Abstract
Using a new combination of previously published techniques, we developed a method for quantitating modified nucleotides in RNAs. First, an RNA is cleaved with RNase H at the 5′ side of a nucleotide of interest. Next, 32P is substituted for the phosphate at the 5′ end of this nucleotide. Finally, after nuclease P1 digestion, the released radiolabeled nucleotide is analyzed by thin layer chromatography and quantitated by PhosphorImager. Using this method, we showed that the analysis of a pseudouridine at a specific site within an in vitro synthesized U2 RNA is indeed quantitative. We also applied this technique to cellular U2 RNA isolated from mouse liver, and showed that position U34 is ~90% pseudouridylated. This method, combined with previously described reverse transcription-based methods, constitutes a powerful tool for detecting and quantifying modified nucleotides in RNAs. With minor modifications, this method can serve as an effective assay to study RNA modifying enzymes.
Keywords: RNA modification, site-specific radiolabeling, U2 snRNA, pseudouridine
INTRODUCTION
A large number of cellular RNAs contain posttranscriptional modifications. Pseudouridylation, 2′-O-methylation, and base methylation are the most abundant internal modifications identified in all three types of stable RNAs, namely, tRNAs (Bjork 1995; Grosjean et al. 1995; Auffinger and Westhof 1998; Sprinzl et al. 1998; Hopper and Phizicky 2003), rRNAs (Maden 1990; Bachellerie and Cavaille 1998; Ofengand and Fournier 1998), and spliceosomal snRNAs (Reddy and Busch 1988; Massenet et al. 1998). In addition to these modifications, tRNAs contain a variety of other base modifications (Bjork 1995; Grosjean et al. 1995; Auffinger and Westhof 1998; Sprinzl et al. 1998; Hopper and Phizicky 2003). Although the function of some of these RNA modifications is known, the significance and/or function of many of the specific modifications within several of the RNA types remains undefined.
Modified nucleotides were identified decades ago in many RNAs, yet functional studies proceeded slowly because of, at least in part, a lack of effective assays for detecting and quantifying the modifications. The classical approach for detecting rRNA modifications involved in vivo radiolabeling of newly synthesized RNAs followed by ribo-nuclease fingerprinting and sequencing of the RNA fragments (Maden 1986, 1988; Maden et al. 1995). The approach for detecting and localizing modified nucleotides in short RNAs, such as tRNAs and snRNAs, also involved radioactive labeling of fragments liberated from the partial cleavage of purified RNA followed by direct RNA sequencing of the fragments by using ribonuclease digestion and thin layer chromatography (TLC; Reddy et al. 1981; Kuchino et al. 1990; Grosjean et al. 1998). Although these approaches have been used extensively, they are laborious and time-consuming. Newer, more effective methods were eventually developed (for review, see Grosjean et al. 1995; Maden et al. 1995), most of which were based on the fact that modified nucleotides can cause stops/pauses during reverse transcription, thereby providing a means to detect the modifications. For instance, 2′-O-methylated nucleotides directly induce reverse transcription stops/pauses at low concentrations of dNTPs (Maden et al. 1995). On the other hand, a pseudouridine that is chemically modified by N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimid-metho-p-toluolsulfonate (CMC) blocks reverse transcription one nucleotide prior to the CMC-modified pseudouridine (Bakin and Ofengand 1993, 1998). The development of such reverse transcription-based approaches has greatly facilitated the detection of modified nucleotides in RNAs and has directly contributed to recent advances in the area of RNA modifications (Jady et al. 2003; Kaya and Ofengand 2003; King et al. 2003; Ma et al. 2003; and for review, see Maden et al. 1995; Massenet et al. 1998; Ofengand and Fournier 1998; Kiss 2002). However, although highly convenient and effective, the reverse transcription-based methods do have limitations. For instance, most base modifications (e.g., pseudouridines) require further chemical alterations to induce stops/pauses during reverse transcription. Because RNAs contain a variety of modified bases (Motorin and Grosjean 1998)—each type requiring a specific chemical modifier—establishing and characterizing such chemical modification reactions for all types of modified nucleotides constitutes a formidable task. Furthermore, in many cases it appears that the reverse transcription-based methods are not quantitative.
Using site-specific RNase H cleavage directed by 2′-O-methyl RNA–DNA chimeras, we previously developed a highly sensitive method for detecting 2′-O-methylated nucleotides in RNAs (Yu et al. 1997). This approach is based on the fact that RNase H cleaves RNA only at sites where the 2′ position of the sugar ring is not modified (2′-OH). Thus, when a sugar ring is 2′-O-methylated, cleavage at that site is completely blocked. Importantly, this method can also be quantitative because the degree of resistance to cleavage quantitatively reflects the level of 2′-O-methylation (Yu et al. 1997). Here, we extend the use of site-specific RNase H cleavage to the detection and quantitation of base modifications such as pseudouridines and base-methylated residues.
RESULTS AND DISCUSSION
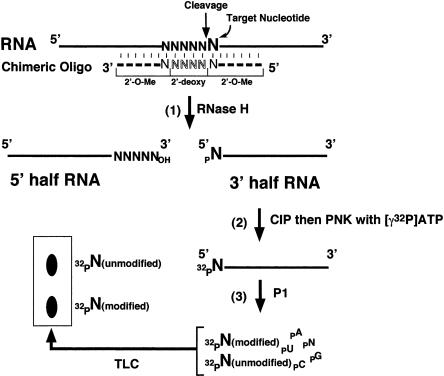
Figure 1 ▶ summarizes the three-step strategy for detecting and quantifying modified bases in RNA. In the first step, the test RNA is specifically cleaved at the 5′ side of the modified nucleotide of interest. This is readily achieved by using RNase H site-specific cleavage directed by 2′-O-methyl RNA–DNA chimeras (Yu 1999, 2000; Lapham and Crothers 2000). RNase H site-specific cleavage is independent of RNA sequence and therefore can be used to cleave any site within any RNA. The cleavage reaction generates a 5′-half RNA with a 3′ hydroxyl terminus and a 3′-half RNA with a 5′ phosphate terminus. In the second step, the 3′-half RNA is purified and dephosphorylated at its 5′ terminus by using alkaline phosphatase (CIP) and subsequently radiolabeled via rephosphorylation with [γ-32P]ATP and polynucleotide kinase (PNK). In the third step, the 5′ radiolabeled RNA is completely digested with nuclease P1, generating a mixture of nucleotides that are subsequently resolved by TLC. Because the nucleotide of interest is the only radiolabeled nucleotide present, both the modified and unmodified forms can be identified and accurately quantitated during the TLC analysis.
FIGURE 1.
The overall scheme for quantitative analysis of modified nucleotides in an RNA involves three basic steps as detailed in the text. (1) The test RNA is site-specifically cleaved by RNase H directed by a complementary 2′-O-methyl RNA–DNA chimera. The cleavage reaction generates 5′-half and 3′-half RNAs. (2) The 3′-half RNA is de-phosphorylated with CIP and subsequently rephosphorylated with PNK and [γ-32P]ATP. As a result, the 5′-terminal nucleotide (target nucleotide, larger-sized N) becomes radiolabeled. (3) The 5′-radiolabeled RNA is digested to completion with nuclease P1, and all nucleotides (including the radiolabeled terminal nucleotide, which may be partially or completely modified) are released from the RNA molecule. The released nucleotides are separated from each other on a TLC plate. Thick lines represent RNA chains. The dashed line indicates the 2′-O-methyl RNA–DNA chimera containing four 2′-deoxynucleotides (outlined letters) flanked by 2′-O-methyl ribonucleotides.
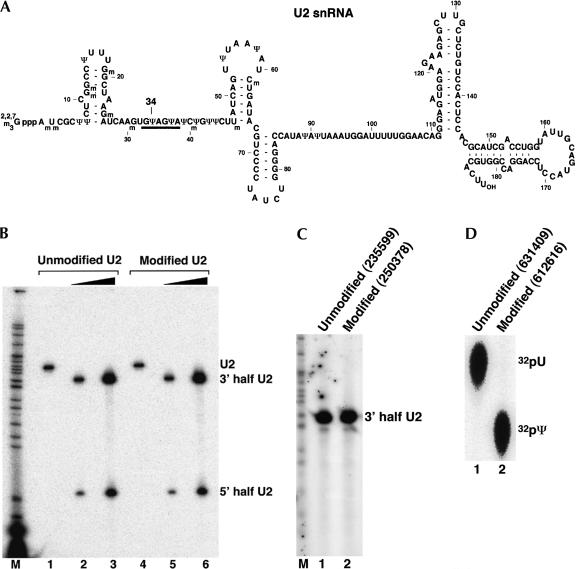
To demonstrate that the method is both sensitive and quantitative, we analyzed every step of the procedure by using U2 snRNA, a molecule in which all base modifications are known (Fig. 2A ▶). By using T7 RNA polymerase, we transcribed Xenopus U2 snRNA in vitro in the presence of ATP, CTP, GTP (with a trace amount of [α-32P]GTP) and either UTP or the modified nucleotide [pseudoUTP (ΨTP)]. Equivalent amounts of trace-labeled U2 transcripts containing either uridine or pseudouridine were subjected to RNase H site-specific cleavage directed by a chimeric oligonucleotide targeting the phosphodiester bond between positions 33 and 34. The cleavage was extremely efficient for both types of U2 snRNA (Fig. 2B ▶) in that only 2 units of RNase H completely cleaved 10 ng (~0.15 pmole; lanes 2,5) or 70 ng (~1.05 pmole; lanes 3,6) of U2 snRNA; no intact U2 was detected (lanes 2,3,5,6). These results indicate that both unmodified U2 (lanes 2,3) and pseudouridine-modified U2 (lanes 5,6) are specifically and quantitatively cleaved by RNase H directed by a 2′-O-methyl RNA–DNA chimera.
FIGURE 2.
Quantitative analysis of U2 snRNA pseudouridylation. (A) The sequence of mammalian U2 snRNA. Position 34 (to be analyzed) is indicated. The thick line denotes the bases involved in base-pairing interactions with the branch site sequence in pre-mRNA. (B) Site-specific RNase H cleavage directed by a 2′-O-methyl RNA–DNA chimera. Lightly radiolabeled unmodified U2 snRNA transcript (unmodified U2; lanes 1–3) and fully pseudouridine-substituted U2 transcript (modified U2; lanes 4–6) were subjected to RNase H cleavage in the presence of a 2′-O-methyl RNA–DNA chimera complementary to nucleotides 19–35 of U2 snRNA. The four 2′-deoxynucleotides in the chimera were designed to hybridize with nucleotides 30–33 of U2, leading to RNase H cleavage of the phosphodiester bond between positions 33 and 34 (target nucleotide). Reactions in lanes 2 and 4 contained 10 ng (0.15 pmole) of U2. Reactions in lanes 3 and 6 contained 70 ng (1.05 pmole) of U2. An overnight PhosphorImager exposure is shown, and the intact and cleaved U2 RNAs are indicated. (C) Replacement of the 5′-terminal phosphate with a radiolabeled phosphate (32P). The 3′-half RNA was treated with CIP and rephosphorylated with PNK and [γ-32P]ATP. The 3′-half RNA in lane 1 was derived from unmodified U2 and the RNA in lane 2 was from fully pseudouridine-substituted U2 (see B). A 1-min PhosphorImager exposure is shown. The numbers in parentheses are original PhosphorImager measurements (in volume). (D) Comparison of the radiolabeled nucleotides released from RNA. A small fraction of the 5′ endradiolabeled RNA derived from unmodified U2 (lane 1) or fully pseudouridine-substituted U2 (lane 2) was completely digested with nuclease P1. The released nucleotides (including the radiolabeled 5′-terminal nucleotide) were subsequently analyzed by TLC. The original numbers of PhosphorImager measurements are shown in parentheses. The positions of uridylate and pseudouridylate are indicated.
It should be noted that RNase H from different sources may exhibit different cleavage specificity (Lapham et al. 1997). RNase H used here was either purchased from Amersham or was a gift from R.J. Crouch. As predicted, the enzyme from both sources cleaved RNA specifically at the site 3′ to the nucleotide that base pairs with the 5′-most deoxynucleotide of the chimera (Lapham et al. 1997; see Fig. 1 ▶). However, caution should be taken when using RNase H from different suppliers. For instance, it was reported that RNase H from Boehringer Mannheim (now Roche) showed a different cleavage specificity—cleavage occurred at the site 5′ to the nucleotide that base pairs with the 5′-most deoxynucleotide of the chimera (Lapham et al. 1997). Hence, it is important to map the cleavage site when using RNase H from a different supplier. Appropriate 2′-O-methyl RNA–DNA chimeras can then be designed accordingly.
The cleaved 3′-half U2 RNAs from lanes 2 and 5 in Figure 2B ▶ were recovered and dephosphorylated with CIP followed by 5′ rephosphorylation with [γ-32P]ATP and PNK. As expected, the 3′-half RNAs derived from unmodified U2 and pseudouridine-modified U2 were labeled to the same extent (Fig. 2C ▶, cf. lanes 1 and 2), demonstrating that the dephosphorylation–rephosphorylation step is also quantitative.
The 5′ end–labeled 3′-half U2 RNAs were then eluted and digested to completion with nuclease P1 and the samples were subsequently analyzed by TLC. As shown in Figure 2D ▶, the respective U2 RNAs contained equivalent amounts of 32P-labeled uridylate (lane 1) and pseudouridylate (lane 2). Thus, we started with the same small amount (~0.15 pmole) of unmodified and pseudouridine-modified U2 and obtained equivalent amounts of uridylate and pseudouridylate, indicating that the method is both sensitive and quantitative.
To further demonstrate the quantitative aspect of the method, we mixed unmodified U2 and modified U2 (fully substituted with pseudouridine) at different ratios (1:9, 2:8, 3:7, 4:6, 5:5, 6:4, 7:3, 8:2, and 9:1, respectively) and performed the entire procedure. The relative amounts of final products (uridylate:pseudouridylate) for each mixed sample accurately reflected the initial ratio of unmodified U2 versus modified U2 (Fig. 3 ▶). Importantly, we also analyzed the pseudouridines at other locations in U2, including Ψ54 and Ψ58 in Stem–Loop II and Ψ89 and Ψ91 near the Sm binding site (see Fig. 2A ▶). Our measurement of pseudouridylation at these sites was also quantitative (data not shown), suggesting that the sequence context and perhaps even the secondary and tertiary structures do not constitute a major obstacle to quantitation. However, this conclusion must be further confirmed in each case.
FIGURE 3.
Unmodified U2 (unmod.-U2) and fully pseudouridine-substituted U2 (mod.-U2) were mixed at different ratios as indicated (lanes 2–10). The mixed U2 RNAs were then quantitated as in Figure 2 ▶ (B–D). PhosphorImager measurements (volume) of the ratio of 32pU:32pΨ are shown in parentheses. Lane 1 contained unmodified U2 only, and lane 11 contained fully pseudouridine-substituted U2 only.
We then applied this method to U2 RNA purified from mouse liver cells. This cellular U2 snRNA was cleaved at the phosphodiester bond just 5′ of position 32 (U32, an unmodified nucleotide) or just 5′ of position 34 (Ψ34, a naturally occurring pseudouridine; see Fig. 2A ▶). The 3′-half RNA was recovered and the 5′ end-labeled via dephosphorylation/rephosphorylation and then subjected to nuclease P1 digestion followed by TLC, as described earlier. Position 34 showed ~90% pseudouridylation (Fig. 4 ▶, lane 4), whereas position 32 was not modified (lane 3), as expected.
FIGURE 4.
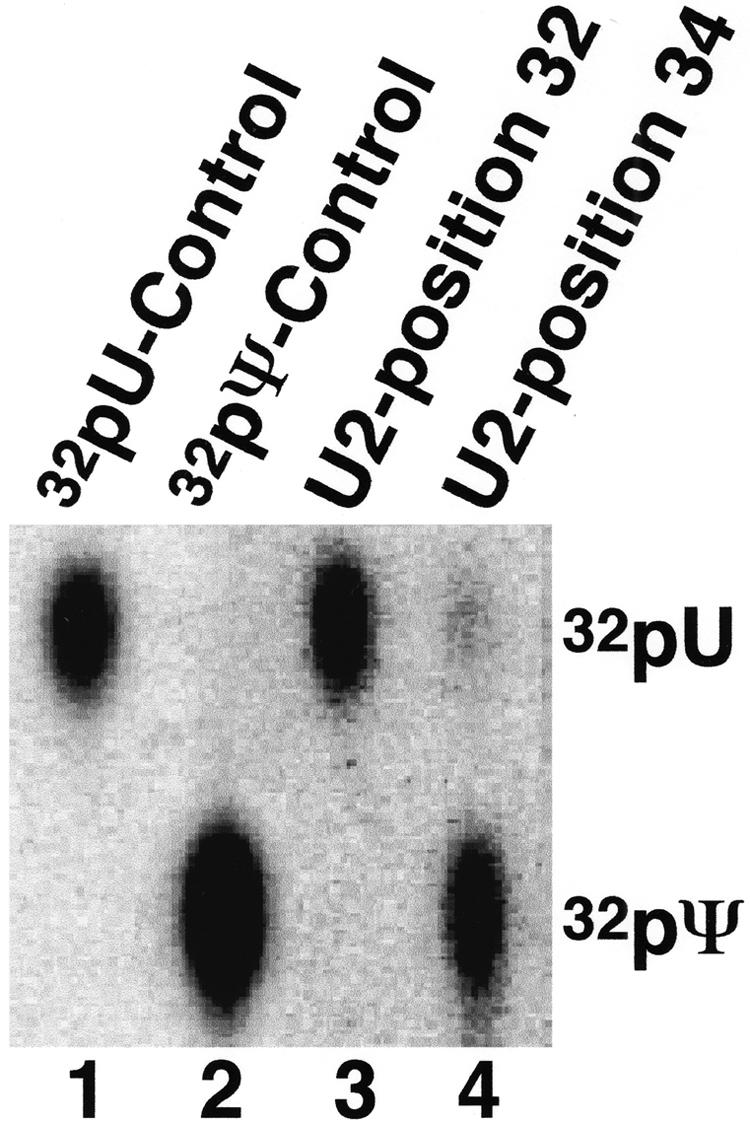
Quantitation of mouse U2 pseudouridylation. U2 RNA isolated directly from mouse liver (lanes 3,4) was subjected to quantitative analysis at positions 32 (lane 3) and 34 (lane 4), as in Figure 2 ▶ (B–D). Lanes 1 and 2 are controls containing unmodified U2 and fully pseudouridine-substituted U2, respectively.
In some cases, RNA ribozymes or even RNA-cleaving DNA enzymes may be used for site-specific cleavage. The advantage of using these enzymes is that the cleaved 3′-half RNA contains a 5′ hydroxyl group that can be directly phosphorylated with [γ-32P]ATP and PNK, thereby eliminating the dephosphorylation step. The disadvantage is that cleavage by most of these enzymes is sequence dependent, and sometimes inefficient. For instance, the RNA-cleaving DNA enzymes 8–17 and 10–23 (Santoro and Joyce 1997) cleave the phosphodiester bond between A and G and between a purine and pyrimidine, respectively. The cleavage efficiency is relatively low compared with RNase H cleavage. However, DNA enzymes can be used for quantitation because many base-modified RNAs do not affect cleavage efficiency (i.e., both unmodified RNA and modified RNA are cleaved equally by the enzymes). To test this idea, we used the DNA 10–23 enzyme to cleave equivalent amounts of unmodified and pseudouridine-modified U2 between G33 and U34/Ψ34. Although not 100%, the cleavage efficiency for both unmodified U2 and modified U2 was identical (data not shown), thus indicating that the DNA enzyme is indeed useful for quantitation.
The applicability of this method is not limited to the quantitation of pseudouridylation. We also tested U2 RNA containing 5-fluorouridines, a different type of base modification, and found that the 5-fluorouridine modification was quantitatively detected as well (data not shown). Because many base modifications do not affect Watson-Crick base pairing, it is unlikely that these modifications have any effect on the reactivity of RNase H directed by 2′-O-methyl RNA–DNA chimeras. However, some modifications may affect base-pairing interactions. One example is the 1-methylation of adenosine, which results in the loss of one hydrogen bond when pairing with uridine. It is thus possible that this modification could negatively impact RNase H cleavage efficiency, and further analysis is necessary to clarify this issue. Also, the other enzymes used in our method, namely, CIP, PNK, and nuclease P1, appear to have no substrate preference with regard to unmodified versus base-modified nucleotides, at least for the two base modifications we examined (pseudouridine and 5-fluorouridine). Hence, our results suggest that these enzymes would not interfere with the quantitation of other base modifications, although this aspect of the procedure must be confirmed in each case. Interestingly, RNA backbone modifications such as 2′-O-methylation block RNase H cleavage. This fact does not represent a limitation of the applicability of RNase H, however, because the lack of specific cleavage at suspected 2′-O-methylation sites defines an assay for detecting and quantitating 2′-O-methylation sites in RNAs (Yu et al. 1997).
Given that our method targets a single specific nucleotide, it is disadvantaged with respect to detecting modified nucleotides in uncharacterized long RNAs. Such instances require a combination of our method and the reverse transcription-based methods. For instance, pseudouridylation could be identified initially via CMC modification followed by reverse transcription (Bakin and Ofengand 1993, 1998). Subsequently, our RNase H-based site-specific analysis could confirm and rigorously quantitate the identified pseudouridines.
With minor alterations, this method may also be applied to modification assays for studying enzyme activities. For this purpose, a synthetic RNA containing a single 32P radiolabel at a specific site of interest must be constructed, a task that is readily achieved by coupling the first two steps described in Figure 1 ▶ (using an in vitro transcribed RNA) with the two-piece ligation technique described previously (Moore and Sharp 1992; Yu 1999, 2000). Upon incubation of this single site-radiolabeled RNA with cell extracts or modifying enzyme preparations, the conversion of the radiolabeled residue into a modified nucleotide can be readily detected by TLC and autoradiography. Over the past few years, the single radiolabeled RNA-based assay has been successfully applied to the study of several different modifications. For instance, a yeast U2 RNA containing a single 32P-radiolabel at position 35 was constructed to detect a Ψ35-specific U2 pseudouridylase activity in yeast cell extracts. Incubation of this site-specifically labeled RNA with the yeast extracts permitted the detection of the pseudouridylase activity (Ma et al. 2003). This method has also facilitated the identification of an H/ACA guide RNA necessary for U2 pseudouridylation at two different sites in Xenopus oocytes (Zhao et al. 2002), as well as a methylase responsible for the formation of 1-methyl guanosine at position 9 in yeast tRNA (Jackman et al. 2003).
In summary, the approach we describe here combined with the reverse transcription-based methods should be very useful in detecting, localizing, and quantifying modified nucleotides in a variety of RNAs.
MATERIALS AND METHODS
In vitro transcription
To generate U2 snRNA as an experimental molecule, we linearized a plasmid containing Xenopus U2 snRNA under the control of a T7 promoter with Sma I (the restriction site had been introduced at the 3′ end of the U2 sequence ) and used it as a template for T7 transcription in vitro. The reaction was performed for 1 h at 37°C in 10 μL containing 1.2 mM each of ATP, CTP, GTP, and UTP (Amersham); 0.005 μCi/μL [α-32P]GTP (Amersham); 40 mM Tris-HCl (pH 7.5); 6 mM MgCl2; 2 mM spermidine; 5 mM DTT; 0.1 μg/μL of linearized T7-U2 plasmid; and 4 U/μL T7 phage polymerase (Epicenter). After the reaction, the trace-labeled U2 snRNA was gel purified and quantified by scintillation counting. The reaction produced ~1.2 μg of U2. To generate U2 snRNA containing pseudouridines (Ψ) or 5-fluorouridines (5FU), we replaced the 1.2 mM UTP in the transcription reaction with 1.2 mM pseudouridine 5′ triphosphate (ΨTP; Sierra Bioresearch) or 5-fluorouridine 5′ triphosphate (5FUTP; Sierra Bioresearch), respectively. In both cases, 1–1.2 μg of modified U2 was produced.
RNase H site-specific cleavage directed by 2′-O-methyl RNA–DNA chimeras
The U2 snRNAs were subjected to RNase H site-specific cleavage directed by 2′-O-methyl RNA–DNA chimeras. To cleave the phosphodiester bond between positions 33 and 34, we mixed 10 ng (~0.15 pmole) or 70 ng (~1.05 pmole) of U2 transcript (with or without Ψ or 5FU modifications) with 5 pmole of the 2′-O-methyl RNA–DNA chimera (UmAmdCdAdCdTUmGmAmUmCmUmUm AmGmCmCm) in 4 μL of water. The resulting mixture was heated for 3 min at 95°C and slowly cooled to room temperature. The U2 snRNA that became hybridized to the chimera in the mixture was then subjected to RNase H cleavage for 1 h at 37°C in a final reaction volume of 10 μL containing 10 ng or 70 ng of U2, 5 pmole of 2′-O-methyl RNA–DNA chimera, 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl, 25 mM DTT, 5% sucrose, 20 units RNasin (Promega), and 2 units RNase H (Amersham; and kindly provided by Dr. R.J. Crouch). The resulting 5′- and 3′-half RNAs were visualized by autoradiography and the 3′-half RNA was recovered from the gel.
The 2′-O-methyl RNA–DNA chimera used in this work was purchased from Keck Oligo Synthesis Facility at Yale University for $11/2′-O-methyl residue and $1.60/deoxynucleotide residue (1.0-μmole scale).
Dephosphorylation and rephosphorylation
The 3′-half RNA derived from RNase H cleavage was subjected to dephosphorylation reaction in a volume of 10 μL containing 1× dephosphorylation buffer (Roche), ~0.15 pmole of 3′-half U2, and 1 unit of CIP (Roche) for 45 min at 50°C. The conditions used here were essentially the same as the conditions described by the supplier (Roche). After the reaction, the sample was extracted with phenol/chloroform/isoamyl alcohol (1:1:0.01; PCA) and precipitated with ethanol. The recovered RNA was rephosphorylated at its 5′ terminus for 30 min at 37°C in a 10-μL reaction containing 1× phosphorylation buffer (Amersham), ~0.15 pmole of 3′-half U2, 150 μCi of [γ-32P]ATP (6000 Ci/mmole, DuPont/NEN), and 10 units of T4 PNK (Amersham). The 5′ endradiolabeled 3′-half U2 was recovered by gel purification.
It should be noted that shrimp alkaline phosphatase (SAP), which is sensitive to heat (65°C), could be a better choice for the dephosphorylation reaction. Heat inactivation of SAP after the dephosphorylation reaction could allow the omission of PCA extraction and ethanol precipitation, which would otherwise be needed to remove active phosphatase (see CIP-catalyzed dephosphorylation reaction above).
Nuclease P1 digestion and TLC analysis
The 5′-radiolabeled 3′-half U2 was digested with nuclease P1 (200 μg/mL) in 3 μL of 20 mM sodium acetate (pH 5.2) for 1 h at 37°C. The digested nucleotide mixture was dotted on a cellulose TLC PEI membrane (EM Science) and chromatographed in buffer containing isopropanol/HCl/H2O (70:15:15, v/v/v; Patton 1991; Yu et al. 1998). Labeled uridylate and pseudouridylate were visualized by autoradiography. When the unmodified and modified U2 were mixed first and then quantitated, both uridylate and pseudouridylate were visualized in the same sample. The ratio of uridylate to pseudouridylate was determined using a PhosphorImager (Molecular Dynamics).
Purification of U2 snRNA from mouse brain
Trizol reagent (Invitrogen) was used to isolate RNA from mouse liver essentially as described by the supplier. Briefly, 200 mg of mouse liver was homogenized in 4 mL of Trizol using a Dounce tissue grinder (Wheaton). The homogenized sample was then extracted with 0.8 mL of chloroform and centrifuged at 12,000g for 15 min at 4°C. The upper aqueous phase was recovered and the total RNA was precipitated with isopropanol.
To isolate U2 snRNA, we mixed total RNA isolated from 200 mg of mouse liver with 200 pmole of a biotinylated antisense U2 2′-O-methyl oligonucleotide complementary to nucleotides 158–177 of U2 (UmCmCmUmGmGmAmGmGmUmAmCmUmGmCmAmAm UmAmCmBBB, where B stands for Biotin-TEG) in 50 μL NET-2-MgCl2 buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% (v/v) NP-40, and 2 mM MgCl2. After heating for 2 min at 95°C, the sample was slowly cooled (10 min at 65°C, then another 30 min at 30°C). Streptavidin agarose beads (20 μL; Pierce) were added to the sample and the final volume was brought to 300 μL with NET-2-MgCl2 buffer. This biotinstreptavidin binding reaction was incubated for 1.5 h at room temperature with gentle rotation. The beads were then precipitated (tabletop centrifugation), extensively washed with the NET-2-MgCl2 buffer, and finally mixed with 250 μL of dissociation buffer (10 mM Tris-HCl at pH 7.5, 0.1% SDS, and 0.5 mM EDTA). After a 15-min incubation at 85°C, the beads were precipitated and the supernatant recovered. Following PCA extraction, U2 snRNA was ethanol precipitated and gel purified under 254 nm UV light. The U2 snRNA was then used for pseudouridine quantitation, as described above.
Acknowledgments
We thank Dr. R.J. Crouch for kindly providing the RNase H construct. We also thank members of the Yu laboratory for valuable discussions and comments. This work was supported by grant GM62937 from the National Institute of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7110804.
REFERENCES
- Auffinger, P. and Westhof, E. 1998. Location and distribution of modified nucleotides in tRNA. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 569–576. ASM Press, Washington, DC.
- Bachellerie, J.-P. and Cavaille, J. 1998. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 255–272. ASM Press, Washington, DC.
- Bakin, A.V. and Ofengand, J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: Analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol. 77: 297–309. [DOI] [PubMed] [Google Scholar]
- Bjork, G.R. 1995. Biosynthesis and function of modified nucleotides. In tRNA: Structure, biosynthesis, and function (eds. D. Soll and U. RajBhandary), pp. 165–205. ASM Press, Washington, DC.
- Grosjean, H., Sprinzl, M., and Steinberg, S. 1995. Posttranscriptional modified nucleotides in transfer RNA: Their locations and frequencies. Biochimie 77: 139–141. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Motorin, Y., and Morin, A. 1998. RNA-modifying and RNA-editing enzymes: Methods for their identification. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 21–46. ASM Press, Washington, DC.
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Jackman, J.E., Montange, R.K., Malik, H.S., and Phizicky, E.M. 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady, B.E., Darzacq, X., Tucker, K.E., Matera, A.G., Bertrand, E., and Kiss, T. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22: 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya, Y. and Ofengand, J. 2003. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 9: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, T.H., Liu, B., McCully, R.R., and Fournier, M.J. 2003. Ribosomal structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11: 425–435. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kuchino, Y., Mizushima, H., and Nishimura, S. 1990. Nucleotide sequence analysis and identification of modified nucleotides of tRNA. In Transfer RNAs and other soluble RNAs (ed. J.D. Cherayil), pp. 70–82. CRC Press, Boca Raton, FL.
- Lapham, J. and Crothers, D.M. 2000. Site-specific cleavage of transcript RNA. Methods Enzymol. 317: 132–139. [DOI] [PubMed] [Google Scholar]
- Lapham, J., Yu, Y.-T., Shu, M.-D., Steitz, J.A., and Crothers, D.M. 1997. The position of site-directed cleavage of RNA using RNase H and 2′-O-methyl oligonucleotides is dependent on the enzyme source. RNA 3: 950–951. [PMC free article] [PubMed] [Google Scholar]
- Ma, X., Zhao, X., and Yu, Y.-T. 2003. Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden, B.E.H. 1986. Identification of the locations of the methyl groups in 18S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 189: 681–699. [DOI] [PubMed] [Google Scholar]
- ———. 1988. Locations of methyl groups in 28S rRNA of Xenopus laevis and man. J. Mol. Biol. 201: 289–314. [DOI] [PubMed] [Google Scholar]
- ———. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39: 241–303. [DOI] [PubMed] [Google Scholar]
- Maden, B.E.H., Corbett, M.E., Heeney, P.A., Pugh, K., and Ajuh, P.M. 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77: 22–29. [DOI] [PubMed] [Google Scholar]
- Massenet, S., Mougin, A., and Branlant, C. 1998. Posttranscriptional modifications in the U small nuclear RNAs. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 201–228. ASM Press, Washington, DC.
- Moore, M.J. and Sharp, P.A. 1992. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science 256: 992–997. [DOI] [PubMed] [Google Scholar]
- Motorin, Y. and Grosjean, H. 1998. Chemical structures and classification of posttranscriptionally modified nucleosides in RNA. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 543–549. ASM Press, Washington, DC.
- Ofengand, J. and Fournier, M. 1998. The pseudouridine residues of rRNA: Number, location, biosynthesis, and function. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 229–253. ASM Press, Washington, DC.
- Patton, J.R. 1991. Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Mol. Cell. Biol. 11: 5998–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, R. and Busch, H. 1988. Small nuclear RNAs: RNA sequences, structure, and modifications. In Structure and function of major and minor small nuclear ribonucleoprotein particles (ed. M.L. Birnstiel), pp. 1–37. Springer-Verlag Press, Berlin, Germany.
- Reddy, R., Dale, H., Epstein, P., and Busch, H. 1981. Primary and secondary structure of U2 snRNA. Nucleic Acids Res. 9: 5645–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.-T. 1999. Construction of 4-thiouridine site-specifically substituted RNAs for cross-linking studies. Methods 18: 13–21. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Site-specific 4-thiouridine incorporation into RNA molecules. Methods Enzymol. 318: 71–88. [DOI] [PubMed] [Google Scholar]
- Yu, Y.-T., Shu, M.-D., and Steitz, J.A. 1997. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 3: 324–331. [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Li, Z.-H., Terns, R.M., Terns, M.P., and Yu, Y.-T. 2002. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA 8: 1515–1525. [PMC free article] [PubMed] [Google Scholar]