Abstract
A key bottleneck in RNA structural studies is preparing milligram quantities of RNA, and current techniques have changed little in over a decade. To address this, we have developed an affinity tag-based purification method of RNA oligonucleotides. The tag is attached to the 3′-end of almost any desired RNA sequence, allowing for the rapid and specific removal of the RNA of interest directly from in vitro transcription reactions using an affinity column to which a specific RNA-binding protein has been attached. Following a wash, the RNA of interest is eluted by the addition of imidazole to the column, activating a mutant HδV ribozyme incorporated into the tag. The affinity column can then be rapidly regenerated using conditions that release the protein–RNA tag interaction without denaturing the protein. To demonstrate that this method rapidly generates high-quality RNA, we have transcribed, purified, and generated diffraction-quality crystals of a mutant form of the Tetrahymena thermophila P4–P6 domain in a 48-h time period.
Keywords: RNA purification, affinity tag, X-ray crystallography, NMR spectroscopy, structural genomics
INTRODUCTION
Discoveries of RNA interference (RNAi), small regulatory RNAs, and cis-acting RNA control elements highlight the central role RNA plays in gene expression. Furthermore, in the biotechnology sector RNA remains a focus for therapeutic design, including a new generation of antibiotics that bind the ribosomal RNA, and antiviral agents that target human immunodeficiency virus (HIV) and hepatitis C virus (HCV) RNAs. To understand and to therapeutically exploit these diverse RNAs, we require a much deeper knowledge of RNA structure. Of particular importance are new tools to aid in the synthesis and purification of large quantities of RNA, as this remains a significant bottleneck in many structural and biophysical studies (Doudna 2000).
The most common means of synthesizing RNA is by T7 RNA polymerase-catalyzed in vitro run-off transcription of a DNA template (Milligan et al. 1987; Doudna 1997). Although T7 RNA polymerase tends to add extra nucleotides to the 3′-end of the desired RNA (Milligan et al. 1987; Draper et al. 1988; Pleiss et al. 1998), this problem has been largely overcome through the use of cis-acting ribozymes at the 5′- and 3′-ends of the RNA of interest (Price et al. 1995; Ferre-D’Amare and Doudna 1996) or through the use of synthesized, partially 2′-O-methyl-modified DNA templates (Kao et al. 1999). The transcription product RNAs are purified by preparative denaturing polyacrylamide gel electrophoresis, eluted from the gel matrix, concentrated, and refolded. Using this denaturing method, synthesis and purification of structural quantities of a single RNA sample (10–20 mg) typically requires >1 wk and thus is not well suited to high throughput. For many RNAs, significant time is spent optimizing refolding conditions to minimize unproductive conformations. Some well-known RNAs, such as Escherichia coli tRNAPhe, cannot be refolded into a conformationally homogeneous and active population (Uhlenbeck 1995). In some cases, this is overcome by a native purification technique, usually involving a combination of anion exchange and gel filtration chromatography. Other RNA purification procedures have been developed, including those based on HPLC (Anderson et al. 1996; Shields et al. 1999).
We present a novel protocol for the purification of RNA under nondenaturing conditions using a new RNA affinity tag that addresses many of the above problems and is completely generalized for the production of any desired RNA (Fig. 1 ▶). This technique is rapid, allows for parallel purification of multiple RNA samples, can be used with any size or sequence of RNA, and applies to both small (<1 mL) and large-scale (<10 mL) transcription reactions. The affinity tag contains two elements: a variant of the hepatitis delta virus (HδV) ribozyme that is activated by imidazole and a hairpin loop from a thermostable SRP RNA that forms a high-affinity and kinetically stable complex with the Thermotoga maritima Ffh-M domain protein. The tag is incorporated on the 3′-end of the target RNA during transcription. The target RNA-tag chimera is retained on an affinity column to which the partner protein has been attached, whereas incomplete abortive transcripts, nucleotides, DNA template, and other reaction components pass through. The target RNA is eluted by adding imidazole, which activates the ribozyme and liberates the RNA of interest. To demonstrate the utility of this procedure, we purified a mutant version of the P4–P6 domain of the Tetrahymena thermophila group I intron and readily obtained diffraction-quality crystals.
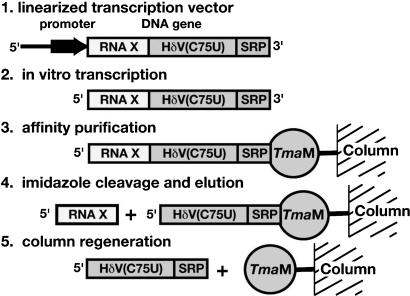
FIGURE 1.
The general scheme for the native purification of the desired sequence (RNA X) using a two-domain affinity tag.
RESULTS
Design of the affinity tag and matrix
We designed a two-domain affinity tag based on a hepatitis delta virus (HδV) ribozyme domain that is activated by imidazole and a well-characterized RNA–protein interaction (Fig. 1 ▶). The HδV ribozyme cleaves at its 5′-end and has no sequence requirements upstream of its cleavage site. For this use, the HδV sequence contains a C75U mutation that inactivates the ribozyme during the transcription reaction, but allows for the affinity tag’s removal during the purification protocol (Perrotta et al. 1999; Nishikawa et al. 2002). This mutant ribozyme is therefore analogous to the intein protein purification tag, which uses a DTT-activated intein to effect simultaneous affinity purification and tag removal (Chong et al. 1998). The second tag domain consists of tandem stem–loop motifs from the T. maritima SRP RNA that specifically and tightly binds the SRP protein, Ffh, which has been chosen for several reasons. First, this binding interaction is both thermodynamically robust and kinetically inert on the time scales of the purification procedure. The placement of two protein-binding sites in the tag enhances the ability of the RNA to remain bound to an affinity column while keeping the tag portion of the RNA transcript a reasonable length. Second, the interaction of this RNA with its cognate protein is highly dependent on both pH and metal ion concentration (Batey and Doudna 2002); therefore, the binding can be modulated with these two parameters. These two domains have been incorporated into a high-copy plasmid vector (Fig. 2A–C ▶) that allows for placement of the tag immediately downstream from any RNA sequence of interest.
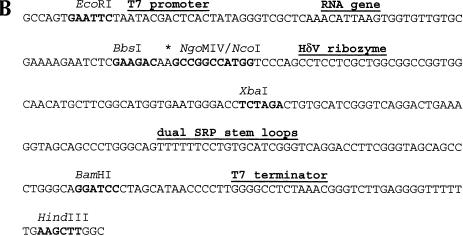
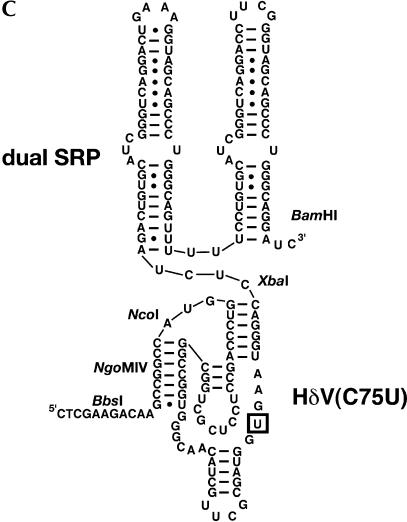
FIGURE 2.
(A) Sequence of the cloning region and affinity tag in pRAV4 (RAV = RNA Affinity Vector). The asterisk denotes the location of the boundary between the RNA of interest and the HδV ribozyme. All unique restriction sites have been denoted in boldface in the vector sequence, and the various functional regions of the vector have been labeled. (B) Sequence of the cloning region and tag of pRAV12. (C) Secondary structure of the RNA affinity tag; the sequence is that of pRAV12. The location of the C75U mutation is boxed.
To create a chromatographic affinity matrix capable of specifically binding the above affinity tag, we coupled the T. maritima SRP Ffh M-domain protein (referred to as TmaM) to an Affigel-10 matrix. This activated chromatographic media contains N-hydroxysuccinamide ester-linked agarose, allowing covalent coupling of proteins through lysine residues. Previously, this resin has been used to covalently couple the MS2 coat protein to create affinity beads for specific RNAs (Bardwell and Wickens 1990). For this application, we have chosen TmaM because unlike many other RNA-binding proteins, the protein’s RNA-binding surface lacks lysine residues. The protein–RNA complex is also readily disrupted under nondenaturing conditions, allowing gentle regeneration of the affinity matrix. TmaM can be expressed in E. coli and purified in large quantities (~70 mg/L culture) with a straightforward purification protocol (Fig. 3 ▶), and ~15 mg of protein can be coupled to 1 mL of resin (corresponding to 1 μmole of potential RNA-binding sites per milliliter of resin) using established methods (Prickett et al. 1989; Bardwell and Wickens 1990).
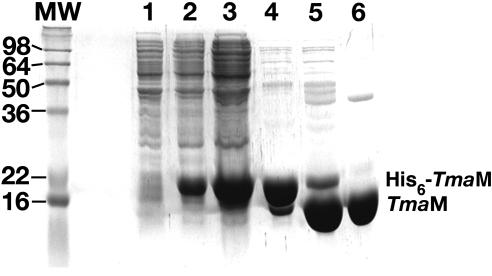
FIGURE 3.
Purification of the T. maritima Ffh M domain (TmaM) as analyzed by a 15% SDS-PAGE gel. (Lane 1) Cells prior to induction with 1 mM IPTG; (lane 2) cells after induction with 1 mM IPTG; (lane 3) supernatant fraction of the cell lysate; (lane 4) fraction of protein eluted from the Ni2+-affinity column; (lane 5) protein following cleavage with TEV protease; (lane 6) peak fraction containing TmaM from the SP-Sepharose column. The major band in each lane (except for lane 1) is TmaM.
Demonstration of the purification scheme
To test the purification scheme, we constructed a plasmid containing a 49-nt sequence from the plautia stali intestinal virus (PSIV) RNA (Sasaki and Nakashima 1999; pRAV4; Fig. 2A ▶). We performed a small (100 μL) 2-h transcription, radioactively labeling the RNA during the reaction. The transcription reaction was diluted with loading buffer, loaded directly onto M-domain affinity matrix, and washed (see Materials and Methods for buffer components). The product RNA was liberated from the column by adding imidazole-containing buffer, incubated for 2 h, and collected by draining the column. Fractions (one column volume each) were desalted and analyzed on a denaturing polyacrylamide gel (Fig. 4 ▶).
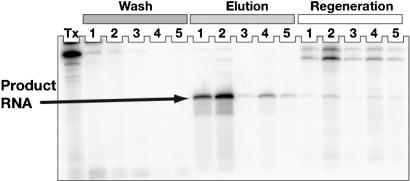
FIGURE 4.
Test purification of RNA transcribed from the linearized pRAV4 vector. The RNA was body-labeled using [α-32P]GTP during transcription. An aliquot of the raw transcription reaction is shown on the left, and wash/elution/regeneration fractions are shown. The pure product RNA is indicated.
Comparison of the raw transcription reaction with the wash fractions reveals almost quantitative uptake of the tagged RNA, and virtually no leaking of tagged RNA from the affinity column. Upon addition of imidazole, the RNA sequence of interest is released. This RNA is virtually the only species liberated from the column (along with minor contaminants, as seen on Fig. 4 ▶), as the uncleaved product and cleaved tag are retained on the column until treated with the regeneration buffer. Transcription and purification of the RNA shown in Figure 4 ▶ required <5 h.
Crystallization of affinity-purified T. thermophila P4–P6 domain
To demonstrate that this method generates high-quality RNA, we purified the ΔC209 mutant of the T. thermophila group I intron P4–P6 domain using our affinity tag and crystallized it. This RNA readily crystallizes under a broad range of conditions. These crystals diffract synchrotron X-ray radiation to 2.2 Å resolution (Juneau et al. 2001). We purified P4–P6 domain RNA from a 10-mL transcription reaction and then concurrently concentrated the RNA and exchanged the buffer in a centrifugal filter device; at no point was the RNA denatured. Single crystals grew in previously reported conditions (Juneau et al. 2001; Fig. 5A ▶) as well as in condition #5 of a commercially available sparse matrix screening kit (Scott et al. 1995). These crystals diffract to ~2.8 Å resolution using a rotating anode home X-ray source (I/σ = 2.1 for the 2.93–2.80-Å resolution bin; Fig. 5B ▶). The space group is P212121 with unit cell dimensions of a = 75.4 Å, b = 125.8 Å, and c = 145.5 Å, values very close to those reported (Juneau et al. 2001). Furthermore, the mosaicity of these crystals is 0.45° on the home source, which is as good, if not better, than crystals of the same RNA purified using traditional techniques (E. Podell, pers. comm.).
FIGURE 5.
(A) Crystals of the T. thermophila ΔC209P4–P6 domain RNA that was transcribed and purified using the affinity-tag protocol. (B) Diffraction pattern of crystals showing clear peaks extending to at least 2.87 Å resolution.
The ability of the affinity-purified RNA to readily crystallize demonstrates several points. First, the time period between the initiation of the in vitro transcription reaction and the first observation of single crystals was 48 h. Thus, the rapid purification did not interfere with the ability of the RNA to crystallize. Second, the lack of a reannealing step did not yield significant quantities of RNA trapped in a non-native conformation. The RNA folded correctly during the transcription reaction, and subsequent purification and concentration prior to crystallization did not change this. Third, despite the fact that well over 50% of the total transcribed RNA was discarded during the purification as the cleaved HδV/SRP tag, the yields of RNA per milliliter of transcription are comparable with traditional gel purification. In fact, preparative polyacrylamide gel electrophoresis routinely results in ~50% loss of target RNA (R.T. Batey and J.S. Kieft, unpubl.).
DISCUSSION
In this work, we describe a nondenaturing method using an imidazole-activated HδV ribozyme coupled to a specific protein–RNA complex to rapidly purify RNA of sufficient quality to crystallize. Although this technique is capable of generating RNA faster and more cheaply than current methods, we designed the system with sufficient flexibility for a diverse set of needs. The pRAV plasmids are completely modular with unique restriction sites defining each segment of the tag (Fig. 2A ▶). Thus, besides cloning RNAs of interest, end users can easily make design changes that suit their particular applications.
Sequence requirements in the RNA of interest
The major advantage of this system is the tremendous flexibility to purify almost any RNA of interest. There are, however, a few RNA sequence design considerations when using this technique. There is the standard requirement for T7 RNA polymerase initiation with a guanine residue. This is circumvented in two ways. First, the use of a 5′-hammer-head ribozyme (vida infra) completely eliminates this requirement. Second, the alternative P25 class T7 promoter uses an adenosine residue at the 5′-end (Huang et al. 2000). Another design requirement lies at the 3′-end of the RNA of interest, at its boundary with the HδV ribozyme. Although the HδV ribozyme has no sequence requirements in the substrate strand, it appears to require at least one unpaired nucleotide at the 3′-end of the substrate for efficient cleavage. Thus, the design of RNAs in which the 3′-end is completely involved in secondary structure will lead to constructs with greatly reduced cleavage rates.
The use of other affinity interactions
In theory, any affinity tag could be used with this protocol, including commercially available matrices. We explored two other affinity tags: a 15-nt poly(A) tag that binds poly(dT) resin and a three-tandem repeat Sephadex G-100 aptamer (Srisawat and Engelke 2001; Srisawat et al. 2001). Both contained the HδV C75U ribozyme 5′ of the affinity tag. The poly(A) tag bound poorly to the column, with unacceptably high amounts of the transcribed material passing through the matrix (data not shown). The Sephadex aptamer tag slowly released from the column during the wash and elution steps, leading to contamination of the target RNA with precursor and tag (data not shown). Future isolation of aptamers with more favorable affinities and interaction kinetics may lead to new tags, but currently the use of highly specific RNA–protein interactions, such as the one described here, seems most appropriate. The commonly used U1A and MS2 coat protein–RNA interactions could be used in place of the TmaM–RNA interaction, with the appropriate RNA element placed between the XbaI and BamHI sites (Fig. 2B ▶). This capability further generalizes the method to RNAs whose purification is incompatible with the TmaM–SRP RNA interaction (e.g., SRP RNAs).
Processing at the 5′-end
A common method in RNA transcription is to use a hammerhead ribozyme at the 5′-end of the transcript. This provides several distinct benefits: chemically homogeneity at the 5′-terminus of the desired product, the use of a strong initiation sequence at the 5′-end of the transcript, and the lack of sequence requirements at the 5′-end of the product RNA. To simplify the development of a working affinity tag, we did not include this feature in our system. However, this method should accommodate a 5′-hammerhead ribozyme. As long as the number of base pairs between the hammerhead and the product RNA is kept to a minimum (3–4 bp), the cleaved hammerhead ribozyme product should dissociate from the product during transcription and subsequently be lost during the wash. Because both the transcription and wash buffers contain magnesium, the hammerhead ribozyme should completely cleave prior to the imidazole incubation step.
Other small-molecule-activated ribozymes
A key feature of this method is the use of a mutant ribozyme that is activated by imidazole. One potential drawback is that imidazole can facilitate the general base-catalyzed hydrolysis of the RNA backbone during prolonged incubations (8–12 h) at 37°C. At 4°C, the imidazole-induced cleavage rates are too slow to be useful (data not shown). Therefore, it may be desirable to use ribozymes activated by other small molecule compounds (Soukup and Breaker 1999). An example of this is the theophylline-activated ribozyme developed by Soukup and coworkers in which the activity of the ribozyme is allosterically controlled through an aptamer (Kertsburg and Soukup 2002). New ribozymes capable of using other small molecules with shorter incubation times, or that cleave efficiently at lower temperatures, will further increase the speed and utility of this technique. In our experience, however, relatively short incubation times (2–4 h at 20°C–37°C) do not cause significant amounts of damage to the RNA.
Conclusions
We present a rapid method for the purification of any given RNA sequence under native conditions. Using this technique, the P4–P6 domain of the T. thermophila group I intron was purified to a sufficient level of homogeneity such that we were able to achieve diffraction-quality crystals. This protocol takes advantage of the use of a removable affinity tag and a reusable affinity matrix, similar to the systems routinely used in protein purification. We believe that this method represents a major advance in the ability to purify large quantities of RNA for structural biology and should also be applicable to a broad range of biochemical applications.
MATERIALS AND METHODS
Expression and purification of T. maritima M domain protein
A domain of the T. maritima Ffh protein (TmaM) corresponding to amino acids 295–423 was cloned from genomic DNA (ATCC 43589) and inserted between the NcoI and BamHI sites of pET15b (Novagen) using standard cloning techniques (Sambrook and Russell 2001). Expression of the TmaM domain was performed by transforming the E. coli strain Rosetta(DE3)/pLysS (Novagen) with pTmaM4. These cells were grown in LB medium in eight 750-mL cultures at 37°C to an absorbance (600 nm) of 0.7–0.8, and expression was induced by the addition of 1 mM IPTG. The cultures were allowed to continue to grow for an additional 4–5 h prior to harvesting by centrifugation. The cell pellets were immediately resuspended in 25 mL Lysis Buffer (300 mM NaCl, 50 mM Tris-HCl at pH 8.0). Cell lysis was performed by three rounds of freeze/thaw in which the cells were frozen in liquid nitrogen and thawed to room temperature. The viscosity of the lysate was reduced by the addition of 20 units of DNase per liter of cell growth (Boehringer Mannheim), 10 mM MgCl2, and 10 mM CaCl2, and incubated at 37°C. The cell lysate was clarified by centrifugation at 30,000g for 30 min at 4°C and the supernatant subjected to further purification.
TmaM domain was initially purified by passing the clarified lysate through a gravity column containing 20 mL of Ni2+-NTA affinity resin (QIAGEN). Following extensive washing with 300 mL of Wash Buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole at pH 8.0), the protein was eluted with Elution Buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole at pH 8.0). Fractions containing the protein were pooled and cleaved with a 1:100 ratio (by mass) of TEV protease:TmaM domain overnight at room temperature (Lucast et al. 2001). It should be noted that the removal of the hexahistidine tag by TEV protease is not likely to be necessary for the successful application of TmaM in this methodology, but we have not specifically tested this. The protein was exchanged into a buffer containing 100 mM NaCl, 10 mM NaMES (pH 6.0) by dialysis in 6–8 kD dialysis membrane and subsequently applied to an SP-Sepharose column. Protein was eluted using a 0.1–1.5 M gradient of NaCl over a 300-mL volume; the protein eluted around 0.55 M NaCl. Fractions containing the protein were pooled and dialyzed into 50 mM K+-HEPES (pH 7.5). The concentration of the protein was assessed by absorbance at 280 nm using an extinction coefficient of 1615 M−1 cm−1 and a molecular weight of 14,975 g/mole. The final yield of protein was 70 mg/L of culture.
Preparation of TmaM4 affinity matrix
TmaM4 was covalently coupled to an activated support, Affigel-10 (BioRad), according to the protocol supplied. In this, 25 mL of beads was washed with 250 mL of ice-cold ddH2O (18 mΩ water; Milli-Q) by vacuum filtration without allowing the beads to completely dry out during the procedure. The beads were then added to 50 mL of a 550 μM protein solution and allowed to incubate for 2 h at 4°C and for 5 h at room temperature with gentle agitation. After coupling, the supernatant containing unreacted protein was removed by placing the slurry in a 20 × 2.5-cm Econo-column (BioRad). The coupled resin was washed twice with 50-mL aliquots of 50 mM K+-HEPES (pH 7.5) followed by 50 mL of 50 mM Tris-HCl (pH 8.0). To block unreacted N-hydroxysuccinamide groups, the column was allowed to incubate overnight in Tris buffer at 4°C. The resin was finally washed and stored in a buffer containing 200 mM NaCl, 10 mM MgCl2, 50 mM Tris-HCl (pH 8.0), and 0.1% Na-azide and stored at 4°C. To test whether the chromatographic media contained residual RNase activity, 200 μL of resin was incubated for 48 h with an RNA at 25°C and the integrity of the RNA was assayed on a denaturing polyacrylamide gel. No significant degradation of the RNA was observed, indicating that the protein preparation was of sufficient quality to yield a chromatographic resin devoid of contaminants that would interfere with the purification protocol.
Construction of the RNA affinity tag vector
Standard PCR and cloning strategies were used to create a DNA insert that contains a T7 RNA polymerase promoter, a 49-nt insert (nucleotides 6157–6195) of the plautia stali intestinal virus IRES RNA, the C75U mutant genomic HδV ribozyme, two T. maritima SRP RNA stem–loops, and a T7 terminator (Fig. 2A ▶). This plasmid, referred to as pRAV4 (RAV for RNA Affinity Vector), was used in the test purification of Figure 4 ▶ and as the basis for further optimization and modification. pRAV4 was subsequently changed to include three Watson-Crick base pairs to the second SRP stem–loop to stabilize the terminal helix and NgoMIV and NcoI restriction sites within the HδV ribozyme (Walker et al. 2003) to facilitate cloning (Fig. 2B ▶) and is referred to as pRAV12.
In vitro transcription of RNA
RNA was transcribed in vitro from linearized plasmid DNA or directly from PCR products using established protocols (Doudna 1997). For reactions from plasmid DNA, the plasmid was linearized with BamHI and used in in vitro transcription reactions at a final concentration of 75 μg/mL. For reactions from PCR products, the reactions were prepared using the QIAGEN PCR cleanup kit. Reactions consisted of 30 mM Tris-HCl (pH 8.0), 10 mM DTT, 0.1% Triton X-100, 0.1 mM spermidine-HCl, 8 mM each NTP (Sigma; pH adjusted to 8.0), 40 mM MgCl2, 50 μg/mL T7 RNA polymerase, 1 unit/mL inorganic pyrophosphatase (Sigma), and template DNA at 75 μg/mL. Reactions were incubated for 1.5 to 2 h (or as indicated in the figures) at 37°C.
Insertion of the ΔC209 variant of the T. thermophila group I intron P4–P6 domain into the affinity vector
A gene corresponding to the (Δ209)P4–P6 domain was cloned using a nested PCR strategy. The gene was amplified with two inner primers (5′-primer, TAATACGACTCACTATAGGAATT GCGGGAAAGGGGT; 3′-primer, CGGGCGGAAGACGCGCCCT GAACTGCATCCATATCA) and two outer primers (5′-primer, GCGCGCGAATTCTAATACGACTCACTATAG; 3′-primer, CCG CGGGCGGAAGACGCGCCC). The resulting product was restriction-digested with EcoRI and BbsI, ligated into pRAV4 digested with the same enzymes, and transformed into E. coli DH5α cells. Individual ampicillin-resistant colonies were picked and screened for the presence of the mutant P4–P6 insert. A single isolate containing the proper insert, pR4P4–P6, was subsequently prepared from 1.5 L of culture to obtain sufficient material for large-scale in vitro transcription reactions. Purified vector was linearized with BamHI, extracted twice with an equal volume of 25:24:1 phenol (pH 8.1):chloroform:isoamyl alcohol, ethanol-precipitated, and brought up in sufficient 1× T.E. buffer to yield a working stock of ~1 mg/mL.
Transcription and purification of the T. thermophila group I intron P4–P6 domain
The (ΔC209)P4–P6-affinity tag fusion was transcribed using standard conditions (see above) in a 10-mL reaction mixture for 1.5 h at 37°C. Following the completion of transcription, 25 mL of Column Wash Buffer (25 mM Tris-HCl at pH 8.0, 250 mM NaCl, 10 mM MgCl2) was added to the reaction and applied to 15 mL of affinity resin in a 20 × 2.5-cm glass column (Econo-column; Bio-Rad).
Binding of the RNA to the column was effected by passing the diluted transcription reaction through the column four times with a flow rate of ~2.0 mL/min at room temperature. Analysis of the flow-through from each passage through the column by gel electrophoresis revealed that the RNA had quantitatively bound to the column after the second time. Following the last reapplication of the flow-through, the column was washed with five 30-mL aliquots of Wash Buffer at a flow rate of 3.5 mL/min. After the last wash, 20 mL of Wash Buffer plus 200 mM imidazole (pH 8.0; Cleavage Buffer) was added and allowed to pass through the column. At this point, the column was stopped, and another 20-mL aliquot of Cleavage Buffer was added and allowed to incubate for 2 h at 37°C to facilitate removal of the P4–P6 domain RNA from the affinity tag. RNA product was recovered by opening the column and collecting five 20-mL aliquots; the product appeared to be completely eluted by the second fraction, as judged by an ethidium bromide-stained 8% denaturing polyacrylamide gel. The chromatographic matrix was regenerated by removal of the cleaved and uncleaved affinity tag by applying five 50-mL aliquots of Regeneration Buffer (1.0 M LiCl, 25 mM Na2EDTA, 174 mM glacial acetic acid). Immediately following regeneration, the column was re-equilibrated in Wash Buffer plus 0.1% Na-azide and stored at 4°C. The quality of the RNA, as judged by an ethidium bromide-stained denaturing polyacrylamide gel, was >95% pure (of similar quality to that shown in Fig. 4 ▶), but some trace contaminants were observed. Although we did not further purify the RNA prior to crystallization, it could easily be further purified using gel filtration chromatography under native conditions to remove other RNA species.
Crystallization of the T. thermophila group I intron P4–P6 domain
To prepare the RNA for crystallization, the two elution fractions containing the product were pooled and concentrated using a centrifuge concentrator (Amicon; Ultra) with a 10,000 MWCO. After concentration down to a volume of 500 μL, the sample was exchanged into a buffer containing 10 mM NaCl, 25 mM MgCl2, and 5 mM K+-HEPES (pH 7.5), with three exchanges against 15 mL of buffer. The final concentration of the RNA stock used for crystallization trials was 5.0 mg/mL as determined using the calculated extinction coefficient based on the nucleotide sequence. This RNA was tested for crystallizability with a highly successful nucleic acid-oriented sparse matrix (Natrix; Hampton) using the hanging-drop method and mixing 2 μL of the RNA solution with 2 μL of the appropriate mother liquor and incubated at 20°C. Crystallization of the mutant P4–P6 domain was also achieved using the exact conditions described by Juneau et al. (2001), except that the RNA was not heat-annealed prior to setting up drops.
Collection of diffraction data
To assess their quality, the crystals were cryoprotected for 1 h in a buffer described by Juneau et al. (2001) and flash-frozen in liquid nitrogen. Data were collected on an R-AXIS IV++ instrument with CuKα X-ray radiation using a 0.5° oscillation angle and 5-min exposures. A 25° wedge of data was reduced with D*TREK (Pflugrath 1999).
Acknowledgments
The authors thank the RNA community at the University of Colorado Health Sciences Center and University of Colorado, Boulder for many stimulating discussions relating to this work and manuscript. In particular, we thank Art Pardi, Tom Cech, Mike Yarus, Norm Pace, Tom Blumenthal, and Judith Jaehning. We also thank Quentin Vicens and Fiona Jucker for critical comments on the manuscript. The plasmid bearing the ΔC209 mutant of P4–P6 was kindly donated by Elaine Podell and Tom Cech. The plasmid bearing the PSIV IRES sequence was kindly donated by Nobuhiko Nakashima. This work was made possible from a Research Innovation Award (R.T.B.), an HHMI Faculty Research Award (J.S.K.), and a generous donation by the Butcher family to the University of Colorado to launch the CU Genomics and Biotechnology Initiative (R.T.B. and J.S.K.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
HδV, hepatitis delta virus
SRP, signal recognition particle
RAV, RNA affinity vector
HCV, hepatitis C virus
IRES, internal ribosomal entry site
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7040604.
REFERENCES
- Anderson, A.C., Scaringe, S.A., Earp, B.E., and Frederick, C.A. 1996. HPLC purification of RNA for crystallography and NMR. RNA 2: 110–117. [PMC free article] [PubMed] [Google Scholar]
- Bardwell, V.J. and Wickens, M. 1990. Purification of RNA and RNA–protein complexes by an R17 coat protein affinity method. Nucleic Acids Res. 18: 6587–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T. and Doudna, J.A. 2002. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry 41: 11703–11710. [DOI] [PubMed] [Google Scholar]
- Chong, S., Montello, G.E., Zhang, A., Cantor, E.J., Liao, W., Xu, M.Q., and Benner, J. 1998. Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acids Res. 26: 5109–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. 1997. Preparation of homogeneous ribozyme RNA for crystallization. Methods Mol. Biol. 74: 365–370. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Structural genomics of RNA. Nat. Struct. Biol. 7 Suppl: 954–956. [DOI] [PubMed] [Google Scholar]
- Draper, D.E., White, S.A., and Kean, J.M. 1988. Preparation of specific ribosomal RNA fragments. Methods Enzymol. 164: 221–237. [DOI] [PubMed] [Google Scholar]
- Ferre-D’Amare, A.R. and Doudna, J.A. 1996. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 24: 977–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F., Bugg, C.W., and Yarus, M. 2000. RNA-catalyzed CoA, NAD, and FAD synthesis from phosphopantetheine, NMN, and FMN. Biochemistry 39: 15548–15555. [DOI] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., and Cech, T.R. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure (Camb) 9: 221–231. [DOI] [PubMed] [Google Scholar]
- Kao, C., Zheng, M., and Rudisser, S. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertsburg, A. and Soukup, G.A. 2002. A versatile communication module for controlling RNA folding and catalysis. Nucleic Acids Res. 30: 4599–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucast, L.J., Batey, R.T., and Doudna, J.A. 2001. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques 30: 544–550. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, F., Shirai, M., and Nishikawa, S. 2002. Site-specific modification of functional groups in genomic hepatitis delta virus (HDV) ribozyme. Eur. J. Biochem. 269: 5792–5803. [DOI] [PubMed] [Google Scholar]
- Perrotta, A.T., Shih, I., and Been, M.D. 1999. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science 286: 123–126. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J.W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55 (Pt 10): 1718–1725. [DOI] [PubMed] [Google Scholar]
- Pleiss, J.A., Derrick, M.L., and Uhlenbeck, O.C. 1998. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 4: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, S.R., Ito, N., Oubridge, C., Avis, J.M., and Nagai, K. 1995. Crystallization of RNA–protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 249: 398–408. [DOI] [PubMed] [Google Scholar]
- Prickett, K.S., Amberg, D.C., and Hopp, T.P. 1989. A calcium-dependent antibody for identification and purification of recombinant proteins. Biotechniques 7: 580–589. [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sasaki, J. and Nakashima, N. 1999. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 73: 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., Grenfell, R., Fogg, J., Smith, T., Gait, M.J., and Klug, A. 1995. Rapid crystallization of chemically synthesized hammerhead RNAs using a double screening procedure. J. Mol. Biol. 250: 327–332. [DOI] [PubMed] [Google Scholar]
- Shields, T.P., Mollova, E., Ste Marie, L., Hansen, M.R., and Pardi, A. 1999. High-performance liquid chromatography purification of homogeneous-length RNA produced by trans cleavage with a hammerhead ribozyme. RNA 5: 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, G.A. and Breaker, R.R. 1999. Engineering precision RNA molecular switches. Proc. Natl. Acad. Sci. 96: 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat, C. and Engelke, D.R. 2001. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA 7: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat, C., Goldstein, I.J., and Engelke, D.R. 2001. Sephadex-binding RNA ligands: Rapid affinity purification of RNA from complex RNA mixtures. Nucleic Acids Res. 29: E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. 1995. Keeping RNA happy. RNA 1: 4–6. [PMC free article] [PubMed] [Google Scholar]
- Walker, S.C., Avis, J.M., and Conn, G.L. 2003. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res. 31: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]