Fedor 2000; Lilley 2001; Rupert and Ferré-D’Amaré 2001). The active center of the hairpin is formed by the close interaction of two internal loops that are presented on two helical arms of a four-way junction (Murchie et al. 1998; Rupert & Ferré-D’Amaré 2001). However, rather like the situation with the hammerhead, the great majority of earlier studies of the hairpin ribozyme employed a simplified version in which the junction was removed and the two loop-carrying helices were connected by a phosphodiester hinge point. Unsurprisingly, however, the junction has a major effect on the folding and activity of the ribozyme. Folding of the natural form occurs at a magnesium ion concentration that is three orders of magnitude lower than that of the simplified hinged form (Zhao et al. 2000), and it thus results in activity under physiological conditions. Time-resolved FRET studies confirmed that the stabilization of the folded form was the result of the four-way junction (Walter et al. 1999), and recent FRET studies on single-ribozyme molecules have demonstrated that the junction accelerates the folding 500-fold by virtue of introducing an additional, distinct intermediate state into the folding process that presents the loops for interaction (Tan et al. 2003). The net result of this is that the folding of the ribozyme is not rate limiting, unlike the case of the minimal, hinged form (Zhuang et al. 2002). Ligation activity of the hairpin ribozyme is significantly enhanced by the presence of the four-way junction (Fedor 1999).
Both the hammerhead and hairpin ribozymes have been oversimplified in early studies, resulting in species that were seriously impaired because of removal of elements that are critical for efficient folding under physiological conditions. We can regard these auxiliary elements as “folding enhancers”; these are essential for efficient folding under physiological conditions, although they do not participate directly in the chemistry of catalysis. This provides an object lesson for the future, in taking care not to remove elements that are vital for the function of the RNA, and further underlines the intimate connection between folding and catalysis in the ribozymes.
MATERIALS AND METHODS
Construction of hammerhead species for fluorescence
The hammerhead species used for analysis of folding were constructed from two RNA species (as in Fig. 2 ▶). The sequences were (all written 5′ to 3′): strand 1, gcgagcagguacauccagcugaugagucccaaauaggacgaaacgccaugg; strand 3, Cy3-ccauggcguccuggauuccacugc(f-du)cg, where F-dU indicates 5-fluorescein deoxyuracil substitution, and 2′-deoxyribose nucleotides are underlined. A variant strand 1 was also prepared: strand 1′, gcgagcaguggaauccagcugaugaguccugcaaaggcaggacgaaacgccaugg, in which the internal loop of helix I was removed by complementation of base-pairing, and stem–loop II was modified.
Strands 1 and 1′ were synthesized by transcription using T7 RNA polymerase from single-stranded DNA templates (Milligan et al. 1987). Strand 3 was chemically synthesized using tBDMS phosphoramidite chemistry (Beaucage and Caruthers 1981), as described previously (Wilson et al. 2001). The fluorophores 2′de-oxyuracil-5-fluorescein and Cy3 were coupled as phosphoramidites (Glen Research). RNA was purified by electrophoresis in 12% polyacrylamide gels containing 7 M urea. RNA was extracted from gel slices by electroelution into 8 M ammonium acetate, recovered by ethanol precipitation, and dissolved in water. The fluorescent strand 3 was further purified by reversed-phase high-pressure liquid chromatography.
Hammerhead constructs were prepared by incubating stoichiometric amounts of the two oligonucleotides in 90 mM Tris-borate (pH 8.3) for 10 min. at 80°C, followed by slow cooling. The hybridized species were purified by electrophoresis in a polyacrylamide gel at 4°C for 22 h at 120 V. The buffer system contained 90 mM Tris-borate (pH 8.3) and was recirculated at >1 L/h. The RNA was excised and electroeluted into 8 M ammonium acetate and recovered by ethanol precipitation.
FRET analysis of folding
Fluorescence spectra were recorded in the steady state at 4°C using an SLM-Aminco 8100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of EFRET were measured using the acceptor normalization method (Clegg 1992). FRET data for the natural form of the ribozyme were fitted to a two-state model:
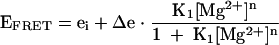 |
(1) |
where ei is the FRET efficiency in the absence of added metal ions, Δe is the change in FRET on addition of magnesium ions (with a negative sign for the I–II vector), K1 is an apparent association constant for magnesium ions, and n is a Hill coefficient. For the folding induced by sodium ions, and the folding of the variant form of the ribozyme, it was necessary to add a further term in the equation:
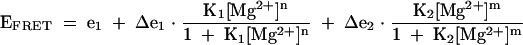 |
(2) |
where K2 and m are the apparent association constant and Hill coefficient for the second transition. Δe1 and Δe2 are changes in FRET efficiency occurring in the two transitions (positive and negative respectively). The half-metal ion concentration ([Mg2+]1/2 or [Na+]1/2) is calculated as (1/K)1/n, where K is the apparent association constant and n the Hill coefficient.
Ribozyme cleavage assay
All RNA for analysis of ribozyme cleavage was chemically synthesized using ACE chemistry (Scaringe 2000) to construct the species shown in Figure 4A ▶. The ribozyme and substrate strands had the following sequences: strand 1, gcagguacauccagcugaugagucccaaauaggacgaaacggg; strand 3, cccguccuggauuccacugc.
A modified substrate strand that removes the internal loop was also synthesized: strand 3′, cccguccuggauguaccugc.
Strands 3 and 3′ were radioactively [5′-32P] labeled using [γ-32P]-ATP and polynucleotide kinase (Maxam and Gilbert 1980). Strand 1 plus strand 3 or 3′ were annealed in 100 mM Hepes (pH 7.0), 5 μM EDTA by heating at 94°C for 1 min, followed by slow cooling to 25°C. The cleavage reaction was initiated by adjusting the solution to final concentrations of 50 mM Hepes (pH 7.0), 100 μM MgCl2, 500 mM NaCl. Substrate and product were separated by polyacrylamide gel electrophoresis, and the extent of cleavage was estimated by exposure to storage phosphor screens and imaging. The first-order rate of cleavage (kobs) was calculated by fitting to
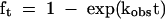 |
(3) |
where ft is the fraction cleaved at time t.
Acknowledgments
We thank Dr. Z. Zhao for chemical synthesis of fluorescent RNA and Cancer Research-UK and the BBSRC for financial support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5268404.
REFERENCES
- Amiri, K.M.A. and Hagerman, P.J. 1996. The global conformation of an active hammerhead RNA during the process of self-cleavage. J. Mol. Biol. 261: 125–134. [DOI] [PubMed] [Google Scholar]
- Bassi, G., Møllegaard, N.E., Murchie, A.I.H., von Kitzing, E., and Lilley, D.M.J. 1995. Ionic interactions and the global conformations of the hammerhead ribozyme. Nat. Struct. Biol. 2: 45–55. [DOI] [PubMed] [Google Scholar]
- Bassi, G.S., Murchie, A.I.H., and Lilley, D.M.J. 1996. The ion-induced folding of the hammerhead ribozyme: Core sequence changes that perturb folding into the active conformation. RNA 2: 756–768. [PMC free article] [PubMed] [Google Scholar]
- Bassi, G.S., Murchie, A.I.H., Walter, F., Clegg, R.M., and Lilley, D.M.J. 1997. Ion-induced folding of the hammerhead ribozyme: A fluorescence resonance energy transfer study. EMBO J. 16: 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi, G.S., Møllegaard, N.E., Murchie, A.I.H., and Lilley, D.M.J. 1999. RNA folding and misfolding of the hammerhead ribozyme. Biochemistry 38: 3345–3354. [DOI] [PubMed] [Google Scholar]
- Beaucage, S.L. and Caruthers, M.H. 1981. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypoly-nucleotide synthesis. Tetrahedron Lett. 22: 1859–1862. [Google Scholar]
- Blount, K.F. and Uhlenbeck, O.C. 2002. Internal equilibrium of the hammerhead ribozyme is altered by the length of certain covalent cross-links. Biochemistry 41: 6834–6841. [DOI] [PubMed] [Google Scholar]
- Bondensgaard, K., Mollova, E.T., and Pardi, A. 2002. The global conformation of the hammerhead ribozyme determined using residual dipolar couplings. Biochemistry 41: 11532–11542. [DOI] [PubMed] [Google Scholar]
- Buzayan, J.M., Hampel, A., and Bruening, G. 1986. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res. 14: 9729–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand, P., Harvey, S.C., Ferbeyre, G., Usman, N., and Cedergren, R. 1995. An oligodeoxyribonucleotide that supports catalytic activity in the hammerhead ribozyme domain. Nucleic Acids Res. 23: 4092–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg, R.M. 1992. Fluorescence resonance energy transfer and nucleic acids. Meth. Enzymol. 211: 353–388. [DOI] [PubMed] [Google Scholar]
- Dahm, S.C. and Uhlenbeck, O.C. 1991. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 30: 9464–9469. [DOI] [PubMed] [Google Scholar]
- De la Pena, M., Gago, S., and Flores, R. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22: 5561–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, C.M., Murray, J.B., and Scott, W.G. 2003. A helical pitch-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 332: 327–336. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Structure and function of the hairpin ribozyme. J. Mol. Biol. 297: 269–291. [DOI] [PubMed] [Google Scholar]
- ———. 2002. The role of metal ions in RNA catalysis. Curr. Opin. Struct. Biol. 12: 289–295. [DOI] [PubMed] [Google Scholar]
- Ferbeyre, G., Smith, J.M., and Cedergren, R. 1998. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell Biol. 18: 3880–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, A.C. and Symons, R.H. 1987. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49: 211–220. [DOI] [PubMed] [Google Scholar]
- Hammann, C. and Lilley, D.M.J. 2002. Folding and activity of the hammerhead ribozyme. ChemBioChem 3: 690–700. [DOI] [PubMed] [Google Scholar]
- Hammann, C., Cooper, A., and Lilley, D.M.J. 2001a. Thermodynamics of ion-induced RNA folding in the hammerhead ribozyme: An isothermal titration calorimetric study. Biochemistry 40: 1423–1429. [DOI] [PubMed] [Google Scholar]
- Hammann, C., Norman, D.G., and Lilley, D.M.J. 2001b. Dissection of the ion-induced folding of the hammerhead ribozyme using 19F NMR. Proc. Natl. Acad. Sci. 98: 5503–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeloff, J.P. and Gerlach, W.L. 1988. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334: 585–591. [DOI] [PubMed] [Google Scholar]
- Hertel, K.J., Pardi, A., Uhlenbeck, O.C., Koizumi, M., Ohtsuka, E., Uesugi, S., Cedergren, R., Eckstein, F., Gerlach, W.L., Hodgson, R., et al. 1992. Numbering system for the hammerhead. Nucleic Acids Res. 20: 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., and Jayasena, S.D. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10: 1–5. [DOI] [PubMed] [Google Scholar]
- Lilley, D.M.J. 2001. Origins of RNA catalysis in the hairpin ribozyme. ChemBioChem 2: 729–733. [DOI] [PubMed] [Google Scholar]
- ———. 2003a. The origins of RNA catalysis in ribozymes. Trends Biochem. Sci. 28: 495–501. [DOI] [PubMed] [Google Scholar]
- ———. 2003b. Ribozymes–A snip too far? Nat. Struct. Biol. 10: 672–673. [DOI] [PubMed] [Google Scholar]
- Maxam, A.M. and Gilbert, W. 1980. Sequencing end-labelled DNA with base-specific chemical cleavages. Meth. Enzymol. 65: 499–560. [DOI] [PubMed] [Google Scholar]
- Menger, M., Tuschl, T., Eckstein, F., and Porschke, D. 1996. Mg2+-dependent conformational changes in the hammerhead ribozyme. Biochemistry 35: 14710–14716. [DOI] [PubMed] [Google Scholar]
- Menger, M., Eckstein, F., and Porschke, D. 2000. Multiple conformational states of the hammerhead ribozyme, broad time range of relaxation and topology of dynamics. Nucleic Acids Res. 28: 4428–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky, P.J. and Feig, A.L. 2002. Cold denaturation of the hammerhead ribozyme. J. Am. Chem. Soc. 124: 890–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherall, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie, A.I.H., Thomson, J.B., Walter, F., and Lilley, D.M.J. 1998. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol. Cell 1: 873–881. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Szöke, H., Szöke, A., and Scott, W.G. 2000. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol. Cell 5: 279–287. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Dunham, C.M., and Scott, W.G. 2002. A pH-dependent conformational change, rather than the chemical step, appears to be rate-limiting in the hammerhead ribozyme cleavage reaction. J. Mol. Biol. 315: 121–130. [DOI] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., and McKay, D.B. 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74. [DOI] [PubMed] [Google Scholar]
- Rueda, D., Wick, K., McDowell, S.E., and Walter, N.G. 2003. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry 42: 9924–9936. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B. and Ferré-D’Amaré, A.R. 2001. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410: 780–786. [DOI] [PubMed] [Google Scholar]
- Scaringe, S.A. 2000. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 317: 3–18. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., and Klug, A. 1995. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Murray, J.B., Arnold, J.R.P., Stoddard, B.L., and Klug, A. 1996. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science 274: 2065–2069. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann, T.K. and Uhlenbeck, O.C. 2001. A covalent crosslink converts the hammerhead ribozyme from a ribonuclease to an RNA ligase. Nature Struct. Biol. 8: 863–867. [DOI] [PubMed] [Google Scholar]
- Takagi, Y., Warashina, M., Stec, W.J., Yoshinari, K., and Taira, K. 2001. Recent advances in the elucidation of the mechanisms of action of ribozymes. Nucleic Acids Res. 29: 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, E., Wilson, T.J., Nahas, M.K., Clegg, R.M., Lilley, D.M.J., and Ha, T. 2003. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. 100: 9308–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck, U.C. 1987. A small catalytic oligoribonucleotide. Nature 328: 596–600. [DOI] [PubMed] [Google Scholar]
- Walter, N.G., Burke, J.M., and Millar, D.P. 1999. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nature Struct. Biol. 6: 544–549. [DOI] [PubMed] [Google Scholar]
- Wang, S., Karbstein, K., Peracchi, A., Beigelman, L., and Herschlag, D. 1999. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry 38: 14363–14378. [DOI] [PubMed] [Google Scholar]
- Wilson, T.J., Zhao, Z.-Y., Maxwell, K., Kontogiannis, L., and Lilley, D.M.J. 2001. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40: 2291–2302. [DOI] [PubMed] [Google Scholar]
- Zhao, Z.-Y., Wilson, T.J., Maxwell, K., and Lilley, D.M.J. 2000. The folding of the hairpin ribozyme: Dependence on the loops and the junction. RNA 6: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X.W., Kim, H.D., Pereira, M.J.B., Babcock, H.P., Walter, N.G., and Chu, S. 2002. Correlating structural dynamics and function in single ribozyme molecules. Science 296: 1473–1476. [DOI] [PubMed] [Google Scholar]