Abstract
Escherichia coli is the best-characterized organism with respect to posttranscriptional modifications of its ribosomal RNA (rRNA). It is presently believed that all the modified nucleotides have been identified, primarily on the basis of two detection methods; modification-induced inhibition of the enzyme reverse transcriptase or analysis by combined HPLC and electrospray ionization mass spectrometry. Comparison of data from these different approaches reveals a disagreement regarding modification of C2501 in E. coli 23S rRNA. A. Bakin and J. Ofengand previously reported the detection of a modification at this site based on a reverse transcriptase assay. J.A. McCloskey and coworkers could not confirm the existence of such a modification using an electrospray ionization mass spectrometry approach. C2501 is therefore generally considered unmodified. We have used a strategy involving isolation of a specific rRNA fragment from E. coli 23S rRNA followed by Matrix Assisted Laser Desorption/Ionization mass spectrometry and tandem mass spectrometry to investigate this controversy. Our data reveal a novel 16-Da partial modification at C2501. We believe that the data reported here clarify the above discrepancy, because a minor partial modification detected in a reverse transcriptase assay would not necessarily be detected by the original mass spectrometry approach. The level of modification was furthermore monitored in different growth situations, and we found a significant positive regulation in stationary phase cells. C2501 is universally conserved and implicated in structure folds very close to the catalytic center of the ribosome. Moreover, several antibiotics bind to nucleotides in this region, which altogether make a modification at this site interesting.
Keywords: E. coli rRNA modification, growth phase dependent, MALDI mass spectrometry, tandem mass spectrometry
INTRODUCTION
Maturation of stable RNA in the cell generally involves a high degree of modification during and following generation of the primary transcript. The type of modifications taking place at the single nucleotide level is termed post-transcriptional modifications. These are especially frequent in tRNA, but also appear in rRNA, and typically involve addition of small chemical groups that change the physical and chemical properties of the nucleotide. Escherichia coli 16S and 23S rRNA are modified at 11 and 23 residues, respectively (Rozenski et al. 2000), and the major part of the modifications are found on highly conserved nucleotides in or near the functional centers of the ribosome (Bakin and Ofengand 1993; Brimacombe et al. 1993). Several studies have shown posttranscriptional modifications to be indispensable for normal ribosomal assembly, translational activity, and fidelity of decoding (e.g., Cunningham et al. 1991; Green and Noller 1996; O’Connor et al. 1997; Caldas et al. 2000), but details in the function of posttranscriptionally modified nucleotides are poorly understood.
The contemporary methods used to detect and characterize modified nucleotides in RNA are reverse transcriptase mapping (e.g., Bakin and Ofengand 1993) and analysis by a combination of HPLC and electrospray ionization mass spectrometry (ESI-MS; Kowalak et al. 1993). HPLC/ESI-MS enables parallel measurement of precise molecular mass and chromatographic mobility, and has therefore proven effective and reliable, not only to verify reverse transcriptase data but also for detection and mapping of novel modifications. The initial step in the LC/MS approach is a total nucleoside analysis to quantify and identify modified nucleosides. Specific chemical modification or tandem mass spectrometry may furthermore be used. Localization in the primary sequence is established by LC/MS on defined oligoribonucleotides produced enzymatically by digestion with the guanosine-specific RNase T1. A subsequent comparison of the measured masses with those expected from rDNA data reveals fragments harboring modified nucleotides.
We have previously demonstrated the use of Matrix Assisted Laser Desorption/Ionization (MALDI) mass spectrometry and tandem mass spectrometry as alternative methods to analyze RNA modifications (Kirpekar et al. 2000; Kirpekar and Krogh 2001). The MALDI technique is generally advantageous in terms of sensitivity, tolerance against impurities, and ability to handle complex mixtures. The latter is a result of a predominant generation of singly charged molecular ions, leading to dramatically reduced complexity of the spectrum. Samples containing even a high number of analyte molecules, such as RNase digestions of large RNAs, are therefore typically resolved into a number of distinct peaks, each representing a single analyte species. The properties of MALDI MS therefore make preanalytic separation steps, such as HPLC, less necessary. MALDI tandem mass spectrometry offers the above-mentioned qualities of the MALDI spectrum and, in addition, enables precursor ion selection and ion fragmentation. It is a means of sequencing and of detailed structural analysis of short stretches of RNA.
In the present work, MALDI MS and MALDI tandem MS were used to detect and localize a previously unknown modification in domain V—the peptidyltransferase center—of E. coli 23S rRNA. We were encouraged to investigate this region by unpublished data from a previous study, which indicated occurrence of a partial 16-Da modification near position 2500 (B.T. Porse and F. Kirpekar, unpubl.). A reverse transcriptase assay published in 1993 by Bakin and Ofengand furthermore indicated an unknown modification at C2501. This suggested modification was later investigated by HPLC/ESI-MS analysis with a negative outcome (Kowalak et al. 1995). No link has afterward been established between the reverse transcription data and a modified residue. Here, we resolve this apparent conflict by showing that a fraction of C2501 in E. coli 23S rRNA is indeed subjected to modification. This was first detected as a 16-Da mass increment of the RNase T1 fragment containing nucleotides 2496-C-A-Cm-C-U-C-G-2502. The modification was subsequently localized to C2501 by a combination of tandem mass spectrometry of the above 7-mer and parallel mass spectrometric analysis of RNase A digests of the purified rRNA segment C2480–C2527.
RESULTS AND DISCUSSION
Detection of a partial modification in the RNase T1 fragment C2496–G2502
A successful MALDI MS analysis of rRNA strongly depends on correctly prepared and purified samples. The preliminary steps of this study therefore included the development of a suitable method to site-specifically isolate rRNA fragments expected to carry novel modifications. For this purpose, we utilized a strategy based on nuclease protection of an rRNA sequence by hybridization to a complementary DNA oligodeoxynucleotide (Maden 1980). This concept has also been adapted for use with mass spectrometry (Kowalak et al. 1995). However, we found it necessary to optimize the procedure with respect to hybridization and digestion. In particular, we found hybridization to be more efficient using a HEPES/KCl annealing buffer in a slow-cool procedure, and the addition of a small amount of RNase A together with the single-strand-specific mung bean nuclease promoted proper digestion. The changes were found to improve yield significantly, and ultimately enabled us to use the method on starting material down to ~10 pmoles.
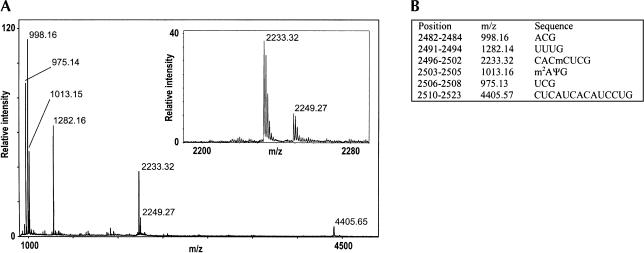
Because we wished to isolate a short segment covering the nucleotides around position 2500 of E. coli 23S rRNA, a DNA oligonucleotide complementary to the region C2480–C2527 was hybridized to E. coli total rRNA. Single-stranded DNA and RNA were subsequently removed by nuclease digestion, and the protected RNA fragment was gel-purified. This RNA fragment was then digested to completion by the guanine-specific RNase T1 and directly analyzed by MALDI time-of-flight (TOF) mass spectrometry (Kirpekar et al. 2000). Figure 1A ▶ displays a typical mass spectrum from an MALDI MS analysis of the RNase T1 digest. Comparison between the expected fragment masses listed in Figure 1B ▶ and those in the spectrum immediately confirms the identity of the sequence C2480–C2527. The high accuracy of mass determination (better than one-tenth of a dalton) as well as a good signal-to-noise ratio additionally allows the detection of a peak at m/z = 2249.27. An enlargement of the m/z = 2190–2290 region (Fig. 1A ▶, inset) shows an isotopic distribution as expected from an RNA fragment of this mass. The presence of this unexpected peak is interpreted as a novel 16-Da partial modification of a nucleotide in the digestion fragment 2496-C-A-Cm-C-U-C-G-2502. The m/z = 2249.3 peak was detected repeatedly in several other preparations, including preparations from a different strain of E. coli. In all samples, similar relative intensities of the m/z 2249.3 and 2233.3 peaks were observed, showing the fraction of 16-Da modified 23S rRNA to be 10%–30%. The low frequency of modification is not due to thermal degradation during the 90°C denaturation step (see Materials and Methods), as significant variations in the times of 90°C treatment did not influence the observed ratio (data not shown). We can also rule out that the 2249.3-Da fragment is an artifact from the nuclease isolation procedure, because the same modified digestion fragment was observed in a previous work where the initial ~50-mer rRNA fragment was purified by means of oligodeoxynucleotide annealing and RNase H digestion (B.T. Porse and F. Kirpekar, unpubl.).
FIGURE 1.
Fragment C2480–C2527 of E. coli 23S rRNA digested with RNase T1. (A) MALDI time-of-flight mass spectrum of the m/z region covering trinucleotides or larger. The inset reveals the +16.0-Da partial modification of the 2496-CACmCUCG-2502 fragment at m/z 2249.27. (B) A list of the expected RNase T1 digestion fragments based on the rRNA sequence with presently known nucleotide modifications. m2A is 2-methyladenosine, and ψ indicates pseudouridine.
Localization of the modified nucleotide
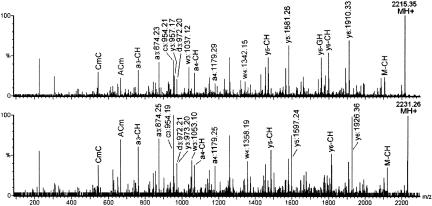
To narrow down the possible sites of modification, we employed tandem mass spectrometry using a MALDI quadrupole-TOF mass spectrometer (Kirpekar and Krogh 2001) to sequence the +16-Da-modified 7-mer. Favorably, two different phosphate forms are often produced by RNase T1 digestion; one with the normal 3′-phosphate and one with its 2′-3′-cyclic precursor (Kirpekar et al. 2000). Tandem mass spectrometry of the 7-mer in both phosphate forms as well as their 16-Da heavier posttranscriptionally modified counterparts generated two sets of comparable data. Supported by tandem mass spectrometry data from the normal 3′-phosphate species, peaks were assigned in the tandem mass spectra of the 2215.35-Da 2′-3′-cyclic phosphate and the +16-Da-modified species as shown in Figure 2 ▶. Essentially all major peaks are accounted for, and the presence of many backbone cleavages facilitates a partial sequencing of the two RNAs. Comparison of the spectra reveals that only some ions appear at equal m/z ratios, whereas the remaining ions display a characteristic 16-Da mass shift to a higher value. This makes it possible to narrow down nucleotides likely to be modified, by excluding nucleotides residing in fragment ions present at the same m/z value in the two spectra.
FIGURE 2.
Tandem mass spectra of the 2496-CACmCUCG-2502 fragment and its 16-Da-larger modified derivative (2215.35 and 2231.26 Da ions; 2′-3′-cyclic phosphate versions of the m/z 2233.32 and 2249.27 ions shown in Fig. 1A ▶). Lowercase letters refer to backbone cleavage fragments, where a, b, c, and d contain the original 5′-end, and w, x, y, and z the original 3′-end. Associated numbers are the length of the fragment in nucleotides. Capital letters refer to the nucleotides of internal fragments or, if preceded by (−) to loss of the indicated nucleobase.
The nucleotides 2496-C-A-Cm-C-2499 at the 5′-end of the oligoribonucleotide are immediately ruled out, as the 5′-sequence ions containing some or all of these nucleotides, that is, a3, c3, d3, a4, appear at the same m/z ratios in the two spectra (a, b, c, and d ions are backbone cleavage ions containing the original 5′-end, whereas w, x, y, and z ions contain the original 3′-end; nomenclature according to McLuckey et al. 1992). This interpretation is supported by the peaks corresponding to the w3, y3, w4, y5, and y6 ions, which all retain a 16-Da increment in the tandem mass spectrum of the modified species. The exclusion of positions 2496-C-A-Cm-C-2499 as putative sites of a 16-Da modification was fully substantiated by tandem mass spectrometry analyses of the modified and unmodified 3′-phosphate species (data not shown). Minor discrepancies between the spectra, as well as the absence of some important sequence ions, unfortunately impeded further analysis of the 3′-end of the 7-mer, and unambiguous assignment of the 16-Da modification to a single of the 3′-proximal nucleotides 2500-U-C-G-2502 was therefore not possible based on tandem mass spectrometry.
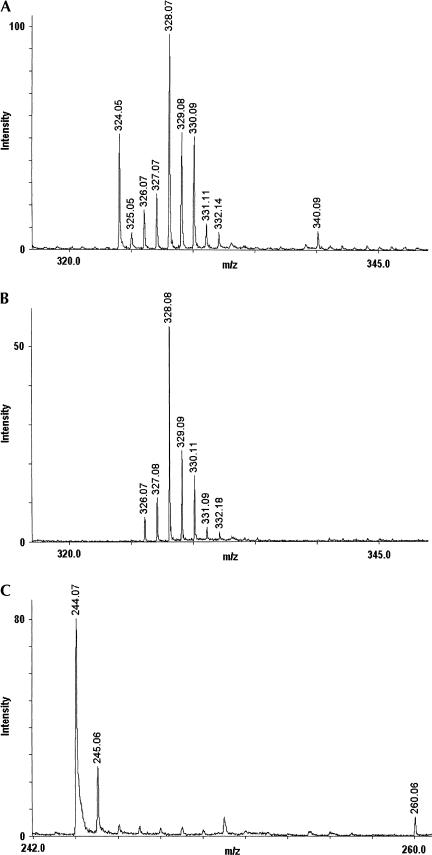
To determine the exact site of modification, U2500, C2501, and G2502 were examined by mass spectrometric analysis of the C2480–C2527 fragment digested with the pyrimidine specific RNase A. This digestion releases U2500 and C2501 as mono-nucleotides, whereas G2502 resides in the trinucleotide 2502-G-m2A-ψ-2504. A modified G2502 would be revealed by a partial mass shift of this latter fragment, as G2502 is the only nucleotide common to the RNase T1 digestion fragment in which the partial modification was initially detected. No 2502-G-m2A-ψ-2504 +16-Da fragment was observed (data not shown), which therefore excludes G2502 as being modified. The m/z region covering single nucleotides is shown for the RNase A digest in Figure 3A ▶. It reveals three nonmatrix peaks at m/z 324.05, 325.05, and 340.09, corresponding to cytidine monophosphate, uridine monophosphate, and a putative +16-Da-modified cytidine monophosphate. The m/z 340.1 peak was repeatedly detected in independent rRNA preparations. We carefully analyzed RNA-free, but otherwise identical, samples to identify signals originating from the matrix, the buffer, and so on (Fig. 3B ▶). Subsequently, we de-phosphorylated the RNase A digest to confirm the identity of the m/z 340.1 signal as a modified cytidine mono-phosphate. As shown in Figure 3C ▶, the modified nucleoside as well as C and U nucleosides appear at m/z values 80 Da lower than in Figure 3A ▶ upon dephosphorylation. Comparison with spectra of RNA-free matrix controls showed that these peaks were specific to the RNase-digested samples (data not shown). This unambiguously demonstrates that the +16-Da posttranscriptional modification resides on C2501.
FIGURE 3.
MALDI time-of-flight mass spectra of the region covering single nucleotides. (A) Fragment C2480–C2527 of E. coli 23S rRNA digested with the pyrimidine specific RNase A. Single cytidine and uridine monophosphates produced appear at the m/z values 324.05 and 325.05, respectively (the relative low intensity of the uridine monophosphate ion is caused by its lower proton affinity; Liguori et al. 2000). A peak corresponding to a 16-Da-modified cytidine monophosphate is furthermore observed at m/z 340.09. (B) The peaks of analyte ions are close to, but do not overlap, seven distinct matrix peaks in this region shown in the RNA-free control. (C) MALDI time-of-flight mass spectrum of a sample prepared as in A, but with an additional treatment with alkaline phosphatase. The peaks corresponding to cytidine, uridine, and 16-Da-modified cytidine are shifted to an 80-Da-lower value, corresponding to loss of phosphate.
Putative roles of the 16-Da modification at C2501
In case of a physiological role, a regulatory dependence would be a likely reason for the partial nature of the modification. Modulation of the rRNA modification level has been suggested as a means to cope with extreme growth temperatures (e.g., Noon et al. 1998). We tested this correlation for the C2501 modification, but observed no significant temperature dependence (data not shown). Another factor known to affect the level of nucleotide modification in RNA is the growth phase at isolation (e.g., Singhal and Vold 1976). We therefore analyzed rRNA extracted from cells harvested at three different time points in the stationary phase. In this case, our measurements showed a significant up-regulation of the 16-Da modification upon transition from exponential to stationary growth (Fig. 4 ▶). The modification level in stationary cells was consistently found to be 40%–60 %, that is, a rise in modification level of ~2× compared with exponentially growing cells. We cannot rule out the possibility of a more pronounced regulation in response to other environmental factors, maybe leading to complete modification. Further knowledge of the modification such as identity and pathway of synthesis is needed to make any biological suggestions from these observations. Nevertheless, the dependence on growth phase reveals an interesting and previously unseen dynamic behavior of nucleotide modifications in E. coli rRNA.
FIGURE 4.
The m/z region 2190–2290 from an MALDI time-of-flight mass spectrum of the C2480–C2527 fragment of E. coli 23S rRNA digested with RNase T1. rRNA in this sample was isolated from stationary-phase cells. The 2496-CACmCUCG-2502 fragment at m/z 2233.34 and its 16-Da-larger derivative at 2249.31 appear at about equal intensities.
It is difficult to judge whether the modification at C2501 is purposely synthesized at this position, or if the modification is a side effect created by, for example, a tRNA-modifying enzyme. Indications for the latter would be its relatively low abundance and the fact that 16-Da cytidine modifications have, as yet, only been detected in tRNA, not in ribosomal RNA. One explanation for the modification may therefore involve recognition of the motif around C2501 by a tRNA-modifying enzyme. However, some aspects of the 16-Da modification, such as its spatial localization and its growth phase dependence, make it worth considering possible structural/functional roles in the local ribosomal environment. C2501 is implicated in maintaining the tertiary structure near the catalytic center, because it forms a conserved wobble pair with A2450, positioning it within 6 Å of the proposed active-site residue A2451 (Ban et al. 2000; Nissen et al. 2000). A role of the modification at C2501 could be to stabilize this H-bonding, and thereby function in the maintenance of local spatial structures.
C2501 is furthermore localized in a hot spot for binding of peptidyl transferase antibiotics, in particular the streptogramin A and B drugs (for review, see Porse et al. 2000). Using a UV-cross-linking procedure, the therapeutically important streptogramin B antibiotic pristinamycin IA was previously found to bind tightly at position U2500/C2501 in authentic 23S rRNA (Porse et al. 1999). Interestingly, natural, posttranscriptionally modified rRNA was required for drug binding, as no cross-link was observed using full-length T7 23S rRNA transcripts. This was interpreted to indicate involvement of one or more posttranscriptional modifications in the sequence 2498-Cm-C-U-C-G-m2A-ψ-G-2505. Our finding that C2501 is, indeed, partially modified itself, suggests that the 16-Da modification at C2501 may, directly or indirectly, be implicated in pristinamycin IA binding. Alternatively, the modification at C2501 may simply render this nucleotide a more potent UV-cross-linking reactant.
Conclusion
By application of the sensitive MALDI MS technique, we investigated the functionally important peptidyltransferase center of E. coli 23S rRNA for the presence of unknown modification. Own unpublished results, as well as other published data, suggested the existence of a modification at or near C2501, making this site the focus of the present study. By combining a preparation method based on nuclease protection with mass spectrometric and tandem mass spectrometric analysis, it was possible to detect and localize a novel 16-Da partial modification at C2501.
Based on the data acquired during this work, we estimate the C2501-modified rRNA from exponentially growing cells to be 10%–30% of total rRNA. The level of modification was found to increase in stationary cells, reaching 40%–60%. A modification at C2501 was suggested earlier from reverse transcriptase mapping of E. coli 23S rRNA (Bakin and Ofengand 1993), but these data were not confirmed by a subsequent screen using the ESI/LC technique (Kowalak et al. 1995). The modification most likely escaped detection because of its presence in relative low amounts, a possibility also discussed by these investigators. The partial nature of the modified form likewise impairs a precise identification of the modification, which we have, as yet, not been able to establish. Not many types of modifications, however, would display the characteristic 16-Da mass displacement. The most evident are a sulfur replacing an oxygen or addition of a hydroxyl group; looking at the current list of known cytidine modifications, only 2-thiocytidine has the characteristic 16-Da mass increment (Rozenski et al. 2000). Partially methylated 23S rRNA nucleotides were recently found in Sulfolobus acidocaldarius and Bacillus subtilis (Hansen et al. 2002) using a similar MALDI mass spectrometry approach. The modification at C2501 reported here is to our knowledge the only reported naturally occurring partial modification found in E. coli rRNA. That it previously has escaped detection in an otherwise thoroughly scrutinized region suggests the presence of other as yet overlooked partially modified residues. As shown by the present study, mass displacement of a minor fraction of the RNA is easily recognizable in the MALDI spectrum. A thorough screen using a similar method may therefore reveal other novel partial modifications.
MATERIALS AND METHODS
Preparation of ribosomal rRNA
E. coli MRE 600 and DH1 strains were grown in liquid LB medium to an optical density of 0.4 at 450 nm, and harvested by centrifugation. Stationary-phase cells were harvested 1 h, 3 h, and 24 h after entrance into stationary phase. Cells were washed and resuspended in TMN buffer (50 mM Tris-HCl at pH 7.8, 10 mM MgCl2, 100 mM NH4Cl). The cells were lyzed by sonication, and cell debris was removed by centrifugation (12 min twice at 6000 rpm). Ribosomes were collected from the supernatants by centrifugation at 18,000 rpm for 16 h at 4°C in a Beckman Ti50 rotor. The pellet was resuspended in TMN buffer and stored at −80°C. Total rRNA was extracted three times with phenol/chloroform and ethanol-precipitated.
Isolation of a defined rRNA sequence by specific hybridization and single-strand digestion
To isolate a defined rRNA sequence, 1000 pmoles of a synthetic oligodeoxynucleotide complementary to C2480–C2527 of E. coli 23S rRNA was incubated with 100 pmoles of total rRNA in 0.3 volumes of hybridization buffer (250 mM HEPES, 500 mM KCl at pH 7). The mixture was incubated for 5 min at 90°C and then allowed to hybridize during slow cooling to 45°C over 3.5 h. Mung bean nuclease buffer was added to a final concentration of 50 mM NaOAc (pH 5) at 25°C, 30 mM NaCl, and 1 mM ZnCl2 together with 30 units of mung bean nuclease (New England Biolabs) and 0.5 μg of RNase A (Sigma-Aldrich), followed by a 50-min incubation at 35°C. The reaction was extracted once with phenol/ chloroform, and the RNA:DNA hybrid was ethanol-precipitated. The precipitate was redissolved in a 1:2 solution of H2O and formamide.
To purify the specific rRNA sequence and remove the DNA part, the sample was run on a 13% polyacrylamide gel containing 7 M urea. Bands were visualized by ethidium bromide staining, and the rRNA band excised and eluted overnight at 4°C in 2 M NH4Ac (pH 5.3). RNA was precipitated from the NH4Ac eluate by adding 1 volume of ethanol and 1 volume of isopropanol.
RNase and phosphatase digestions
For RNase and phosphatase digestions, 0.5 μL of 0.5 M 3-hydroxypicolinic acid (3-HPA) was added to a 1-μL aliquot of 20–30 pmoles of the isolated 48-mer, and the RNA was digested with either 20 units of RNase T1 (USB) or 0.25 μg of RNase A. RNase T1 digestions were performed for 3–4 h, RNase A digestions for 2–3 h at 37°C. Dephosphorylated samples were prepared by a further adding of 0.5 μL of alkaline phosphatase buffer (0.5 M Tris-HCl, 1 mM EDTA at pH 8.5 and 20°C) and 0.1 units of alkaline phosphatase (Boehringer Mannheim) followed by 1 h of incubation at 37°C.
MALDI TOF mass spectrometry
When linear phosphates were needed, the cyclic phosphates from the RNase T1 digestions were hydrolyzed with HCl by adding 0.25 volumes of 0.5 M HCl with incubation for 30 min at room temperature. The sample was dried under vacuum and redissolved in H2O. MALDI TOF mass spectrometry was carried out as described previously (Kirpekar et al. 2000). In brief, 10 pmoles of the RNase digest, 0.7 μL of 0.5 M 3-HPA matrix, and a small amount of suspended ammonium-loaded ion exchange beads were mixed on the target plate. The sample was allowed to dry and beads were subsequently removed. Spectra were recorded in reflector and positive ion mode on a PerSeptive Voyager STR mass spectrometer (Applied Biosystems). All spectra were smoothed using the software ‘m/z’ (Proteometrics Inc.)
MALDI tandem mass spectrometry
Samples were prepared as for MALDI TOF mass spectrometry. The tandem mass spectra were recorded in positive ion mode on a MicroMass MALDI Q-TOF Ultima mass spectrometer. To obtain exclusive ion selections, the parent ion window for the m/z 2215, 2231, and 2233 ions was set to two m/z values. To efficiently isolate the scarce m/z 2249 ion, this window was set to approximately four m/z values (no other interfering ions occurred near m/z 2249). The collision energy used for tandem mass spectrometry varied between 60 and 70 eV. All spectra were smoothed using the MassLynx software supplied by the manufacturer.
Acknowledgments
This work was supported by the Danish Biotechnology Instrument Center and the Danish Natural Science Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5259404.
REFERENCES
- Bakin, A. and Ofengand, J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Brimacombe, R., Mitchell, P., Osswald, M., Stade, K., and Bochkariov, D. 1993. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 7: 161–167. [DOI] [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., and Richarme, G. 2000. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271: 714–718. [DOI] [PubMed] [Google Scholar]
- Cunningham, P.R., Richard, R.B., Weitzmann, C.J., Nurse, K., and Ofengand, J. 1991. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S ribosomal subunit of Escherichia coli. Biochimie 73: 789–796. [DOI] [PubMed] [Google Scholar]
- Green, R. and Noller, H.F. 1996. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA 2: 1011–1021. [PMC free article] [PubMed] [Google Scholar]
- Hansen, M.A., Kirpekar, F., Ritterbusch, W., and Vester, B. 2002. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA 8: 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirpekar, F. and Krogh, T.N. 2001. RNA fragmentation studied in a matrix-assisted laser desorption/ionization tandem quadrupole/orthogonal time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 15: 8–14. [DOI] [PubMed] [Google Scholar]
- Kirpekar, F., Douthwaite, S., and Roepstorff, P. 2000. Mapping post-transcriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA 6: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak, J.A., Pomerantz, S.C., Crain, P.F., and McCloskey, J.A. 1993. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 21: 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak, J.A., Bruenger, E., and McCloskey, J.A. 1995. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J. Biol. Chem. 270: 17758–17764. [DOI] [PubMed] [Google Scholar]
- Liguori, A., Napoli, A., and Sindona, G. 2000. Survey of the proton affinities of adenine, cytosine, thymine and uracil dideoxyribo-nucleosides, deoxyribonucleosides and ribonucleosides. J. Mass Spectrom. 35: 139–144. [DOI] [PubMed] [Google Scholar]
- Maden, B.E. 1980. Methylation map of Xenopus laevis ribosomal RNA. Nature 288: 293–296. [DOI] [PubMed] [Google Scholar]
- McLuckey, S.A., Van Berkel, G.J., and Glish, G.L. 1992. Tandem mass spectrometry of small multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 3: 60–70. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Noon, K.R., Bruenger, E., and McCloskey, J.A. 1998. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyper-thermophile Sulfolobus solfataricus. J. Bacteriol. 180: 2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor, M., Thomas, C.L., Zimmermann, R.A., and Dahlberg, A.E. 1997. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 25: 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse, B.T., Kirillov, S.V., Awayez, M.J., and Garrett, R.A. 1999. UV-induced modifications in the peptidyl transferase loop of 23S rRNA dependent on binding of the streptogramin B antibiotic, pristinamycin IA. RNA 5: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse, B.T., Kirillov, S.V., and Garrett, R.A. 2000. Antibiotics and the peptidyltransferase center. In The ribosome: Structure, function, antibiotics, and cellular interactions (eds. R.A. Garrett et al.), pp. 441–449. American Society for Microbiology Press, Washington, DC.
- Rozenski, J., Crain, P.F., and McCloskey, J.A. 2000. The RNA Modification Database: 2000 update. Nucleic Acids Res. 27: 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal, R.P. and Vold, B. 1976. Changes in transfer ribonucleic acids of Bacillus subtilis during different growth phases. Nucleic Acids Res. 3: 1249–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]