Abstract
CHAT and Cox type 1 live-attenuated poliovirus strains were developed in the 1950s to be used as vaccines for humans. This paper describes their characterization with respect to virulence, sensitivity for growth at high temperatures, and complete nucleotide and amino acid sequences. The results are compared to those for their common parental wild virus, the Mahoney strain, and to those for two other poliovirus strains derived from Mahoney, the Sabin 1 vaccine strain and the mouse-adapted LS-a virus. Analysis of four isolates from cases of vaccine-associated paralytic poliomyelitis related to the CHAT vaccine revealed genetic and phenotypic properties of the CHAT strain following replication in the human gut. CHAT-VAPP strain 134 contained a genome highly evolved from that of CHAT (1.1% nucleotide differences), suggesting long-term circulation of a vaccine-derived strain in the human population. The molecular mechanisms of attenuation and evolution of poliovirus in humans are discussed in the context of the global polio eradication initiative.
Oral live-attenuated poliovirus vaccines have been the main tool in the program for the global eradication of poliomyelitis (13). Poliovirus is a member of the Picornaviridae family, a group of nonenveloped positive-strand RNA viruses (40). The coding region of the genome is translated as a single polyprotein and is then processed to generate the viral capsid (VP1 to VP4 proteins) and the nonstructural proteins. The coding region is preceded by a 5′ noncoding region (NCR) of approximately 740 nucleotides organized in structural stem-loop domains important for RNA synthesis and translation initiation (53). The 3′ end of the genome consists of an NCR of about 34 nucleotides which is also implicated in RNA replication.
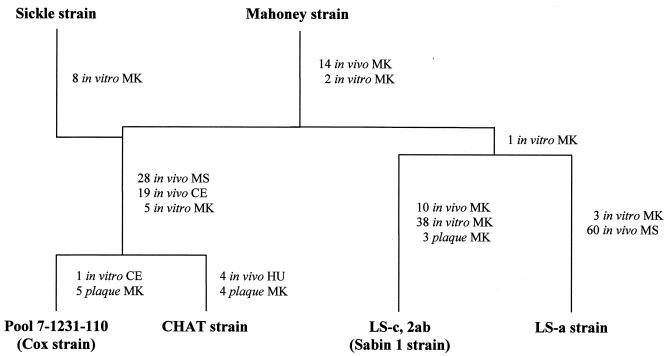
Different strategies were used to produce early versions of poliovirus live-attenuated strains, starting from either virulent or naturally attenuated poliovirus isolates obtained from humans (5, 19, 27, 53). Attenuated vaccine candidates for type 1 poliovirus were derived from the wild Mahoney strain. The Mahoney strain was originally isolated at the University of Michigan in 1941 from a pool of stool specimens of three healthy children, 10, 8, and 3 years of age (19). It was shown to be virulent in monkeys by the intraspinal and intracerebral routes and in humans, during an episode known as the Cutter incident, when a number of children immunized with an inactivated poliovirus vaccine preparation containing inadequately inactivated Mahoney virus developed paralytic poliomyelitis (47). During the 1950s, several virus variants were derived from the Mahoney strain by successive passages in various in vivo and in vitro cell substrates (Fig. 1). Sabin 1, CHAT, and Cox are all attenuated strains derived from Mahoney and selected on the basis of their lack of neurovirulence in monkeys to be used as vaccines (5, 19, 27, 53). LS-a is a mouse-adapted variant, one of the few PV1 strains that have been shown to cause disease in mice, and its entire nucleotide sequence has been determined (30).
FIG. 1.
Passage histories of CHAT, Cox, Sabin 1, and LS-a from the wild-type 1 poliovirus Mahoney strain. MK, monkey; MS, mouse; CE, chicken embryo; HU, human. The number of passages in each substrate is shown. For details, see the text.
As shown in Fig. 1, Sabin 1 arose from passage in monkey tissues exclusively, whereas the CHAT and Cox strains involved passages in monkey, mouse, and chicken embryo cell substrates. Due to their superior properties in terms of attenuation of neurovirulence in monkeys and data from large field trials in humans (36, 46, 50), the Sabin strains of all three serotypes were eventually selected for licensing and have been used almost universally since then, while the use of Cox was not extensive. CHAT, which had shown optimal characteristics in terms of its restricted capacity to propagate from vaccinees, was used to immunize millions of individuals in several countries (2, 19, 50, 51). These immunization campaigns involved people of all ages, including a significant number of polio-seronegative individuals. There was no evidence of vaccine-associated disease in any of these campaigns. The CHAT vaccine was used in monovalent form, together with monovalent versions of Sabin 2 and Sabin 3, for regular immunization of children in a country of Europe during 1970 and 1971. During that time, this country was included in a World Health Organization (WHO) study to monitor the safety of polio vaccination by determining the rate of vaccine-associated paralytic poliomyelitis (VAPP) (22, 59). A few VAPP cases were associated with the use of CHAT, although most were found to be related to either type 2 or type 3 vaccine polioviruses (59). Similarly, the occurrence of VAPP due to the Sabin 1 virus is far less frequent than for the other two Sabin serotypes (56).
The attenuation phenotype of Sabin 2 has been ascribed to only two mutations at nucleotides 481 in the 5′ NCR sequence and 2908 in the sequence corresponding to capsid residue 143 of VP1 (VP1-143) (39). For Sabin 3, mutations at positions 472 (5′ NCR), 2034 (VP3-91), and 2493 (VP1-6) have been identified as determinants of attenuation (39). In contrast, the genome of Sabin 1 differs from that of its wild parental virus, Mahoney, in 54 point mutations, 20 of which are coding changes (48). The large number of differences has usually been thought to explain the relatively lower rate of VAPP with the type 1 Sabin. cDNA-derived infectious clones containing different combinations of the mutations found in Sabin 1 have been used to investigate the determinants of attenuation of Sabin 1 virus in laboratory animals. The studies suggested that attenuating mutations are located in different regions of the genome, including the 5′ and 3′ NCRs, VP1, VP3, and VP4 capsid proteins as well as in the polymerase 3D nonstructural protein (3, 8, 23, 26, 35, 49, 57). The experiments also revealed that different animals such as monkeys and various strains of normal and transgenic mice showed different sensitivity to certain attenuating mutations of Sabin 1. Molecular characterization of Sabin 1-derived VAPP strains confirmed that several of Sabin 1 mutations reverted to wild-type Mahoney sequences following replication in humans but indicated that reversion at only few of those mutations, which in all cases included the mutation at position 480 in the 5′ NCR, seemed to be sufficient for reversion to neurovirulence (16, 29).
In this paper we report the extensive molecular and biological characterization of CHAT and Cox type 1 live-attenuated poliovirus strains. Our studies included the analysis of four isolates from VAPP cases related to the CHAT strain. The results provide new insights into the molecular mechanisms of poliovirus attenuation and poliovirus evolution following vaccination in humans and are discussed in relation to future decision-making on how to stop polio immunization with live vaccines once wild poliovirus has been eradicated (61).
MATERIALS AND METHODS
Virus preparations.
The Cox strain (pool 7-1231-114), also named Lederle SM strain, was developed at Viral and Rickettsia Research, Lederle Laboratory Division, American Cyanamid Co., Pearl River, N.Y. (5). The CHAT vaccine was originally prepared by Hilary Koprowski at the Wistar Institute of Anatomy and Biology, Philadelphia, Pa., from a precursor of the Cox strain, the SM N-90 strain (27). Several batches of the vaccine were prepared from the original CHAT strain to be used in humans. CHAT 10A-11 corresponds to an original vaccine batch from the Wistar Institute used in several countries around the world. CHAT Yugo (National Institute for Biological Standards and Control designation) most likely corresponds to a CHAT vaccine batch originally named CHAT 23 or CHAT 24. It was prepared at the Institute of Immunology, Zagreb, Croatia, and used in several countries of eastern Europe. All results referring to the CHAT strain in this paper correspond to the CHAT 10A-11 batch if not specified otherwise. Type 1 poliovirus isolates from VAPP cases 8/70, 12/70, 14/70, and 15/70 represented the original virus samples isolated at the National Institute for Biological Standards and Control during the 1970 WHO collaborative studies (see Table 3) (22). They were assigned WHO reference numbers 134, 135, 136, and 137, respectively. Working virus preparations were collected by growth of the viruses in HEp-2C cells at 35°C in modified Eagle medium without fetal calf serum. Virus stocks were stored at −70°C.
TABLE 3.
Epidemiological data of CHAT-VAPP cases
| Case no. and year | WHO strain | Age of patient (mo) | Association with vaccinationsa | Antibody response to poliovirus type
|
Time of isolation (days postillness) | ||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||
| 8,1970 | 134 | 11 | No known contact | + | − | − | 7 |
| 12,1970 | 135 | 11 | Recipient (19) | + | − | − | 11 |
| 14,1970 | 136 | 10 | Possible contact (21) | + | − | − | 7 |
| 15,1970 | 137 | 17 | Recipient (27) | + | − | − | 17 |
The interval between vaccination or contact with vaccinee and onset of illness (days) is in parentheses.
Cells.
HEp-2C cells were used in different assays described in this paper and were grown in culture as described previously (33).
Reverse transcription, PCR, and nucleotide sequencing of poliovirus genomes.
Poliovirus RNA was purified from HEp-2C cell culture supernatants and used for reverse transcription and PCRs by standard procedures. DNA fragments containing the 5′ end of the viral cDNAs were obtained with the 5′/3′ rapid amplification of cDNA ends kit (Boehringer) according to the manufacturer's instructions. Sequencing of the purified viral RT-PCR DNA products was carried out with a ABI Prism 310 genetic analyzer as specified by the manufacturer. Primers were designed by the “primer walking” strategy. Sequence data were stored as standard chromatogram format files and analyzed with the Wisconsin Package version 10.0-UNIX (Genetics Computer Group), AlignIRV11 (LI-COR), and ClustalW (58) software.
Temperature sensitivity.
Temperature sensitivity was assayed by comparison of plaque formation on HEp-2C cells at 35.0, 39.0, 39.5, and 40.0°C as described before (33).
Neurovirulence.
Viruses were assayed by the standard WHO-approved test for vaccine safety (4, 62), except that fewer animals were used per virus.
In vivo labeling of viral proteins.
Viral polypeptides were labeled with 35S as described previously (38). Tran35S-label and methionine- and cysteine-free medium (ICN) were used. Infected HEp-2C cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited with the EMBL Data Library under accession no. AJ416942 (CHAT) and AJ430385 (Cox).
RESULTS
Genomic analysis of CHAT and Cox strains.
The complete nucleotide sequences of the CHAT and Cox genomes were determined and analyzed to establish their respective predicted amino acid sequences. CHAT and Cox sequences were compared to those from Mahoney, Sabin 1, and LS-a (30, 52). Table 1 shows the differences in the sequences of CHAT and Cox with respect to their common wild parental virus, Mahoney. Table 2 shows the percentage of nucleotide and amino acid sequence differences between CHAT, Cox, Sabin 1, and LS-a and their parental virus, Mahoney.
TABLE 1.
Nucleotide and amino acid differences between CHAT and Cox strains and Mahoney virusa
| Region | Nucleotide
|
Amino acid
|
||||||
|---|---|---|---|---|---|---|---|---|
| Positionb | Mahoney | CHAT | Cox | Position | Mahoney | CHAT | Cox | |
| 5′ NCR | 42 | U | C | |||||
| 189 | C | U | U | |||||
| 649 | C | U | U | |||||
| 652 | A | G | G | |||||
| 674 | C | U | U | |||||
| 675 | C | U | U | |||||
| 709 | U | C | ||||||
| 716 | U | C | C | |||||
| VP4 | 921 (2) | U | C | C | 60 | Val | Ala | Ala |
| VP2 | 1014 (3) | A | G | G | ||||
| 1111 (3) | A | U | U | |||||
| 1285 (3) | U | C | C | |||||
| 1321 (3) | A | G | ||||||
| 1373 (1) | C | U | U | 142 | His | Tyr | Tyr | |
| 1429 (3) | G | A | A | |||||
| 1490 (1) | C | U | U | 181 | Leu | Phe | Phe | |
| 1597 (3) | C | G | G | |||||
| 1639 (3) | A | G | G | |||||
| 1657 (3) | A | G | G | |||||
| 1678 (3) | C | U | U | |||||
| 1708 (3) | A | G | 253 | Ile | Met | |||
| VP3 | 1795 (3) | C | U | U | ||||
| 1885 (3) | A | U | U | |||||
| 1927 (3) | C | U | ||||||
| 2213 (1) | U | C | ||||||
| 2339 (1) | G | A | 192 | Val | Ile | |||
| 2353 (3) | U | C | C | |||||
| VP1 | 2545 (3) | A | G | G | ||||
| 2585 (1) | A | G | G | 36 | Thr | Val | Val | |
| 2586 (2) | C | U | U | |||||
| 2606 (1) | G | A | 43 | Ala | Thr | |||
| 2762 (1) | C | U | U | 95 | Pro | Ser | Ser | |
| 2784 (2) | A | G | G | 102 | Asp | Gly | Gly | |
| 2812 (3) | U | C | C | |||||
| 2815 (3) | U | C | ||||||
| 2869 (3) | U | C | C | |||||
| 2879 (1) | C | U | U | 134 | Leu | Phe | Phe | |
| 2891 (1) | G | A | 138 | Val | Ile | |||
| 3034 (3) | C | U | ||||||
| 3058 (3) | G | A | A | |||||
| 3140 (1) | U | C | 221 | Ser | Pro | |||
| 3163 (3) | U | C | C | |||||
| 3220 (3) | U | C | C | |||||
| 3353 (1) | A | G | G | 292 | Thr | Ala | Ala | |
| 2A | 3445 (3) | C | U | U | ||||
| 3460 (3) | U | A | A | 25 | Asp | Glu | Glu | |
| 3463 (3) | U | C | C | |||||
| 3492 (2) | G | A | A | 36 | Ser | Asn | Asn | |
| 3685 (3) | C | U | U | |||||
| 3766 (3) | C | A | A | |||||
| 2B | 3880 (3) | A | G | |||||
| 3896 (1) | A | G | G | 22 | Ser | Gly | Gly | |
| 3898 (3) | C | A | A | |||||
| 4078 (3) | A | G | G | |||||
| 2C | 4274 (1) | A | C | |||||
| 4405 (3) | C | U | U | |||||
| 4444 (3) | U | C | C | |||||
| 4729 (3) | C | U | ||||||
| 4741 (3) | G | C | C | |||||
| 5044 (3) | U | C | C | |||||
| 5051 (1) | A | G | G | 310 | Ile | Val | Val | |
| 5098 (3) | G | A | A | |||||
| 5107 (3) | U | C | C | |||||
| 3A | 5264 (1) | A | G | G | 52 | Thr | Ala | Ala |
| 3B | 5420 (3) | C | A | A | ||||
| 3C | 5440 (3) | A | G | G | ||||
| 5605 (3) | C | U | U | |||||
| 5840 (1) | A | G | G | 131 | Thr | Val | Val | |
| 5841 (2) | C | U | U | |||||
| 5899 (3) | U | C | C | |||||
| 5968 (3) | A | G | ||||||
| 3D | 6002 (1) | A | G | G | 6 | Met | Val | Val |
| 6025 (3) | A | G | G | |||||
| 6046 (3) | G | A | A | |||||
| 6096 (2) | U | C | 37 | Val | Ala | |||
| 6214 (3) | G | A | ||||||
| 6616 (3) | G | A | A | |||||
| 6679 (3) | U | C | C | |||||
| 6761 (1) | G | A | 209 | Gly | Arg | |||
| 6937 (3) | U | C | C | |||||
| 6985 (3) | C | U | ||||||
| 7243 (3) | U | A | A | |||||
| 3′ NCR | 7441 | A | G | |||||
Mutations also found in Sabin 1 are underlined.
Numbers in parentheses show position within the codon.
TABLE 2.
Percentage of nucleotide and amino acid sequence differences between CHAT, Cox, Sabin 1, LS-a, and Mahoney type 1 poliovirus strains
| Strain | % Difference from straina
|
||||
|---|---|---|---|---|---|
| Mahoney | Sabin 1 | LS-a | CHAT | Cox | |
| Mahoney | 0.91 | 0.91 | 0.96 | 0.73 | |
| Sabin 1 | 0.71 | 1.09 | 1.28 | 1.05 | |
| LS-a | 0.71 | 0.73 | 1.32 | 1.09 | |
| CHAT | 1.04 | 1.10 | 1.21 | 0.32 | |
| Cox | 0.90 | 0.97 | 1.08 | 0.27 | |
The lower left and upper right portions of the table show nucleotide and amino acid differences, respectively.
As expected, CHAT and Cox showed the two most similar sequences (Tables 1 and 2), differing in 20 nucleotide positions, 7 of them resulting in coding changes. CHAT and Cox viruses were found to be closely related to Mahoney, although a significant number of mutations were identified in both viruses. The CHAT genome contained 77 nucleotide changes with respect to Mahoney, 7 in the 5′ NCR and 70 in the coding region resulting in 21 coding changes. The Cox genome had 68 nucleotide changes, 7 in the 5′ NCR, 60 in the coding region resulting in 16 coding changes, and 1 in the 3′ NCR. Sixty-two mutations were common to both CHAT and Cox (6 in the 5′ NCR and 53 in the coding region, 15 of them nonsynonymous and 38 synonymous). Sabin 1, CHAT, Cox, and LS-a all shared 15 nucleotide mutations with respect to Mahoney at positions thought not to be under selective pressure, 3 of them located in the hypervariable region in the 5′ NCR and 12 located at synonymous coding positions distributed throughout the genome. These changes were most likely acquired during the first passages of Mahoney in monkey cells before the separation of the four lineages (Fig. 1) and were maintained during successive passages through different cell substrates.
Of the seven changes identified in the 5′ NCR of CHAT and Cox genomes, only that at nucleotide 189 was located in a predicted stem-loop structure (in domain III of the 5′ NCR) (54). The same mutation is present in the Sabin 1 strain. The mutation from C to U results in a predicted weak base pair, G·U, instead of the G·C pair present in Mahoney. Most of the other mutations in the 5′ NCR of CHAT and Cox strains were located in the hypervariable region outside the structural domains (at nucleotides 649, 651, 674, 675, 709, and 716). Mutation at nucleotide 42 in CHAT mapped between domains I and II and would not be expected to affect base pairing (54). Only one change was found in the 3′ NCR of Cox virus, at nucleotide 7441, also present in Sabin 1 genome.
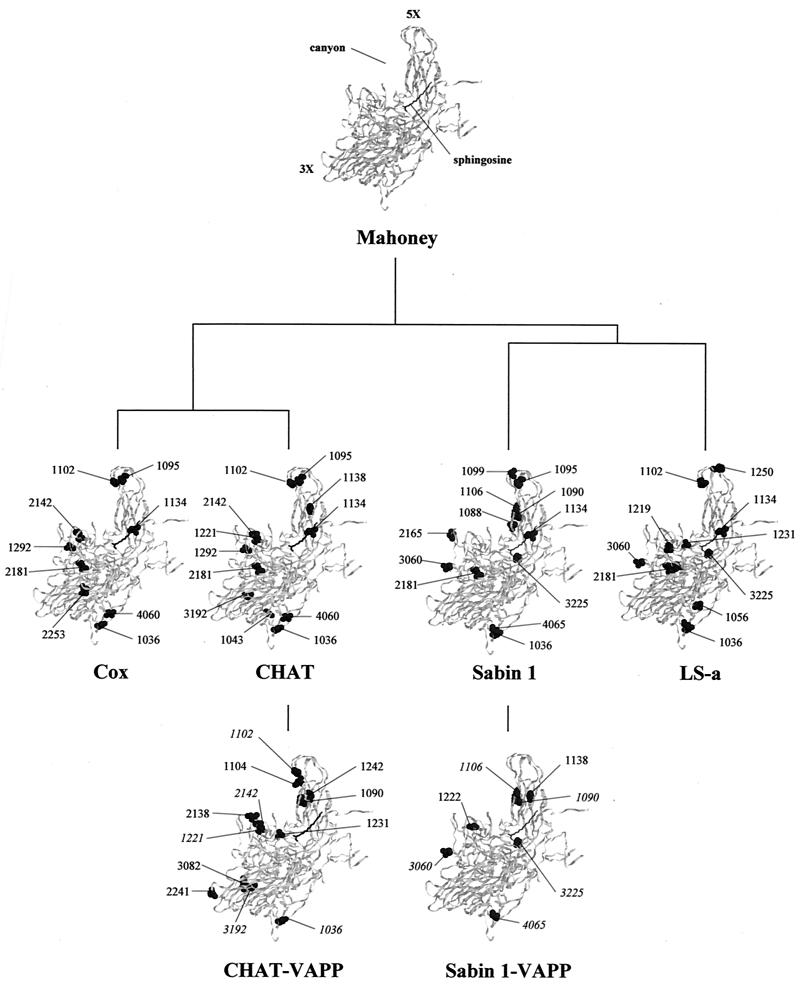
Changes in the amino acid sequences of CHAT and Cox with respect to the Mahoney strain are shown in Table 1. A total of 12 and 9 amino acid changes were identified in the capsid regions of CHAT and Cox strains, respectively. Most of them, 8 in CHAT and 6 in Cox, were located in VP1, the most exposed of the capsid proteins (21). As shown in Fig. 2, capsid mutations involved different regions in the virion, including the BC loop in VP1 (VP1-95 and VP1-102), the canyon (VP2-142), antigenic site 2a (VP1-221), the hydrocarbon-binding pocket (VP1-134), and the inner capsid surface (VP1-36, VP1-43, and VP4-60). Several amino acid mutations were scattered through the genomic regions coding for nonstructural proteins and included changes in proteins 2A, 2B, 2C, 3A, 3C, and 3D (Table 1). Mutations at capsid residues VP1-36, VP1-134, and VP2-181 and at nonstructural sites 2A-25, 2A-36, and 2B-22 in CHAT and Cox were also present in Sabin 1 and LS-a.
FIG. 2.
Ribbon diagrams of the α-carbon trace of the wild-type 1 poliovirus Mahoney strain protomer (21), viewed from the side of the virion (the outside is toward the top left of the image and the inside is toward the bottom right). The virus particle consists of 60 protomers, each containing a single copy of VP1, VP2, VP3, and VP4, arranged in icosahedral symmetry. 3X and 5X, threefold and fivefold axes of symmetry. The location of the canyon and the sphingosine in the hydrocarbon-binding pocket are also indicated. Amino acid mutations found in CHAT and Cox strains are shown and compared to those reported for Sabin 1 and LS-a strains (30, 48). Mutations identified in CHAT-VAPP isolates and those previously published for Sabin 1-VAPP strain 1-IIs (16) are also displayed. Reversions to wild Mahoney amino acid sequences are shown in italics.
Genomic analysis of type 1 isolates from VAPP cases.
Four type 1 isolates from VAPP cases were selected from those available from a WHO collaborative study on the basis that they were from poliomyelitis cases temporally and geographically associated with the use of the CHAT live-attenuated vaccine (22). Epidemiological data of these cases are presented in Table 3. All four poliovirus strains were isolated from stool samples. Isolates 135 and 137 were from children who had received monovalent CHAT vaccine 30 and 44 days earlier, respectively. Strain 136 was from a possible contact of a CHAT vaccinee(s) taken 28 days after vaccination day. The case yielding isolate 134 was classified as a community case, since the patient was not given the vaccine and no association with vaccinees or contacts could be identified. The viruses represented the original virus samples isolated during a WHO collaborative study (22) in 1970 by two serial passages of clarified fecal material on primary rhesus monkey kidney cells.
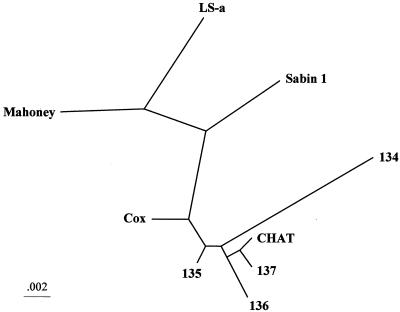
The complete nucleotide sequences of the four CHAT-VAPP strains were determined and compared with those of CHAT, Cox, Sabin 1, LS-a, and Mahoney. Table 4 shows the differences found in the amino acid sequences of the four VAPP isolates with respect to the CHAT strain. Phylogenetic relationships between the CHAT-VAPP isolates and the Mahoney, CHAT, Cox, Sabin 1, and LS-a strains were estimated by comparing the nucleotide sequences coding for the VP1 capsid protein. The sequences were aligned by using ClustalW (version 1.75) (58) and analyzed by the neighbor-joining method using the Advanced Genebee ClustalW 1.75 online facility (http://www.genebee.msu.su). The unrooted phylogenetic tree resulting from this analysis is shown in Fig. 3.
TABLE 4.
Amino acid changes in CHAT-VAPP isolatesa
| Region | Position | Amino acid in strain:
|
||||
|---|---|---|---|---|---|---|
| CHAT | 134 | 135 | 136 | Mahoney | ||
| VP1 | 36 | Val | Ala | Thr | ||
| 90 | Met | Leu | Met | |||
| 102 | Gly | Asp | Asp | |||
| 104 | Leu | Gln | Leu | |||
| 221 | Pro | Ser | Ser | Ser | Ser | |
| 231 | Ala | Val | Ala | |||
| 242 | Val | Ala | Val | |||
| VP2 | 138 | Thr | Ser | Thr | ||
| 142 | Tyr | His | His | |||
| 241 | Ser | Asn | Ser | |||
| VP3 | 82 | Ile | Val | Ile | ||
| 192 | Ile | Val | Val | Val | ||
| 2A | 25 | Glu | Asp | |||
| 32 | Asn | Ser | Asn | |||
| 36 | Asn | Asp | Ser | |||
| 59 | Ala | Thr | Ala | |||
| 87 | Asn | Asp | Asn | |||
| 2B | 54 | Ile | Val | Ile | ||
| 3A | 86 | His | Tyr | Tyr | Tyr | His |
| VPg | 21 | Val | Ala | Ala | ||
| 3C | 131 | Val | Thr | Thr | Thr | |
| 3D | 6 | Val | Met | Met | ||
| 12 | Val | Ala | Val | |||
| 21 | Ser | Thr | Ser | |||
| 37 | Ala | Val | Val | |||
| 143 | Lys | Arg | Lys | |||
| 147 | Thr | Ala | Thr | |||
| 209 | Arg | Gly | Gly | |||
Reversion to wild-type Mahoney sequences is indicated by italics. No changes were detected in strain 137.
FIG. 3.
Neighbor-joining phylogenetic tree showing the genetic relationships between the wild-type 1 poliovirus strain Mahoney, CHAT, Cox, Sabin 1, and LS-a, all derived from the Mahoney virus, and strains 134, 135, 136, and 137, all isolated from CHAT-VAPP cases. The 906-nucleotide VP1 coding sequences were compared.
The sequences of all four VAPP strains very closely resembled that of CHAT. This was particularly obvious when the distribution of synonymous nucleotide mutations was examined with respect to the parental Mahoney strain. All such changes previously identified in the CHAT genome (Table 1) were present in the genomes of the four VAPP isolates (data not shown). There were very few nucleotide differences between most of the VAPP strains (135, 136, and 137) and CHAT. Three mutations were found in the entire genome of strain 137, all of them synonymous (C1250U, C3316U, and C6901U). The genome of virus 135 contained six nucleotide mutations, five resulting in coding changes and one synonymous (A3896C). Isolate 136 had nine nucleotide changes, five coding and four synonymous (C1090U, G1105A, U2950C, and U3640C). In contrast, the genome of strain 134 was rather different from that of CHAT. As shown in Fig. 4, sequences specific for Sabin type 3 vaccine strain were identified in the 3′ region of the 134 genome, most likely as a result of a recombination event(s). In addition, the genome of strain 134 contained 63 nucleotide changes with respect to CHAT, 5 in the 5′ NCR (A157G, U337C, U649C, A711U, and A737G) and 58 in the coding region, 23 of them resulting in amino acid changes (Table 4). Strain 134 contained only five nucleotide changes with respect to Sabin 3 in the Sabin 3-derived region, one resulting in an amino acid change at 3C-131, a reversion to the sequence present in the Sabin 3 wild parent, the Leon strain (55).
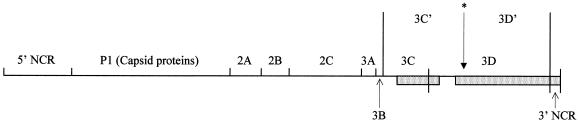
FIG. 4.
Genomic structure of CHAT-VAPP strain 134. Sabin 3 sequences are indicated by shaded boxes. Crossover points were located at nucleotides 5633 to 5637, 6236 to 6237, and 6350 to 6357. The protease (3C) and polymerase (3D) can undergo an alternative cleavage within 3D (∗) mediated by the viral protease 2A to give polypeptides 3C′ and 3D′. This alternative cleavage site is absent in Sabin 3 and was lost in isolate 134.
Direct reversions to Mahoney amino acid sequences occurred in one or more of the four VAPP strains at capsid residues VP1-102 (VP1 BC loop), VP1-221 (antigenic site 2a), VP2-142 (canyon), and VP3-192 (internal) and at nonstructural sites 3C-131, 3D-6, 3D-37, and 3D-209. Mutations at VP1-90, VP1-104, and VP1-242 at or close to the VP1 BC loop and changes at VP1-138 and VP1-231 in the canyon were identified in different VAPP isolates and could have been incorporated as secondary site reversions to mutations present in CHAT.
Temperature sensitivity.
The results for temperature sensitivity of CHAT, Cox, and the CHAT-VAPP strains are shown in Table 5. CHAT and Cox viruses exhibited a significant degree of temperature sensitivity for growth in HEp-2 cells compared to the wild strain Mahoney. However, the temperature sensitivity phenotype of CHAT and Cox was less severe than that of the Sabin 1 strain. Strains 134, 135, and 136 showed partial loss of the temperature sensitivity phenotype of CHAT, but none of them reverted to the non-temperature-sensitive phenotype of the Mahoney strain. As expected, strain 137, with only three silent mutations with respect to CHAT, showed a temperature sensitivity phenotype very similar to that of the vaccine strain.
TABLE 5.
Temperature sensitivity of CHAT, Cox, and CHAT-VAPP strains
| Virus | Log10 titer reduction
|
||
|---|---|---|---|
| 35 vs 39.0°C | 35 vs 39.5°C | 35 vs 40.0°C | |
| Sabin 1 | 2.1 | >5.0 | >5.0 |
| CHAT | 0.9 | 3.1 | >5.0 |
| Cox | 0.3 | 2.3 | >5.0 |
| 134 | −0.1 | 0.0 | 2.5 |
| 135 | 0.1 | 0.3 | >5.0 |
| 136 | 0.5 | 0.8 | >5.0 |
| 137 | 1.0 | 2.9 | >5.0 |
| Mahoney | 0.0 | 0.1 | 0.1 |
Neurovirulence.
As shown in Table 6, CHAT and Cox strains exhibited a higher degree of neurovirulence than Sabin 1. The results for CHAT 10A-11 and CHAT Yugo batches were very similar. CHAT-VAPP strains 134 and 136 showed an increase in neurovirulence with respect to CHAT, but they were still less pathogenic than Mahoney virus.
TABLE 6.
Neurovirulence of CHAT, Cox, and CHAT-VAPP strains
| Strain | No. of clinically affected animals/total | Mean histological lesion score |
|---|---|---|
| Sabin 1 | 0/4 | 0.56 |
| CHAT 10A-11 | 2/4 | 1.22 |
| CHAT Yugo | 1/3 | 1.27 |
| Cox | 2/4 | 1.55 |
| 134 | 4/4 | 2.01 |
| 136 | 2/3 | 1.85 |
| Mahoney | 4/4 | 2.5 |
Alternative cleavage of 3CD polyprotein.
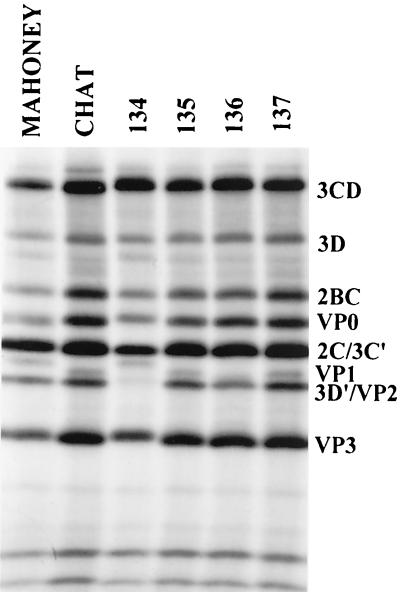
Due to its recombination with the Sabin 3 genome, CHAT-VAPP isolate 134 acquired an amino acid change at 3D-147, resulting in the loss of a recognition site for cleavage by the viral protease 2A, absent in the Sabin 3 strain (Fig. 4). To confirm the loss of this alternative cleavage site in the 3CD polypeptide of strain 134, the protein synthesis of this virus in tissue culture was studied and compared to that of the Mahoney, CHAT, and the three other CHAT-VAPP strains. As shown in Fig. 5, the 3D′ protein, a product of the alternative cleavage of 3CD by protease 2A, was not detected in cells infected with CHAT-VAPP strain 134 but was present in all cell extracts infected with the other strains studied. Obvious differences in the migration of VP1 were observed among the different strains. These differences are likely to be due to amino acid sequence differences in the strains. The mutation of a negatively charged aspartic acid at position VP1-102, in Mahoney, to a glycine in CHAT may explain, at least in part, the differences in electrophoretic behavior between VP1s. The aspartic acid at VP1-102 was restored in CHAT-VAPP strain 134, and the VP1 of this strain showed electrophoretic migration similar to that of Mahoney VP1. The slight difference in mobility and radioactive signal of the VP1 of strain 136 may be explained by the loss of a methionine at VP1-90, which changed to a leucine in this strain.
FIG. 5.
Autoradiography of sodium dodecyl sulfate-polyacrylamide protein gel of 35-S-labeled cell extracts infected with CHAT-VAPP strains 134, 135, 136 and 137.
DISCUSSION
The mechanisms of attenuation of Sabin type 1 vaccine strain appear to involve several mutations scattered throughout its genome, as concluded from studies in animal models (3, 8, 23, 26, 35, 49, 57). Some of those mutations reverted to wild-type Mahoney sequences in Sabin 1-VAPP isolates, suggesting their relevance for replication and/or virulence in humans, although only a few Sabin 1-VAPP isolates have been fully characterized to date (16, 29). Here we report a detailed study of the genetic and phenotypic characteristics of CHAT and Cox type 1 live-attenuated poliovirus vaccine strains which, like Sabin 1, were developed from the wild Mahoney strain but with rather different passage schedules (Fig. 1).
As shown in this paper, CHAT and Cox strains displayed lower degrees of temperature sensitivity for growth in HEp-2c cells and attenuation of neurovirulence in primates than Sabin 1 virus with respect to the wild Mahoney strain. These observations confirmed early reports from several laboratories that showed increasing levels in the attenuation of Cox, CHAT, and Sabin 1 strains, in this order, after direct comparisons of their neuropathogenicity in monkeys (36, 46).
Nucleotide sequence analysis confirmed the phylogenetic relationships between the four Mahoney-derived strains and their respective passage histories relative to each other (Tables 1 and 2 and Fig. 1 to 3).
The CHAT vaccine strain was similar to the Sabin 1 vaccine strain in that a number of differences from the wild parental Mahoney virus were identified in the CHAT genome and seemed to be implicated in its attenuation phenotype. As for Sabin 1-VAPP isolates (16, 29), reversion at some of these positions in CHAT-VAPP strains resulted in an increase of neurovirulence and a loss of temperature sensitivity with respect to the vaccine viruses, linking them to the paralytic disease. Although few mutations have been shown to contribute to the temperature sensitivity of Sabin 1 but not to its attenuation phenotype (3), a good correlation between both phenotypes has been observed for all three Sabin vaccine strains of the three poliovirus serotypes (39). CHAT-VAPP isolate 137 had only three nucleotide changes with respect to the CHAT strain, all at synonymous codon positions. However, the sequence determined might not represent that of the true etiological agent of the neurological disease. Differences in viral sequences have been found in isolates from stool and cerebrospinal fluid samples of the same patient in VAPP cases related to the Sabin vaccine, particularly in cases involving vaccine recipients, such as 137 (17).
Contrary to what was found for the Sabin strains, CHAT and Cox viruses did not seem to contain important attenuation mutations in structural domains of the 5′ NCR. Only one mutation identified in this region, at position 189 in domain II of the internal ribosome entry site, results in the weakening of a predicted base pair (54). This mutation is also present in the genome of Sabin 1 and has been associated with a limited role in the attenuation of this strain (16). However, the mutation at nucleotide 189 did not revert to the Mahoney sequence in any of the CHAT-VAPP isolates investigated. It is striking that, in contrast to the strains studied here and elsewhere (41), the three Sabin strains of poliovirus all have important attenuation mutations within a relatively short sequence in domain V of the internal ribosome entry site in the 5′ NCR (42).
Comparison of mutations in the capsid regions of CHAT, Cox, Sabin 1, and LS-a relative to their parental virus, Mahoney, identified regions in the capsid in which mutations were located more often and which were therefore likely to be important for adaptation for growth in the different in vivo and in vitro substrates used during the preparation of the four strains (Fig. 1). Analysis of the sequences found in VAPP isolates from either Sabin 1 (16, 29) or CHAT vaccinees revealed capsid mutations in the vaccine strains that were relevant for replication and/or virulence in humans.
As a result of this analysis, relevant mutations were identified in CHAT and Cox at capsid residues VP1-95 and VP1-102 in the BC loop of VP1, a highly exposed region on the virion surface near the fivefold axis of symmetry (21) and at position VP2-142, located in the south rim of the canyon, a surface depression that encircles the fivefold axis, in a region that contacts the cell receptor (1, 20). Similar mutations have been associated with mouse adaptation (32, 45), ability to use mutated receptors (9), and ability to persist in neuroblastoma cells (11) in different Mahoney variants.
Comparison of the sequences and X-ray three-dimensional structures of Mahoney, Sabin 3, and the Lansing/Mahoney BC loop chimeric virus indicated that amino acid variations at positions VP1-95 and VP1-102 were greatly responsible for the dramatic structural differences in the VP1 BC loop (63). Mutations at VP1-90 and at VP1-106, at the base of the BC loop, are known to be important determinants of attenuation of Sabin 1 (3, 8, 16, 29). Apart from direct reversions of the mutations at VP1-102 and VP2-142 found in CHAT-VAPP isolate 134, mutations were also identified at VP1-90 in isolate 136, at VP1-104 (adjacent to VP1-102) in isolate 135, and at VP1-242 (next to VP1-90 and VP1-106) in isolate 134. Alterations in the BC loop, located outside the footprint of the receptor, are associated with significant changes in thermal stability and the ability of the virus to undergo receptor-mediated conversions, and so mutations in the BC loop most likely affect events that follow receptor binding and lead to virus cell entry (1, 60).
CHAT and Cox strains also had changes in two other regions of the virion believed to be involved in the control of receptor-induced conformational alterations essential for virus cell entry and in capsid assembly. The first region involved the inner surface of the capsid shell at the interface between protomers formed by the N-terminal extension of VP1 and VP4 residues, where important host range determinants of poliovirus infectivity have been identified (10, 43). This region undergoes dramatic structural rearrangements after virus attachment to susceptible cells, during which VP4 is detached from the virion and the N-terminal region of VP1 becomes extruded from its buried location (14, 15). The second region concerned a conserved hydrocarbon-binding pocket in VP1 located underneath the canyon, which is normally occupied by an endogenous lipid and where drugs that prevent virus uncoating are inserted (14, 44). Mutations in both regions have been shown to contribute to the attenuation of Sabin 1 vaccine strain (3).
Two mutations were found in the CHAT strain only, at capsid amino acids VP1-221 and VP3-192. VP1-221 is located on the surface of the virion at the south rim of the canyon close to the receptor footprint and forms part of antigenic site 2a, fairly conserved among type 1 human poliovirus isolates (21, 37). Mutations at antigenic site 2a have also been seen in Sabin1-VAPP strains and in the mouse-adapted LS-a virus (16, 30). The second mutation exclusive to the CHAT strain was found at residue VP3-192, inaccessible in the interior of the virion (21). Peptides containing poliovirus sequences in that region have been identified as possible T-cell epitopes in in vitro assays (31). Mutations at VP3-192 and VP1-221 appeared to be unstable in humans, since both reverted to the wild Mahoney sequence in two and three of the four CHAT-VAPP isolates, respectively.
Several mutations were identified in nonstructural amino acids of CHAT and Cox strains with respect to Mahoney (Table 1). Two mutations were found in the protease 2A at positions 2A-25 and 2A-36, also present in Sabin 1 and LS-a. These two amino acid mutations plus an additional change at 2A-70 have been shown to contribute to the mouse neurovirulence of the LS-a strain (30). Other mutations of possible significance found in CHAT and Cox were located at the protease 3C and the polymerase 3D. A mutation at residue 73 in 3D has been associated with the temperature sensitivity of Sabin 1 and, to a limited extent, to its attenuation phenotype (3, 8, 35, 49, 57).
The genomic structure of isolate 134, shown in Fig. 4, was the result of at least two recombination events between CHAT and Sabin 3 genomes. As a consequence, isolate 134 incorporated a change at amino acid 3D-147 resulting in the loss of a recognition site (Thr-Tyr/Gly) for cleavage by the viral protease 2A that is conserved among most poliovirus strains but absent in Sabin 3 and its wild parent, Leon (39). Cleavage at this site results in the alternative processing of the protease-polymerase 3CD polypeptide, but its biological significance has not yet been determined (28, 33). This observation confirms that natural recombination can rapidly and significantly change the structure of the virus genome and consequently the virus phenotype (12, 18).
CHAT-VAPP strain 134 contained a viral genome highly evolved from CHAT showing 1.1% nucleotide changes in the VP1 region. This difference corresponds to approximately 1 year of replication and circulation in humans, based on the molecular clock of poliovirus evolution (24, 25, 33). CHAT-VAPP case 134 was classified as a community case, i.e., no contact with a vaccinee or any of his/her contacts was identified, and therefore it may have involved at least two steps of human-to-human transmission. Similarly, a Sabin 1-VAPP isolate from Romania (1-IIs strain) has been reported to contain 0.95% nucleotide changes in the VP1 region with respect to the Sabin 1 genome (16).
The identification of highly drifted vaccine-derived poliovirus isolates such as 134 from CHAT and 1-IIs from Sabin 1 (16) is clear evidence that these strains can survive in the human population for long periods, possibly even within well-immunized communities. These strains are a potential source of poliomyelitis epidemics, particularly in populations with low polio immunity and in the absence of wild poliovirus competitors. Examples of such events, although rare, have been reported. An outbreak of type 3 poliomyelitis in Poland in 1968 was associated with poliovirus strains derived from the USOL-D-bac vaccine (34). Sabin-derived type 2 strains circulated in Egypt during 1982 to 1993 and were responsible for at least 32 poliomyelitis cases (6). More recently, an outbreak of type 1 poliomyelitis in 2000 to 2001 in the island of Hispaniola (Dominican Republic and Haiti), free of wild poliovirus since 1989, has been related to evolved Sabin 1 strains (7). Maintenance of high levels of polio immunization and surveillance of the paralytic disease is essential before global eradication of wild poliovirus is achieved. It will then be necessary to carefully devise the best strategies to stop polio immunization in order to minimize the risks of a polio comeback (61).
Acknowledgments
We thank Geoffrey Schild, Andy Macadam, and Glynis Dunn for helpful advice. Sophie Reid, Ghazi Auda, Robin Hull, and Gary Beaven are also acknowledged for their contributions.
REFERENCES
- 1.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and, A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottiger, M., S. Gard, and R. Lagercrantz. 1966. Vaccination with attenuated type I poliovirus, the Chat strain. I. A study of 20 families. Acta Paediatr. Scand. 55:405-415. [DOI] [PubMed] [Google Scholar]
- 3.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulger, L. R., S. C. Arya, P. Ahourai, and S. A. Marsden. 1978. International comparison of species of monkey used for the neurovirulence test for oral poliomyelitis vaccine. J. Biol. Stand. 6:233-242. [DOI] [PubMed] [Google Scholar]
- 5.Cabasso, V. J., G. A. Jervis, A. W. Moyer, M. Roca-Garcia, E. V. Orsi, and H. R. Cox. 1959. Cumulative testing experience with consecutive lots of oral poliomyelitis vaccine, p. 102-134. In Live poliovirus vaccines. First International Conference on Live Poliovirus Vaccines. Scientific publication no. 44. Pan American Sanitary Bureau, Washington, D.C.
- 6.Centers for Disease Control and Prevention. 2001. Circulation of a type 2 vaccine-derived poliovirus--Egypt, 1982-1993. JAMA 285:1148-1149. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2001. Outbreak of poliomyelitis—Dominican Republic and Haiti, 2000-2001. JAMA 285:1438.. [PubMed] [Google Scholar]
- 8.Christodoulou, C., F. Colbere-Garapin, A. Macadam, L. F. Taffs, S. Marsden, P. Minor, and F. Horaud. 1990. Mapping of mutations associated with neurovirulence in monkeys infected with Sabin 1 poliovirus revertants selected at high temperature. J. Virol. 64:4922-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colston, E. M., and V. R. Racaniello. 1995. Poliovirus variants selected on mutant receptor-expressing cells identify capsid residues that expand receptor recognition. J. Virol. 69:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couderc, T., F. Delpeyroux, H. Le Blay, and B. Blondel. 1996. Mouse adaptation determinants of poliovirus type 1 enhance viral uncoating. J. Virol. 70:305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couderc, T., N. Guedo, V. Calvez, I. Pelletier, J. Hogle, F. Colbere-Garapin, and B. Blondel. 1994. Substitutions in the capsids of poliovirus mutants selected in human neuroblastoma cells confer on the Mahoney type 1 strain a phenotype neurovirulent in mice. J. Virol. 68:8386-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and, F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowdle, W. R., D. A. Featherstone, M. E. Birmingham, H. F. Hull, and R. B. Aylward. 1999. Poliomyelitis eradication. Virus Res. 62:185-192. [DOI] [PubMed] [Google Scholar]
- 14.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgescu, M. M., J. Balanant, A. Macadam, D. Otelea, M. Combiescu, A. A. Combiescu, R. Crainic, and F. Delpeyroux. 1997. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 71:7758-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgescu, M. M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and, R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, E. E. A. 1972. Polioviruses, p. 155-176. In S. A. Plotkin (ed.), Strains of human viruses. S. Karger AG, Basel, Switzerland.
- 20.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 22.Journal of Biological Standardization. 1981. Markers of poliovirus strains isolated from cases temporally associated with the use of live poliovirus vaccine: report on a W. H. O. collaborative study. J. Biol. Stand. 9:163-184. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kew, O. M., M. N. Mulders, G. Y. Lipskaya, E. E. da Silva, and M. A. Pallansch. 1995. Molecular epidemiology of polioviruses. Semin. Virol. 6:401-414. [Google Scholar]
- 25.Kew, O. M., R. W. Sutter, B. K. Nottay, M. J. McDonough, D. R. Prevots, L. Quick, and M. A. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohara, M., T. Omata, A. Kameda, B. L. Semler, H. Itoh, E. Wimmer, and A. Nomoto. 1985. In vitro phenotypic markers of a poliovirus recombinant constructed from infectious cDNA clones of the neurovirulent Mahoney strain and the attenuated Sabin 1 strain. J. Virol. 53:786-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koprowski, H. 1958. Vaccination with modified active viruses, p. 112-123. In Poliomyelitis. Papers and discussion presented at the Fourth International Poliomyelitis Conference. J. B. Lippincott Company, Philadelphia, Pa.
- 28.Lee, C. K., and E. Wimmer. 1988. Proteolytic processing of poliovirus polyprotein: elimination of 2Apro-mediated, alternative cleavage of polypeptide 3CD by in vitro mutagenesis. Virology 166:405-414. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., L. B. Zhang, T. Yoneyama, H. Yoshida, H. Shimizu, K. Yoshii, M. Hara, T. Nomura, H. Yoshikura, T. Miyamura, and A. Hagiwara. 1996. Genetic basis of the neurovirulence of type 1 polioviruses isolated from vaccine-associated paralytic patients. Arch. Virol. 141:1047-1054. [DOI] [PubMed] [Google Scholar]
- 30.Lu, H. H., C. F. Yang, A. D. Murdin, M. H. Klein, J. J. Harber, O. M. Kew, and E. Wimmer. 1994. Mouse neurovirulence determinants of poliovirus type 1 strain LS-a map to the coding regions of capsid protein VP1 and proteinase 2Apro. J. Virol. 68:7507-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahon, B. P., K. Katrak, and K. H. Mills. 1992. Antigenic sequences of poliovirus recognized by T cells: serotype-specific epitopes on VP1 and VP3 and cross-reactive epitopes on VP4 defined by using CD4+ T-cell clones. J. Virol. 66:7012-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, A., C. Wychowski, T. Couderc, R. Crainic, J. Hogle, and M. Girard. 1988. Engineering a poliovirus type 2 antigenic site on a type 1 capsid results in a chimaeric virus which is neurovirulent for mice. EMBO J. 7:2839-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, J., G. L. Ferguson, D. J. Wood, and P. D. Minor. 2000. The vaccine origin of the 1968 epidemic of type 3 poliomyelitis in Poland. Virology 278:42-49. [DOI] [PubMed] [Google Scholar]
- 35.McGoldrick, A., A. J. Macadam, G. Dunn, A. Rowe, J. Burlison, P. D. Minor, J. Meredith, D. J. Evans, and J. W. Almond. 1995. Role of mutations G-480 and C-6203 in the attenuation phenotype of Sabin type 1 poliovirus. J. Virol. 69:7601-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melnick, J. L., and J. C. Brennan. 1959. Monkey neurovirulence of attenuated poliovirus vaccines being used in field trials, p. 65-100. In Live poliovirus vaccines. First International Conference on Live Poliovirus Vaccines. Scientific publication no. 44. Pan American Sanitary Bureau, Washington, D.C.
- 37.Minor, P. D. 1990. Antigenic structure of picornaviruses. Curr. Top. Microbiol. Immunol. 161:121-154. [DOI] [PubMed] [Google Scholar]
- 38.Minor, P. D. 1980. Comparative biochemical studies of type 3 poliovirus. J. Virol. 34:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minor, P. D. 1992. The molecular biology of poliovaccines. J. Gen Virol. 73:3065-3077. [DOI] [PubMed] [Google Scholar]
- 40.Minor, P. D. 1997. Poliovirus, p. 555-577. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 41.Minor, P. D., and G. Dunn. 1988. The effect of sequences in the 5′ non-coding region on the replication of polioviruses in the human gut. J. Gen. Virol. 69:1091-1096. [DOI] [PubMed] [Google Scholar]
- 42.Minor, P. D., A. J. Macadam, D. M. Stone, and J. W. Almond. 1993. Genetic basis of attenuation of the Sabin oral poliovirus vaccines. Biologicals 21:357-363. [DOI] [PubMed] [Google Scholar]
- 43.Moss, E. G., and V. R. Racaniello. 1991. Host range determinants located on the interior of the poliovirus capsid. EMBO J. 10:1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosser, A. G., J. Y. Sgro, and R. R. Rueckert. 1994. Distribution of drug resistance mutations in type 3 poliovirus identifies three regions involved in uncoating functions. J. Virol. 68:8193-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray, M. G., J. Bradley, X. F. Yang, E. Wimmer, E. G. Moss, and V. R. Racaniello. 1988. Poliovirus host range is determined by a short amino acid sequence in neutralization antigenic site I. Science 241:213-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray, R., R. Kirchstein, G. Van Hoosier Jr., and S. Baron. 1959. Comparative virulence for rhesus monkeys of poliovirus strains used for oral administration, p. 39-64. In Live poliovirus vaccines. First International Conference on Live Poliovirus Vaccines. Scientific publication no. 44. Pan American Sanitary Bureau, Washington, D.C.
- 47.Nathanson, N., and A. D. Langmuir. 1995. The Cutter incident. Poliomyelitis following formaldehyde-inactivated poliovirus vaccination in the United States during the spring of 1955. II. Relationship of poliomyelitis to Cutter vaccine. 1963. Am. J. Epidemiol. 142:109-140. [DOI] [PubMed] [Google Scholar]
- 48.Nomoto, A., T. Omata, H. Toyoda, S. Kuge, H. Horie, Y. Kataoka, Y. Genba, Y. Nakano, and N. Imura. 1982. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc. Natl. Acad. Sci. USA 79:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omata, T., M. Kohara, S. Kuge, T. Komatsu, S. Abe, B. L. Semler, A. Kameda, H. Itoh, M. Arita, E. Wimmer, et al. 1986. Genetic analysis of the attenuation phenotype of poliovirus type 1. J. Virol. 58:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Payne, A. M. 1960. Oral immunization against poliomyelitis. Bull. W. H. O. 23:695-703. [PMC free article] [PubMed] [Google Scholar]
- 51.Plotkin, S. A., A. Lebrun, G. Courtois, and H. Koprowski. 1960. Vaccination with the CHAT strain of type 1 attenuated poliomyelitis virus in Leopoldville, Belgian Congo. III. Safety and efficacy during the first twenty-one months of study, p. 466-473. In Live poliovirus vaccines. First International Conference on Live Poliovirus Vaccines II. Scientific publication no. 50. Pan American Sanitary Bureau, Washington, D.C.
- 52.Racaniello, V. R., and D. Baltimore. 1981. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc. Natl. Acad. Sci. USA 78:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabin, A. B., and L. R. Boulger. 1973. History of Sabin attenuated poliovirus oral live vaccines. J. Biol. Stand. 1:115-118. [Google Scholar]
- 54.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 55.Stanway, G., P. J. Hughes, R. C. Mountford, P. Reeve, P. D. Minor, G. C. Schild, and J. W. Almond. 1984. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc. Natl. Acad. Sci. USA 81:1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strebel, P. M., R. W. Sutter, S. L. Cochi, R. J. Biellik, E. W. Brink, O. M. Kew, M. A. Pallansch, W. A. Orenstein, and A. R. Hinman. 1992. Epidemiology of poliomyelitis in the United States one decade after the last reported case of indigenous wild virus-associated disease. Clin. Infect. Dis. 14:568-579. [DOI] [PubMed] [Google Scholar]
- 57.Tardy-Panit, M., B. Blondel, A. Martin, F. Tekaia, F. Horaud, and F. Delpeyroux. 1993. A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J. Virol. 67:4630-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.WHO Consultative Group. 1982. The relation between acute persisting spinal paralysis and poliomyelitis vaccine--results of a ten-year enquiry. Bull. W. H. O. 60:231-242. [PMC free article] [PubMed] [Google Scholar]
- 60.Wien, M. W., S. Curry, D. J. Filman, and J. M. Hogle. 1997. Structural studies of poliovirus mutants that overcome receptor defects. Nat. Struct. Biol. 4:666-674. [DOI] [PubMed] [Google Scholar]
- 61.Wood, D. J., R. W. Sutter, and W. R. Dowdle. 2000. Stopping poliovirus vaccination after eradication: issues and challenges. Bull. W. H. O. 78:347-357. [PMC free article] [PubMed] [Google Scholar]
- 62.World Health Organization. 1983. Requirements for poliomyelitis vaccine (oral). W. H. O. Tech. Rep. Ser. 687:107-175. [Google Scholar]
- 63.Yeates, T. O., D. H. Jacobson, A. Martin, C. Wychowski, M. Girard, D. J. Filman, and J. M. Hogle. 1991. Three-dimensional structure of a mouse-adapted type 2/type 1 poliovirus chimera. EMBO J. 10:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]