Abstract
ATP(CTP):tRNA nucleotidyl transferases, tRNA maturing enzymes found in all organisms, and eubacterial poly(A) polymerases, enzymes involved in mRNA degradation, are so similar that until now their biochemical functions could not be distinguished by their amino acid sequence. BLAST searches and analysis with the program “Sequence Space” for the prediction of functional residues revealed sequence motifs which define these two protein families. One of the poly(A) polymerase defining motifs specifies a structure that we propose to function in binding the 3′ terminus of the RNA substrate. Similar motifs are found in other homopolyribonucleotidyl transferases. Phylogenetic classification of nucleotidyl tranferases from sequenced genomes reveals that eubacterial poly(A) polymerases have evolved relatively recently and are found only in a small group of bacteria and surprisingly also in plants, where they may function in organelles.
Keywords: nucleotidyl transferase, ATP(CTP):tRNA nucleotidyl transferase, poly(A) polymerase, RNA modification, RNA processing
INTRODUCTION
ATP(CTP):tRNA nucleotidyltransferases (we also use the synonyms CCA-adding enzymes, CCA transferases, or the abbreviation CCAtrs) are nucleotidyl transferases (Ntrs) responsible for the synthesis or repair of the 3′ terminal CCA sequence of tRNA molecules. They are coded by essential genes in almost all organisms and are able to add the three ribonucleotides C, C, and A to a tRNA’s 3′ end in a sequential order. CCAtrs were also identified and cloned from archaea (Yue et al. 1996; Seth et al. 2002). In addition, it was shown recently that in some eubacteria, two separate nucleotidyl transferases collaborate to build or repair a CCA tag by adding either CC or A to the 3′ ends of tRNA precursors (Tomita and Weiner 2001, 2002). Crystal structures of eubacterial, archaeal, and human CCAtrs have been solved recently (Li et al. 2002; Augustin et al. 2003; Okabe et al. 2003; Xiong et al. 2003).
Eubacterial poly(A) polymerases (eubPAPs, to distinguish them from eukaryotic poly(A) polymerases or PAPs) are RNA polymerases that add multiple AMPs to the 3′ ends of messenger RNAs. These poly(A) tails promote the degradation of the attached mRNAs by 3′-5′-exonucleases (Symmons et al. 2002). In Escherichia coli, eubPAP (Cao and Sarkar 1992) is a nonessential enzyme and its function can be replaced by polynucleotide phosphorylase (PNP; Mohanty and Kushner 2000), an enzyme which can both synthesize and degrade poly(A). PNP was also found to be responsible for poly(A) addition to mRNAs in spinach chloroplasts and cyanobacteria (Yehudai-Resheff et al. 2001; Rott et al. 2003).
eubPAPs and CCAtrs belong to a superfamily of nucleotidyl transferases (Martin and Keller 1996; Aravind and Koonin 1999), members of which share sequence homology mainly in the catalytic domain. They have been divided into class I and II according to specific sequence motifs in the catalytic domain (Yue et al. 1996). All class I Ntrs (Yue et al. 1996) which share the same active site signature, including archaeal CCAtrs, eukaryotic, nuclear, and regulatory poly(A) polymerases, and related enzymes such as DNA polymerase β(Pol β), terminal deoxynucleotidyl transferase (TdT), and 2′-5′ oligo(A) synthase, must share a common ancestor (Holm and Sander 1995; Martin and Keller 1996; Yue et al. 1996; Aravind and Koonin 1999). Class II Ntrs must have branched very early from class I Ntrs (Yue et al. 1996), and the two classes may even have evolved independently twice (Aravind and Koonin 1999). This is very likely if one considers the fact that the crystal structures of class I and class II CCAtrs differ quite extensively (Li et al. 2002; Augustin et al. 2003; Okabe et al. 2003; Xiong et al. 2003). For example, one of the obvious differences between the two classes of enzymes is that the helix that interacts with the phosphates of the incoming nucleotides has a single turn in class I Ntrs and two turns in class II enzymes (see below).
RESULTS AND DISCUSSION
Search for protein sequences with homology to bona fide eubacterial poly(A) polymerases and CCA transferases
We first conducted an extensive database search with the BLAST program for protein sequences with homology to experimentally verified CCAtrs or eubPAPs as query. Sequences with a probability threshold (E-value) of better than 10−7 were included for further analysis, although most positives had E-values of better than 10−10. Resulting multiple sequence alignments were then subjected to analysis by the computer program “Sequence Space,” which allows the determination of residues which should specify functional differences between protein families (Casari et al. 1995). This program designates the entire protein and sequence residues as vectors in a generalized sequence space. Projection of these vectors onto a lower-dimensional space reveals protein subfamilies as clusters and highlights characteristic residues.
We expected the Sequence Space program to identify conserved motifs that were characteristic for either CCAtrs or eubPAPs. Figure 1A ▶ depicts an example of an alignment displayed in the viewer window of the Sequence Space program, where motif-specific residues are highlighted in red (columns) and rows depict sequences (in blue) that contain motifs complying with eubPAP criteria (summarized in Table 1 ▶). Other Sequence Space viewer windows display either the spatial distribution of individual residues (Fig. 1B ▶) or of the protein sequences (Fig. 1C ▶). The figures illustrate how residues specific for CCAtrs (Fig. 1B ▶) and a cluster of protein sequences of CCAtrs (Fig. 1C ▶) are located to the left in both windows, whereas eubPAP-specific residues and sequences form clusters to the right in these windows. In a further step we systematically searched the entire collection of sequenced genome databases for the presence of eubPAPs and CCAtrs (for details see Materials and Methods). Table 1 ▶ contains a list of sequence signatures that were applied for the classification of positives, and the results are listed in Table 2 ▶.
FIGURE 1.
Analysis of multiple sequence alignments with the program Sequence Space. (A) Part of a multiple sequence alignment of the Java viewer window of the Sequence Space program with eubPAPs and CCAtrs from a BLAST output with E. coli eubPAP sequence as query (top line). Only columns of the MSF alignment are displayed which contain the catalytic site region and motifs of interest. Some blocks of rows with very similar proteins were left out. Numbers at the top correspond to residues in the E. coli eubPAP protein sequence (Swiss-Prot accession pcnb_ecoli). Rows in blue indicate predicted eubPAP sequences, and rows in red highlight conserved residues in the upstream and the downstream motif. Swiss-Prot or TrEMBL accessions are listed to the left. At the bottom, asterisks (*) mark the three catalytic Asp or Glu residues, and # marks other invariant residues in Ntrs; $ indicates residues that belong to the upstream and downstream motifs, and the color code red is given for eubPAP-specific residues and black for residues invariant in both CCAtrs and eubPAPs. α-helices and β-strands corresponding to the B. stearothermophilus structure (PDB accession 1MIW) are indicated by gray bars and arrows at the top. (Left column) Species of the organism (first two letters as abbreviation of species name) and classification of the protein (third letter; P for eubPAP and C for CCAtr). Ah, Aeromonas hydrophila; At, Arabidopsis thaliana; Ba, Bacillus anthracis; Bb, Borrelia burgdorferi; Cc, Chlamydophila caviae; Ce, Clostridium perfringens; Cm, Chlamydia muridarium; Cp, Chlamydia pneumoniae; Ct, Chlamydia trachomatis; Cl, Clostridium tetani; Ec, Escherichia coli; Hi, Haemophilus influenzae; Li, Listeria interrogans; Ll, Lactococcus lactis; Lm, Listeria monocytogenes; Lp, Lactobacillus plantarum; Mm, Mus musculus; Ne, Nitrosomonas europaea; Os, Oryza sativa; Rl, Rhizobium loti; Sm, Streptococcus mutans; So, Shewanella oneidensis; Si, Salmonella typhi; St, Salmonella typhimurium; Tp, Treponema pallidum; Vc, Vibrio cholerae; Vp, Vibrio parahaemolyticus; Yp, Yersinia pestis. Note that in Swiss-Prot and TrEMBL file annotations the assignments of eubPAPs and CCAtrs are not reliable. Also, files of these proteins are not frequently updated for newer publications; for example, the PAPS_BACSU Swiss-Prot file classified a protein as a poly(A) polymerase; however, when this protein was cloned and expressed, it was found to be a CCAtr (Raynal et al. 1998). (B) Display of the residues window of the Sequence Space Java viewer with a 3D view of the results of the alignment in A. To the left, the red * (circled and named E/251) indicates the position of the residues only found in CCAtrs corresponding to residue 251 in E. coli eubPAP in A. (C) The protein window of the Java viewer displays the proteins from the alignment in red if they contain a eubPAP-specific motif (blue rows in A), found in the same location in 3D as eubPAP-specific residues in B.
TABLE 1.
Motifs that distinguish nucleotidyl transferases
| Specificity | Upstream motif | Downstream motif |
| EubPAP | [LIV][LIV]G[R/K][R/K]Fx-[LIV]h[HQL][LIV] | sRxxxExxxhh |
| CCAtr | n.a. | eRxxxExxxhha |
| CC-addingb | n.a. | ERxxxExxxhh |
| A-addingb | n.a. | sRxxxExxxhh |
aA few exceptions were found to have N, Q, or small residues at the first position; see text.
bValid if found together.
TABLE 2.
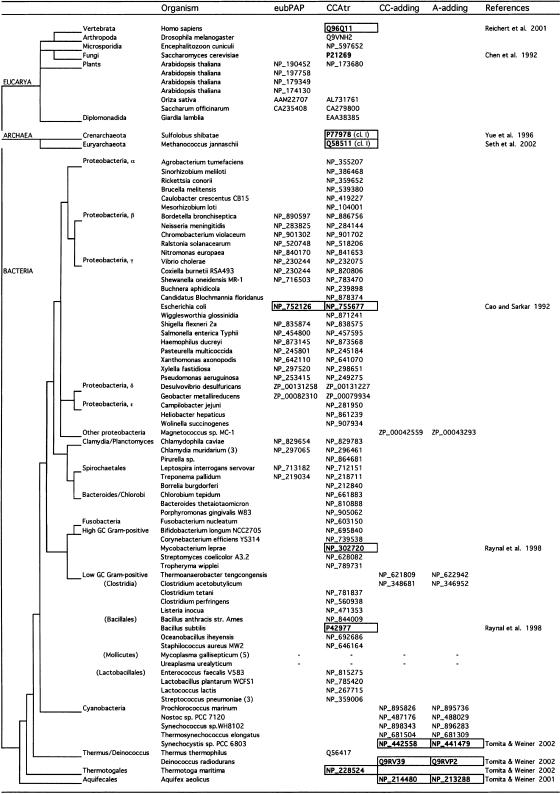
Phylogenetic distribution of eubPAPs, CCAtrs, CC- and A-adding enzymes
Distribution of Ntrs in a phylogenetic tree based on rRNA sequence analysis (distances not drawn to scale; Olsen et al. 1994; Woese 2002).
Accession numbers in bold and framed indicated tested proteins (see references).
Numbers in parentheses indicate the total number of organisms of the same genus that contain an identical type of Ntr.
Accession numbers of the type NP_xxxxxx are from completed genomes; accessions ZP_xxxxxx are from unfinished genomes.
Two distinct signatures emerged in the Sequence Space analysis (Table 1 ▶): The first corresponded to amino acids 108–118 of the E. coli eubPAP and had the consensus [LIV]-[LIV]-G-[RK]-[RK]-F-x-[LIV]-h-[HLQ]-[LIV], where x is any and h is a hydrophobic residue. This consensus signature is only found in bona fide eubPAPS but not in CCAtrs. Because there are no crystal structures of eubPAPs available, we tried to locate this motif in the closely related structures of bacterial (Li et al. 2002) and human CCAtr (Augustin et al. 2003). The corresponding sequence in these CCAtrs forms a turn or loop of variable size followed by β-strand 4 or 3, respectively. Since no structures of CCAtrs in complex with a primer are known, we superimposed the catalytic region of the class I enzyme Pol β including a primer (Sawaya et al. 1997) with the corresponding structure of the B. stearothermophilus CCAtr (Fig. 2A ▶). The resulting model shows a close contact between the terminal three nucleotides of the primer and the loop between β-strands 3 and 4 (“loop 3/4”) of the CCAtr structure.
FIGURE 2.
(A) Superposition of CCAtr and Pol β catalytic sites. Residues 21–86 of the B. stearothermophilus CCAtr (orange) including ATP and Mg2+ ions (in orange, pdb accession 1MIW) and residues 174–260 of the human Pol β structure including nucleotides 7–10 of the primer, ddCTP, and Mg2+ (all in marine, pdb accession 1BPY) were included for alignment of the catalytic Asp or Glu residues (D190/192 and D256 of Pol β with D40/42 and E79 of B. stearothermophilus, respectively). The proposed 3′ primer binding domain in the B. stearothermophilus CCAtr is depicted in yellow. Molecular graphics were done with the program PyMol (www.pymol.org). (B) Multiple protein sequence alignment of the loop 3/4 region in different classes of Ntrs. (Left column) Accession numbers of the Swiss-Prot (sp), TrEMBL (tr), TrEMBL-new (tn), and PDB (pdb) databases. In the bovine PAP features row, + and o indicate results of kinetics tests with mutants of bovine PAP: + is a strong effect and o is no effect on primer binding upon mutation; $ marks the catalytic Asp or Glu in all proteins. Protein sequences that form β -strands in crystal structures are framed by black boxes and are numbered in the consensus at the bottom. If available, Swiss-Prot IDs of the format CCA_SULSH are given in the same line; otherwise confirmed or predicted enzyme activity is indicated together with the species names. P.aeroph, Pyrobaculum aerophilum; T.volcanic, Thermoplasma volcanicum; T.brucei, Trypanosoma brucei; N.meni, Neisseria meningitidis; Ch.tra, Chlamydia trachomatis; A.thali, Arabidopsis thaliana; O.sati, Oryza sativa; P.aerugin, Pseudomonas aeruginosa; Aeh1, Aeromonas hydrophila bacteriophage Aeh1.
A motif forming a similar loop 3/4 structure has been proposed to be important in primer binding in class I eu-karyotic PAPs and in terminal uridylyl transferases (Keller and Martin 2002). Therefore, it appears that two groups of Ntrs possess a loop 3/4 motif with a strongly conserved sequence signature, whereas most of the remaining Ntrs have a loop 3/4 whose amino acid sequence is much less well conserved. Close inspection of these enzymes reveals that the two groups of proteins consist of Ntrs that synthesize homopolyribonucleotides (hprNtrs) and that the two groups belong to either class I or class II Ntrs. We designate these two types of loop 3/4 motifs according to the two classes of Ntrs “hprNS-I” and “hprNS-II”, for “homopolyribonucleotidyl transferase signature I and II.” An alignment with a representative selection of sequences belonging to the two classes of loop 3/4 motifs is shown in Figure 2B ▶. All motifs contain a confirmed or predicted loop 3/4 structure about 14 residues upstream of the third catalytic Glu or Asp residue, except for terminal uridylyl transferase from T. brucei, where the distance is larger. The crystal structures of five proteins in the list are solved and result in a consensus of structural elements (Fig. 2B ▶, bottom). Therefore, we predict that the group of hprNS-II proteins (the eubPAPs) contain a loop structure at the same position. Although sequences between groups are not strongly conserved for reasons of group specificity, hydrophobic residues or Pro/Gly are frequently conserved and are an indication for structure conservation. Loop 3/4 structures often feature residues with a ring-containing side chain (Phe or His) or the basic residues Lys or Arg. The conserved features may be specific for the type of primer involved and could be an indication that these side chains engage in stacking interactions with the primer via the ring or the aliphatic central part of lysine and arginine. Mutagenesis of Phe153, located on the loop, and Val 156 and Lys158 on strand 4 of the hprNS-I of bovine PAP resulted in a strongly increased KM when titrated with oligo (A)15 primer, an indication that several residues of the hprNS-I are involved in primer binding (G. Martin and W. Keller, unpubl.). A domain close to the catalytic site (which most likely corresponds to an hprNS-I structure) was also found to be responsible for binding the three terminal nucleotides of the primer in yeast poly(A) polymerase (Zhelkovsky et al. 1998). The combined results suggest that the hprNS-I and hprNS-II motifs are responsible for binding of the 3′ end of the RNA substrate.
It is not clear whether or not the loop 3/4 structures in proteins that do not belong to the hprNtrs are also involved in binding of the RNA 3′ terminus. However, in the B. stearothermophilus CCAtr structure, a primer was modeled into the space between loop 3/4 and residue R194 on the opposite helix J, and it was proposed that the state of the primer strand could be communicated via R194 and helix J to helix G, where protein templating would then switch from C to A recognition (Li et al. 2002). Furthermore, it has been postulated that all DNA and RNA polymerases catalyze nucleotide addition by a unified mechanism, and that catalysis can only proceed when the two metal ions and the 3′ terminus of the primer strand are positioned in a unique orientation (Steitz et al. 1994). Thus, our prediction of the position of the 3′ terminus of the RNA substrate in CCAtrs and eubPAPs conforms to this general principle. Superposition of class I and class II Ntrs (Fig. 2A ▶) illustrates this conservation of the catalytic domain structure. The loop 3/4 has a different angle in the two proteins to accomodate the either single- or double-stranded nucleic acid substrates. Docking experiments have suggested that the axes of tRNA or a tRNA minihelix do not enter the active site from the same direction in class I and class II CCAtrs (Okabe et al. 2003; Xiong et al. 2003), although this does not necessarily imply that the orientations of the 3′ terminal nucleotides differ. In eubPAPs and other nucleotidyl transferases that synthesize long polynucleotides, the 3′ terminal nucleotides of the primer are presumably linearly stacked on each other. Therefore, a primer binding structure in these enzymes may facilitate rapid translocation of the emerging polynucleotide chain. In contrast, in the case of CCAtrs the primer’s 3′ end is held in a fixed position to force scrunching of the growing CCA tail into a mold or cavity of the catalytic site (Shi et al. 1998). In summary, we assume that there are specific differences between the loop 3/4 structures in CCAtrs and in eubPAPs, and that the upstream sequence signature that we detected in eubPAPs corresponds to a specific loop 3/4 structure that is involved in primer binding and is present only in polyribonucleotide polymerases.
With the help of the Sequence Space program we identified a second eubPAP-or CCAtr-specific motif about 125 amino acids downstream of the eubPAP-specific motif, conforming to either the consensus ERxxxExxxhh or sRxxxExxxhh (“s” is a small residue, “h” is a hydrophobic and “x” is any amino acid), here termed “downstream motif.” Interestingly, the two variants of this motif could in most cases be assigned specifically to either the CCAtrs (ERxxxExxxhh) or to the eubPAP group (sRxxxExxxhh; Fig. 1A ▶; Table 1 ▶). Arg194 of motif E, described in the B. stearothermophilus CCAtr structure (the R at the second position in the two motifs) was suggested to play a critical role in the templating specificity to generate CCA (Li et al. 2002). In addition, the CCAtr-specific residue Glu193 (E/251 in Fig. 1B ▶) may also have a role in the nucleotide selection mechanism for either CC or A. Analysis of appropriate mutants could shed light on this question.
Some early branching eubacteria employ two separate enzymes to collaboratively add either CC or A to tRNAs (Tomita and Weiner 2001, 2002). These enzymes conform to a modified rule, whereby CC-adding proteins contain an ERhxxExxxhh motif and A-adding proteins carry an sRhxxExxxhh signature. Thus, a glutamic acid at the N-terminal end of the downstream motif is only present if the enzyme adds CC or CCA to tRNA precursors, whereas if only A has to be added as in eubPAPs or A-adding tRNA-specific Ntrs, Glu is replaced by a small inert residue. However, there are also exceptions to this rule: For example, the Thermotoga maritima nucleotidyl transferase contains the downstream motif PRxxxExxxhh (a signature for eubPAP or A-adding enzymes) but has recently been determined to be a CCAtr (Tomita and Weiner 2001). Therefore, the fact that the Glu in the downstream motif is replaced by a Pro in Thermotoga CCAtr could be an indication that this residue is not essential for catalysis. In addition, the α-proteobacterium Buchnera aphidicola and the early branching protist Giardia lamblia have Glu replaced by the functional analogs Asn or Gln in the downsteam motif (Table 2 ▶), and also several protozoa carry small residues at this position. In summary, our rule strictly applies to the upstream eubPAP-specific signature, where no exceptions were found so far. Although the downstream motif is less well conserved, it is still useful to distinguish between CC- and A-adding enzymes.
Phylogenetic assignment of eubacterial poly(A) polymerases and CCA transferases
Integrating our results into a phylogenetic tree based on rRNA sequence analysis (Olsen et al. 1994; Woese 2002) which was adapted to include selected organisms according to the results of database searches, disclosed several new findings (Table 2 ▶). First, eubPAPs are only detected in the β, γ, and δ subdivisions of proteobacteria and in some Chlamydiales and Spirochaetales but were not found in the α- and ɛ-proteobacteria subdivisions. This might indicate that eubPAPs evolved in an ancestor of these closely related bacteria and may have been lost in more derived lineages. With the exception of plants, no eubPAPs could be detected in archaea and eukaryotes. All Gram-positive bacteria and eubacteria that have diverged before the Gram-positives do not contain eubPAPs. The parasitic bacteria Mycoplasma and Ureaplasma (six genomes) remain the only organisms with no CCA transferase or eubPAP (Mushegian and Koonin 1996). These bacteria contain a minimal gene set, and all tRNA genes code for 3′ terminal CCA. Nevertheless, it is surprising that Mycoplasma does not need a CCA transferase, because many other organisms also code for CCA in their tRNA genes but do require a CCA-transferase as tRNA repair enzymes.
Early lineages of eubacteria, and in particular the cyanobacteria, carry two different enzymes for the synthesis or repair of the tRNAs’ CCA ends (Tomita and Weiner 2001, 2002). The Thermus and Thermotoga lineages were among the first to acquire a single CCAtr. However, a few Gram-positives and even one of the proteobacteria kept a system with separate CC- and A-adding enzymes.
Because eubPAPs are found in eubacteria and in plants, two separate branches of the phylogenetic tree (Table 2 ▶), we can consider a scenario for their origin where a first eubPAP was derived in a common ancestor of proteobacteria and Chlamydiales and Spirochaetales. Some bacterial species or entire groups, such as the α- and ɛ-proteobacteria, eventually lost eubPAP. If loss of eubPAP is “easy” for the cell, then many losses can be accepted on parsimony grounds, considering the fact that other enzymes, such as polynucleotide phosphorylase (PNP), take over the function of eubPAP (Yehudai-Resheff et al. 2001; Rott et al. 2003). In a second event, eubPAPs and CCA-adding enzymes were transmitted from eubacteria to an early eukaryote by endosymbiosis. Endosymbiosis of α-proteobacteria is generally considered to be the origin of mitochondria, whereas chloroplasts are thought to originate from the endosymbiosis of cyanobacteria (Burger et al. 2003).
Do the plant eubPAPs indeed descend from eubPAPs of a proteobacterial ancestor, or do they originate from plant CCAtrs? Sequence relationships between eubPAPs and CCAtrs from plants and eubPAPs of proteobacteria (as displayed in Fig. 1C ▶) reveal that, although plant eubPAPs are close to a cluster of CCAtrs, they share a cluster with eubPAPs of Spirochaetales, a lineage which has branched before the proteobacteria, and also with γ-proteobacteria. It is therefore likely that plants inherited eubPAPs from eubacteria and that the enzymes were not reinvented by conversion of plant CCAtrs. Furthermore, CC- and A-adding enzymes were found in the same cluster as CCAtrs in the Sequence Space protein viewer window (data not shown).
We found that all known plant genomes contain at least one eubPAP and one CCAtr. For instance, we identified cDNA sequences for one CCAtr and four eubPAPs, each coded by different genes on nuclear chromosomes in the Arabidopsis thaliana databases. Interestingly, the prediction programs for sorting signals Predotar (http://www.inra.fr/predotar/), TargetP (Emanuelsson et al. 2000), Mitoprot (Claros and Vincens 1996), and iPSORT (Bannai et al. 2002) revealed that all eubPAPs and CCAtrs tested and listed in Table 3 ▶ are predicted to contain either mitochondrial or chloroplast targeting sequences, an indication for transport to these organelles. In the yeast Saccharomyces cerevisiae, protein products of a single CCAtr gene were found to be targeted to the nucleus, the cytosol, and to mitochondria (Chen et al. 1992), and mammalian CCAtrs were also found to be imported into mitochondria (Nagaike et al. 2001; Reichert et al. 2001). Interestingly, these enzymes are coded not in the mitochondrial or plastid genomes but rather in the nucleus. It has been reported that ~18% of protein-coding genes in the Arabidopsis nuclear genome are derived from cyanobacteria but that gene origin and compartmentation do not strictly correlate (Martin et al. 2002). It is an intriguing possibility that the eubPAPs of proteobacterial origin in Arabidopsis (and possibly other plants) which are encoded in the nucleus, are targeted to the cytoplasm, to chloroplasts, or to mitochondria.
TABLE 3.
Prediction of transit peptides
| Predotar | TargetP | Mitoprot II | iPSORT | ||||
| Accession | Species | Protein | Result | Result | Mito score | Result | Consensus |
| NP_173680 | A. thaliana | CCAtr | — | Plastid | 0.9688 | Plastid | Plastid |
| NP_197758 | A. thaliana | eubPAP | Mito | Mito | 0.7029 | Mito | Mito |
| NP_174130 | A. thaliana | eubPAP | Plastid | (Plastid) | 0.648 | Plastid | Plastid |
| NP_190452 | A. thaliana | eubPAP | Mito | Sig-Pep | 0.7269 | Sig-Pep | Sig-Pep |
| AAM22707 | Oryza sativa | eubPAP | Mito | Mito | 0.946 | Plastid | Mito |
| P21269 | S. cerevisiae | CCAtr | Mito | Mito | 0.9561 | Mito | Mito |
| Q96Q11 | H. sapiens | CCAtr | Mito | Mito | 0.9991 | Mito | Mito |
| Controls: | |||||||
| p25500 | Bos taurus | nucPAP-α | — | — | 0.0333 | — | none |
| Q10569 | Bos taurus | CPSF-160 | Mito | — | 0.0391 | — | none |
| P79101 | Bos taurus | CPSF-73 | — | — | 0.0721 | — | none |
Mito, mitochondrial; Sig-Pep, signal peptide. As controls for proteins that are not expected to contain target peptides, sequences of bovine nuclear poly(A) polymerase α(nucPAP-α) and two subunits of the cleavage and polyadenylation specificity factor CPSF (CPSF-73 and CPSF-160) were submitted.
In conclusion, the sequence signatures reported here are useful to assign functions to nucleotidyl transferase sequences present in existing sequence databases and those emerging from the rapidly growing number of new genome-sequencing projects.
MATERIALS AND METHODS
BLAST searches were conducted in the genome databases (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) or other nonredundant or EST databases at the Swiss EMBnet node (Falquet et al. 2003) at http://www.ch.embnet.org/software/BottomBLASTadvanced.html or at the NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). The Sequence Space program (Casari et al. 1995) was downloaded from http://industry.ebi.ac.uk/SeqSpace/. For Table 2 ▶, a phylogenetic tree describing archaea and bacteria (Olsen et al. 1994) was modified according to results from database searches and was extended to include the eucarya according to the universal phylogenetic tree determined by rRNA sequence analysis (Woese 2002).
Acknowledgments
We thank Lorenza Bordoli for help with the Sequence Space program and for critically reading the manuscript, David Rand (Brown Univ.) for his invaluable expert advice, Stepanka Vanacova for improving the manuscript, and an anonymous referee for many useful suggestions. This work has been supported by the University of Basel and the Swiss National Science Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5242304.
REFERENCES
- Aravind, L. and Koonin, E.V. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27: 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin, M.A., Reichert, A.S., Betat, H., Huber, R., Mörl, M., and Steegborn, C. 2003. Crystal structure of the human CCA-adding enzyme: Insights into template-independent polymerization. J. Mol. Biol. 328: 985–994. [DOI] [PubMed] [Google Scholar]
- Bannai, H., Tamada, Y., Maruyama, O., Nakai, K., and Miyano, S. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305. [DOI] [PubMed] [Google Scholar]
- Burger, G., Gray, M.W., and Lang, B.F. 2003. Mitochondrial genomes: Anything goes. Trends Genet. 19: 709–716. [DOI] [PubMed] [Google Scholar]
- Cao, G.J. and Sarkar, N. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. 89: 10380–10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casari, G., Sander, C., and Valencia, A. 1995. A method to predict functional residues in proteins. Nat. Struct. Biol. 2: 171–178. [DOI] [PubMed] [Google Scholar]
- Chen, J.Y., Joyce, P.B., Wolfe, C.L., Steffen, M.C., and Martin, N.C. 1992. Cytoplasmic and mitochondrial tRNA nucleotidyltransferase activities are derived from the same gene in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 267: 14879–14883. [PubMed] [Google Scholar]
- Claros, M.G. and Vincens, P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241: 779–786. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Falquet, L., Bordoli, L., Ioannidis, V., Pagni, M., and Jongeneel, C.V. 2003. Swiss EMBnet node web server. Nucleic Acids Res. 31: 3782–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1995. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 20: 345–347. [DOI] [PubMed] [Google Scholar]
- Keller, W. and Martin, G. 2002. Gene regulation: Reviving the message. Nature 419: 267–268. [DOI] [PubMed] [Google Scholar]
- Li, F., Xiong, Y., Wang, J., Cho, H.D., Tomita, K., Weiner, A.M., and Steitz, T.A. 2002. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell 111: 815–824. [DOI] [PubMed] [Google Scholar]
- Martin, G. and Keller, W. 1996. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 15: 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martin, G., Keller, W., and Doublié, S. 2000. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 19: 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., and Penny, D. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. 99: 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K. and Kushner, S.R. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. 97: 11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian, A.R. and Koonin, E.V. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. 93: 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike, T., Suzuki, T., Tomari, Y., Takemoto-Hori, C., Negayama, F., Watanabe, K., and Ueda, T. 2001. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J. Biol. Chem. 276: 40041–40049. [DOI] [PubMed] [Google Scholar]
- Okabe, M., Tomita, K., Ishitani, R., Ishii, R., Takeuchi, N., Arisaka, F., Nureki, O., and Yokoyama, S. 2003. Divergent evolutions of trinucleotide polymerization revealed by an archaeal CCA-adding enzyme structure. EMBO J. 22: 5918–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, G.J., Woese, C.R., and Overbeek, R. 1994. The winds of (evolutionary) change: Breathing new life into microbiology. J. Bacteriol. 176: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal, L.C., Krisch, H.M., and Carpousis, A.J. 1998. The Bacillus subtilis nucleotidyltransferase is a tRNA CCA-adding enzyme. J. Bacteriol. 180: 6276–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert, A.S., Thurlow, D.L., and Mörl, M. 2001. A eubacterial origin for the human tRNA nucleotidyltransferase? Biol. Chem. 382: 1431–1438. [DOI] [PubMed] [Google Scholar]
- Rott, R., Zipor, G., Portnoy, V., Liveanu, V., and Schuster, G. 2003. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 278: 15771–15777. [DOI] [PubMed] [Google Scholar]
- Sawaya, M.R., Prasad, R., Wilson, S.H., Kraut, J., and Pelletier, H. 1997. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry 36: 11205–11215. [DOI] [PubMed] [Google Scholar]
- Seth, M., Thurlow, D.L., and Hou, Y.M. 2002. Poly(C) synthesis by class I and class II CCA-adding enzymes. Biochemistry 41: 4521–4532. [DOI] [PubMed] [Google Scholar]
- Shi, P.Y., Maizels, N., and Weiner, A.M. 1998. CCA addition by tRNA nucleotidyltransferase: Polymerization without translocation? EMBO J. 17: 3197–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T.A., Smerdon, S.J., Jäger, J., and Joyce, C.M. 1994. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science 266: 2022–2025. [DOI] [PubMed] [Google Scholar]
- Symmons, M.F., Williams, M.G., Luisi, B.F., Jones, G.H., and Carpousis, A.J. 2002. Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem Sci. 27: 11–18. [DOI] [PubMed] [Google Scholar]
- Tomita, K. and Weiner, A.M. 2001. Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science 294: 1334–1336. [DOI] [PubMed] [Google Scholar]
- Tomita, K. and Weiner, A.M. 2002. Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystis sp. and Deinococcus radiodurans. J. Biol. Chem. 277: 48192–48198. [DOI] [PubMed] [Google Scholar]
- Woese, C.R. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. 99: 8742–8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y., Li, F., Wang, J., Weiner, A.M., and Steitz, T.A. 2003. Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes. Mol. Cell 12: 1165–1172. [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., and Schuster, G. 2001. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21: 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, D., Maizels, N., and Weiner, A.M. 1996. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: Characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 2: 895–908. [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky, A., Helmling, S., and Moore, C. 1998. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol. 18: 5942–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]