Abstract
The pseudouridine (Ψ) synthases Pus7p and TruD define a family of RNA-modifying enzymes with no sequence similarity to previously characterized Ψ synthases. The 2.2 Å resolution structure of Escherichia coli TruD reveals a U-shaped molecule with a catalytic domain that superimposes closely on that of other Ψ synthases. A domain that appears to be unique to TruD/Pus7p family enzymes hinges over the catalytic domain, possibly serving to clasp the substrate RNAs. The active site comprises residues that are conserved in other Ψ synthases, although at least one comes from a structurally distinct part of the protein. Remarkably, the connectivity of the structural elements of the TruD catalytic domain is a circular permutation of that of its paralogs. Because the sequence of the permuted segment, a β-strand that bisects the catalytic domain, is conserved among orthologs from bacteria, archaea and eukarya, the permutation likely happened early in evolution.
Keywords: RNA modification, X-ray crystallography, SIRAS, TruD, Pus7p
INTRODUCTION
Pseudouridine (Ψ) synthases are responsible for the most abundant posttranscriptional nucleobase modification of cellular RNAs. These enzymes are found in all free-living organisms whose genomes have been sequenced. In some cases, pseudouridylation has been shown to play biochemically important roles. For instance, pseudouridylation of U2 snRNA has been shown to be essential for efficient assembly of the corresponding ribonucleoprotein (Yu et al. 1998; Zhao and Yu 2004). Biophysical analyses suggest that the modification at position 35 of this snRNA facilitates extrusion of the branch-point adenosine of the intron during pre-mRNA splicing (Newby and Greenbaum 2002).
Until recently, all Ψ synthase sequences could be classified into four families named after the Escherichia coli proteins TruA, TruB, RsuA, and RluA (Koonin 1996). Structure determinations of representatives of each (Foster et al. 2000; Hoang and Ferré-D’Amaré 2001; Sivaraman et al. 2002, 2004; Del Campo et al. 2004) revealed that despite a lack of sequence similarity, proteins from all four families adopt superimposable folds with equivalent connectivity and share a characteristic set of active site residues (Mueller 2002;Ferré-D’Amaré 2003). Enzymes from the different families have accessory domains inserted into different parts of the conserved catalytic domain or appended to it. These insertions and appendices are thought to play a role in substrate recognition. In the case of TruB, this has been demonstrated structurally (Hoang and Ferré-D’Amaré 2001; Pan et al. 2003; Phannachet and Huang 2004).
The proteins Pus7p (from baker’s yeast; Ma et al. 2003) and TruD (from E. coli; Kaya and Ofengand 2003) were identified biochemically as the enzymes responsible for Ψ35 and Ψ13 in U2 snRNA and tRNAGlu, respectively. These novel enzymes had no sequence similarity to previously characterized Ψ synthases, but database searches revealed the presence of close Pus7p/TruD homologs in archaea, bacteria, and eukarya. The yeast enzyme was subsequently shown to have multisite specificity, being capable of modifying both U2 snRNA and tRNAs. Because metazoans are thought to modify U2 snRNA through a guide-RNA–dependent mechanism, this specificity would account for the ubiquity of Pus7p orthologs in multicellular eukaryotes (Behm-Ansmant et al. 2003).
To determine whether enzymes of this family represent a structurally distinct class of Ψ synthases, and as a first step in understanding their substrate specificity, we have now solved the structure of E. coli TruD by X-ray crystallography and refined the model at 2.2 Å resolution. We find that despite the lack of any meaningful sequence similarity (Sander and Schneider 1991), the catalytic domain of TruD superimposes closely on that of other Ψ synthases. After our manuscript was submitted, an independent structure determination of TruD, in a different crystal form, was reported (Kaya et al. 2004). Superposition of the two TruD molecules in the asymmetric unit of our crystal form with the single TruD molecule in the other crystal form shows that the arms of the U-shaped Ψ synthase can flex by 18°, possibly to accommodate substrate. Unexpectedly, the connectivity of the structural elements of the TruD catalytic domain is a circularly permuted variation of the conserved Ψ synthase fold. We also find that although the functional groups that decorate the active site of TruD and the paralogous Ψ synthases are similar, some of these come from structurally nonequivalent parts of the conserved folds. The overall structural similarity and the clear divergence of Ψ synthases into at least two superfamilies (TruD on one side and all other known families on the other) argue for the great antiquity of these RNA-modifying enzymes.
RESULTS and DISCUSSION
Structure determination and overview
Initial crystallization screens (Jancarik and Kim 1991) of N-terminally His-tagged TruD resulted in monoclinic crystals that diffracted X-rays to 2.9Å but suffered from disorder, making them unsuitable for structure determination. We had synthesized an RNA heptanucleotide 5′-CGUC[5FU]AG-3′ (5FU indicates 5-fluorouridine) for biochemical studies of TruD. The RNA sequence is from the D-arm of E. coli tRNAGlu, with 5FU at the site of modification of the only known substrate of TruD in this bacterium. Some 5FU-containing substrates have been shown to inhibit Ψ synthases, either by formation of a covalent adduct or through tight binding of the isomerized 5FU (Spedaliere and Mueller 2004). Binding of this heptanucleotide to TruD was undetectable biochemically (data not shown). Nonetheless, because crystallization screens are carried out at near-millimolar concentrations and thus may be able to capture even very weak interactions, we carried out crystallization screens of mixtures of TruD with this RNA. This produced a new crystal form of greatly improved quality (Materials and Methods).
The structure was solved by the single isomorphous replacement with anomalous scattering (SIRAS) method using diffraction data collected from native and selenomethionyl TruD crystals using a copper rotating anode X-radiation source. The SIRAS phases, improved by density modification (mean overall figure of merit after modification = 0.84), produced an electron density map (Fig. 1A ▶) that allowed the crystallographic model to be built unambiguously. Previously, an asymmetric unit with 455 amino acid residues (16 of which were methionines) was phased by using this technique (Lemke et al. 2002). Our structure determination shows that in-house sulfur/selenium SIRAS can produce high-quality phases for an asymmetric unit containing ~740 residues, only eight of which are methionine, even though the mean B-factor of the selenium atoms is moderately high (57.9 Å2). Presumably, the high degree of isomorphism of the native and derivative crystals helps overcome the modest difference in scattering between sulfur and selenium. The model has good stereochemistry and an Rfree factor of 25.4% for all data between 30 and 2.2 Å (Table 1 ▶; Materials and Methods).
FIGURE 1.
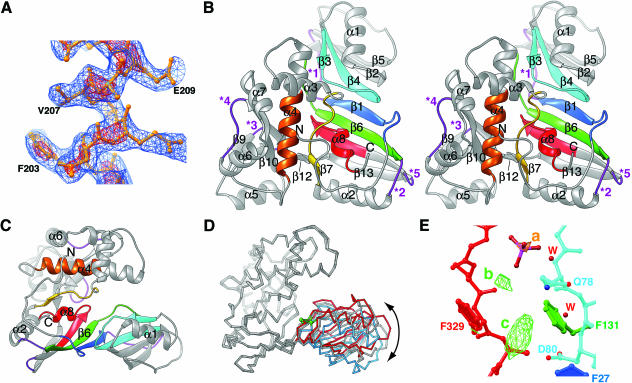
Overall structure of TruD. (A) Experimental electron density. The blue and red meshes depict a portion the “solvent flattened” SIRAS electron density map contoured at 1.0 and 2.5 SD above mean peak height, respectively. The map was drawn around amino acids 200–212 of the refined model. (B) Stereo ribbons representation of TruD. The colors (N- to C-terminal, blue to red in rainbow order) denote the six conserved sequence segments of TruD/Pus7 proteins from all phyla (Kaya and Ofengand 2003). The five purple loops (*) are where insertions exist in different TruD orthologs (e.g., a 136-amino-acid insertion in loop *1 in the yeast Pus7p). The active site aspartate is in the loop between β3 and β4. (C) View perpendicular to B emphasizing the two-domain structure of TruD. (D) Superposition (using 148 Cα atoms of the TruD/Pus7p family insertion domain; root mean square deviation ~0.35 Å for this domain) of the two TruD molecules in the crystallographic asymmetric unit of our structure (gray, and gray with blue right-hand side) and the single TruD molecule in the structure of Kaya et al. (2004; Protein Data Bank accession 1SI7, red). Green spheres denote the Cα of the active site aspartates. The angular displacement is 11° between the blue and gray molecules and 18° between the blue and red molecules. (E) View of the active site of TruD (gray molecule in panel D, colored as in panels B, C) with electron density features that correspond to poorly ordered bound RNA. Red spheres (W) are water molecules. The yellow mesh (feature a) depicts an anomalous difference Fourier synthesis calculated with crystal I data, contoured at 6 SD, that has been modeled as the phosphorus atom of a phosphate. The green mesh depicts a residual (|Fo| − |Fc|) synthesis, contoured at 3 SD. Features b and c may correspond to poorly ordered ribose and nucleobase moieties, respectively.
TABLE 1.
Crystallographic statistics
| Diffraction data | Crystal I (SeMet) | Crystal II (Native) | ||
| Data set | ||||
| Unit cell a, b, c (Å) | 63.67, 108.78, 111.89 | 63.70, 108.82, 111.99 | ||
| Resolution range (Å) | 30–2.2 | 2.28 – 2.2 | 30–2.25 | 2.33–2.25 |
| Reflections | ||||
| observed | 431,614 | 40,762 | 288,667 | 19,524 |
| unique | 40,114 | 3941 | 37,455 | 3454 |
| Completeness (%) | 99.7 | 99.6 | 99.1 | 92.7 |
| 〈I〉/〈σ(I) 〉 | 43.1 | 7.2 | 30.6 | 4.0 |
| Rsyma (%) | 0.07 | 0.414 | 0.062 | 0.395 |
| SIRAS analysis | ||||
| Resolution range (Å) | 30–2.2 | 2.28–2.2 | ||
| Rderivb | 0.062 | 0.155 | ||
| Mean figure of meritc | 0.35 | 0.20 | ||
| Phasing power (centric)d | 1.04 | 0.44 | ||
| Phasing power (acentric) | 1.23 | 0.52 | ||
| Refinement | ||||
| Resolution range (Å) | 30–2.2 | 2.28–2.2 | ||
| Rfreee | 0.254 | 0.324 | ||
| Rwork | 0.216 | 0.274 | ||
| RMSD bonds/anglesf | 0.0059 Å/1.23° | |||
| Coordinate errorg | 0.27 Å |
aRsym = ∑|I − 〈I〉|/∑ I where I is the observed intensity and 〈I〉 is the statistically weighted absolute intensity of multiple measurements of symmetry related reflections.
bRderiv = ∑||FPH| − | Fp||/∑|Fp|, where |FPH| and |Fp| are the heavy-atom derivative and protein structure factor amplitudes, respectively.
cMean figure of merit = 〈∑ P(α)eiα/∑P(α) >, where α is the phase and P(α) is the phase-probability distribution.
dPhasing power = 〈|FH|〉/〈||Fp + FH| − |FPH||〉, where |FH| is the heavy atom structure factor amplitude; reported for centric and acentric reflections.
eThe free-R factor was calculated with a randomly selected 10% of the reflections.
fRoot mean square deviations (RMSD) from ideal bond lengths and angles.
gCross-validated σA coordinate error.
TruD adopts a mixed α-β fold with two distinct domains (Figs. 1 ▶, 2 ▶) arranged into a “U” shape. One domain consists primarily of a large, curved, eight-stranded β-sheet. The other domain (itself a mixed α-β fold) protrudes from a corner of the sheet and hinges over it (Fig. 1C ▶). Comparison of the two crystallographically independent TruD molecules in our crystals and the single copy in the structure of Kaya et al. (2004) shows that the two arms of the U-shaped enzyme can flex toward each other by at least 18° (Fig. 1D ▶).
FIGURE 2.
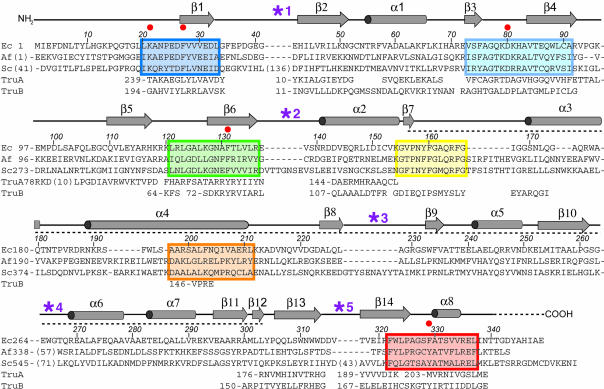
Sequence alignment of TruD orthologs and structure-based alignment of paralogs. The sequences of E. coli TruD and its orthologs from the archaeon Archaeoglobus fulgidus (accession no. O28596) and baker’s yeast (accession no. CAA99464) are aligned with the conserved sequence blocks described by Kaya and Ofengand (2003), color coded to correspond to Figure 1 ▶. Numbers in parentheses indicate the number of residues omitted either in insertions into loop regions (purple asterisks) or at the N termini. Secondary structure and amino acid numbering is that of E. coli TruD. The dashed line indicates the extent of the insertion domain. Red dots denote conserved active site residues of the TruD family (also see Fig. 3B ▶). Sequences of segments of E. coli TruA and TruB that correspond structurally to TruD (according to DALI; Holm and Sander 1993) have been aligned. Numbers before these segments, as well as at the start of each line, indicate the residue number of the first amino acid shown for each segment. Note that the segments of TruA and TruB that align with the β1 segment of TruD are C-terminal in the former two protein families.
A conserved catalytic domain
A search for similar structures using the program DALI (Holm and Sander 1993) shows that the β-sheet domain of TruD is most similar to the Ψ synthase TruA (Fig. 3A ▶), and to the catalytic domains of the Ψ synthases TruB, RsuA, and RluD (Z-scores of 7.7, 7.1, 5.8, and 4.1, respectively; a Z-score above 2 indicates similarity). TruB family enzymes have a PUA domain (Aravind and Koonin 1999) C-terminal to the catalytic domain (Ferré-D’Amaré 2003), whereas RsuA (Sivaraman et al. 2002) and RluA (Del Campo et al. 2004; Sivaraman et al. 2004) family enzymes have an N-terminal domain that resembles the ribosomal protein S4. The divergent domain of TruD family Ψ synthases is an insertion into the catalytic domain (Figs. 1B,C ▶, 2 ▶) and, in that sense, is more similar to the “thumb” insertion of TruB synthases that is responsible for substrate binding (Hoang and Ferré-D’Amaré 2001) in that family of enzymes. The DALI search suggests that the insertion domain of TruD is not significantly similar to any previously characterized protein fold.
FIGURE 3.
Comparison of TruD with other Ψ synthases. (A) Similarity of TruD (yellow) and TruA (pink). In this stereoview, conserved active site aspartate, basic, and aromatic residues are also shown. Dissimilar regions are omitted for clarity. Asterisk indicates the central β-strand of the sheet. The position of the active site lysine of TruD (K21) is indicated. (B) Superposition of active site residues of TruD (yellow), TruA (red; Foster et al. 2000), and TruB (blue; Hoang and Ferré-D’Amaré 2001). In gray is the unnatural nucleotide in the active site of the TruB-RNA cocrystal (Hoang and Ferré-D’Amaré 2001). This stereoview also depicts a conserved active site hydrophobic residue (F27 in TruD) and a partially conserved active site aromatic (F329 in TruD). (C) Conserved structural core of TruD. The central β-strand and the loop preceding it from the nearby N terminus are yellow. Gray dots and asterisk denote the location of the insertion domain (omitted). Green asterisk denotes the active site aspartate. (D) Schematic structure of TruA (Foster et al. 2000) with central β-strand and loop following it out to the nearby C terminus in red.
Alignment of TruD family sequences revealed six segments with high conservation (Kaya and Ofengand 2003). These segments (Fig. 2 ▶) line the inside of the U-shaped TruD (Fig. 1B,C ▶). This inside surface is strongly basic (Kaya et al. 2004) and is the likely tRNA-binding site of the enzyme. Consistent with this, one of the conserved segments includes an absolutely conserved aspartate residue that when mutated abolishes enzyme activity (Kaya and Ofengand 2003). The structure shows that this aspartate lies in the loops connecting strands β3 and β4 (Figs. 1B,C ▶, 2 ▶). The catalytic aspartate of TruA lies in the structurally equivalent loop (Fig. 3A ▶) as do the aspartates of enzymes of the three other Ψ synthase families (data not shown). Thus, similar to enzymes of the TruB, RsuA, and RluA families, TruD consists of a conserved catalytic domain characteristic of all Ψ synthases and a divergent insertion domain.
Active site and substrate recognition
Superposition of the active sites of TruD and other Ψ synthases shows that the catalytic aspartate, and aromatic and basic residues (D80, F131, and K21 in TruD, respectively), are positioned in equivalent positions relative to the putative substrate (Fig. 3B ▶). In addition to these residues, which are present with the equivalent spatial arrangement in the active sites of all Ψ synthases whose structures have been determined, there are two partially conserved active site residues. A leucine (L245 and L200, respectively, in TruA and TruB) is replaced by a phenylalanine (F27) in TruD. A tyrosine (Y179) that was found to stack on the substrate nucleotide in the TruB-RNA complex structure (Hoang and Ferré-D’Amaré 2001) is replaced by a phenylalanine (F329) in the equivalent location in the TruD active site (Fig. 3B ▶).
An interesting difference between TruD and the other enzymes is the location in the protein chain of the active site basic residue that makes a salt bridge with the catalytic aspartate. The basic residue of the TruD active site (K21) is grafted on a structural element that is different and distant from the location of the basic residue in other Ψ synthases (the loop preceding the central strand of the domain versus the helix at the bottom of the domain in Fig. 3A ▶). This phenomenon, the nonequivalence of residues with the same catalytic role in homologs, has been documented for other enzyme families (Todd et al. 2002).
The RNA heptanucleotide that was added to TruD during crystallization is not tightly bound. Weak residual electron density features are present in the active site cleft of the copy of TruD that adopts a more closed conformation (Fig. 1D,E ▶). The active site loop of this molecule is better ordered than that of the other molecule in the asymmetric unit (mean Cα B-factors for the loops are 47.1 Å2 and 57.9 Å2 for the closed and open molecules, respectively). A strong electron density feature in the anomalous difference Fourier synthesis is also present in the active site of the more closed molecule (Fig. 1E ▶) and has been modeled as the phosphorus atom of a phosphate that is present with an occupancy of 0.3. The location of the residual features (Fig. 1E ▶, b,c) is compatible with a ribose and a nucleobase, respectively. The putative nucleobase feature is located adjacent to the catalytic aspartate. Unfortunately, the electron density features are too weak for reliable model building. Thus, analysis of TruD substrate interaction must await further studies.
Although sequences corresponding to the catalytic core are highly conserved between the tRNA-specific E. coli TruD and the multisite, multisubstrate yeast enzyme Pus7p, structure-based sequence alignment demonstrates that a large insertion (Fig. 1B ▶, *1) is present in the eukaryotic enzyme. This insertion is conserved between yeast and human (Fig. 7 of Ma et al. 2003). These residues would lie to one side of the active site cleft, and they may play a role in recognizing the stem–loops IIa and IIb that lie distal from the site of U2 snRNA modification and that have been shown to be essential for Pus7p activity (Ma et al. 2003).
Circular permutation of a conserved domain
An unexpected difference between TruD and the other structurally characterized Ψ synthases is topological. The spatial arrangement of structural elements of the catalytic domain is the same in TruD and other Ψ synthases, and the central β-strand of the sheet (Fig. 1B ▶, β1) is in equivalent positions in TruD and its paralogs (Fig. 3A ▶, *). However, this strand is the most N-terminal structural element of TruD (Fig. 3C ▶) and the most C-terminal in other Ψ synthases (Fig. 3D ▶). The connectivity of the rest of the secondary structural elements of the catalytic domains is the same. The permutation replaces the N and C termini of the conventional Ψ synthase with a loop that connects strands β1 and β2 (Figs. 1B ▶, 3C ▶). Interestingly, this is the site of the large insertion (Figs. 1B ▶, 2 ▶, *1) that distinguishes the Pus7 proteins, capable of U2 snRNA modification, from the TruD proteins that appear to modify tRNA only.
The sequences that correspond to β1 are conserved among all TruD family members (Fig. 2 ▶). All these enzymes, therefore, are circularly permuted (Lindqvist and Schneider 1997; Jung and Lee 2001; Uliel et al. 2001) relative to members of the TruA, TruB, RluA, and RsuA families. The permuted strand (β1), which lies at the heart of the catalytic domain and is part of the floor of the active site cleft, has two of the conserved active site residues (K21 and F27) of the TruD/Pus7p family of proteins (Figs. 2 ▶, 3B ▶). Although the corresponding permuted (C-terminal) strands of TruA and TruB have the conserved hydrophobic residue (L245 and L200, respectively) in the same place as F27 of TruD, there is otherwise no sequence conservation in the permuted segment. Thus, at least with the currently available sequences, it may be impossible to determine the connectivity of the ancestral Ψ synthase from which the two superfamilies of enzymes diverged. Nonetheless, the presence of TruD orthologs in all phyla suggests that the permutation was already present in the last common ancestor several billion years ago.
Conclusions
The structure determination of a protein of the divergent TruD/Pus7p family of Ψ synthases reveals that this group of enzymes adopts the same catalytic domain architecture as all other structurally characterized Ψ synthases. The conservation extends to active site moieties, suggesting that the catalytic mechanism is also conserved. In addition, this work delineates the structure of a novel insertion domain that appears to play a role in substrate binding. The discovery that TruD/Pus7p family Ψ synthases share a topology that is distinct from all other Ψ synthases supports their early divergence. The structure opens the way to biochemical and genetic investigation of the role that the circularly permuted fold and the unique insertion domain of these enzymes play in substrate recognition.
MATERIALS AND METHODS
Protein and RNA preparation
Plasmid pECTruD is a pET15b (Novagen) derivative that encodes the E. coli pseudouridine synthase TruD (residues 1–349; E.C. 4.2.1.70) preceded by a Histag of sequence MGSSHHHHHHSS GLVPRGSH. The TruD gene insert was amplified from E. coli DH5α and cloned between the NdeI and BamHI sites of pET15b. Selenomethionyl protein was expressed in the auxotroph E. coli B834 (DE3) as described (Doublié 1997). The proteins were purified as previously described (Hoang and Ferré-D’Amaré 2001), except that all chromatography buffers had 10 mM DTT, and glycerol was omitted from the chromatography and storage buffers. Matrix-assisted laser desorption-ionization mass spectrometry confirmed absence of the initiation methionine; the protein is otherwise intact. The 7-mer RNA 5′-CGUCU[5F]AG-3′ was purchased from Dharmacon, deprotected according to the manufacturer’s instructions, and used without further purification.
Crystallization and data collection
An initial crystal form of TruD was obtained from hanging drops prepared by mixing protein at 0.25 mM and a reservoir solution consisting of 0.25 M Li2SO4, 27% (w/v) PEG4000, and 100 mM Tris-HCl (pH 8.5). This form (space group P2 or P21, a = 53Å, b = 103Å, c = 68Å, β= 98.5°) suffered from disorder and was not pursued further. A second crystal from of TruD was obtained when the protein was crystallized in the presence of the 7-mer RNA. These crystals were grown a 1:1 protein/RNA ratio at a final concentration of 0.24 mM. Hanging drops consisting of 1.5 μL each of the protein–RNA mixture in 20 mM Hepes-KOH (pH 7.5), 150 mM KCl, 10 mM DTT, 0.5 mM EDTA, and a reservoir solution (0.1 mM spermine, 1 mM MgCl2, 0.1 M Hepes-KOH at pH 7.5, 5% isopropanol, 16% PEG4000) were equilibrated against 0.5 mL reservoir solution at 22°C. Crystals grew as plates to typical dimensions of 280 × 200 × 20 μm3 during the course of a week. For data collection, crystals were briefly transferred to a solution containing 0.1 M Hepes-KOH (pH 7.5), 0.15 M KCl, 5 mM MgCl2, 5% isopropanol, 17% PEG4000, and 15% glycerol and flash-cooled in liquid nitrogen. Selenomethionine–TruD cocrystals were obtained in the same manner as for native TruD, except that the reservoir solution was 20 mM MgCl2, 0.1 M Hepes-KOH (pH 7.5), 5% isopropanol, and 10% PEG4000. These crystals typically grew to 360 × 280 × 20 μm3. The crystals (space group P212121, cell parameters in Table 1 ▶) contain two TruD molecules per asymmetric unit and have a solvent content of ~46%. Analysis of washed crystals in denaturing polyacrylamide gels demonstrated the presence of the 7-mer RNA in the crystals (data not shown). While our manuscript was under review, an independent report of the crystallization of TruD was published (Ericsson et al. 2004). Those investigators reported obtaining crystals with cell parameters virtually identical to ours under different crystallization conditions. Diffraction data were measured with CuKα X-radiation from a rotating anode equipped with multilayer optics and were reduced (Table 1 ▶) with the HKL package (Otwinowski and Minor 1997).
Phase determination and structure refinement
Eight selenium sites were located, heavy-atom parameters were refined, and SIRAS phases were calculated to 2.2 Å (Table 1 ▶) with SOLVE (Terwilliger and Berendzen 1999). Heavy-atom sites were confirmed by inspection of isomorphous difference Patterson syntheses. Density modification (Brünger et al. 1998) of the SIRAS phase probability distributions against structure factor amplitudes from crystal I produced an electron density map into which an essentially complete crystallographic model was built by using program O (Jones et al. 1991). Rounds of manual rebuilding, interspersed with torsion angle simulated annealing, conjugate gradient energy minimization, and restrained individual B-factor refinement with the program CNS (Brünger et al. 1998) produced the final model, which consists of residues 0–341 for TruD molecule A and (−4)-(−1) and 1–340 for molecule B (residues with negative and zero numbers are from the Histag), 258 water molecules, six glycerol molecules, and one phosphate ion. Refinement was against all crystal I structure factor amplitudes (|F|> 0) and SIRAS phase-probability distributions using a maximum likelihood target (Brünger et al. 1998). Throughout refinement, a solvent mask and an overall anisotropic temperature factor correction were used. The mean B-factors for protein and water are 39.69 Å2 and 39.17 Å2, respectively. The mean real-space R-factors (in a phase-combined composite annealing omit 2|Fo| − |Fc| map) are 5.0% and 5.8% for TruD molecules A and B. In the crystallographic model, residues H0 (the last residue of the Histag), K64, R92, Q105, E107, K154, Q163, N182, R186, K190, E239, E264, K293, and T341 in molecule A and E3, K67, R71, K96, E97, Q105, E107, K190, E239, R246, K293, and R321 in molecule B are missing parts of their side-chains. Residues N6, R47, E97, and E145 in molecule A and N6, H69, Q146, C152, D187, and N311 in molecule B have been built in two conformations. The model has good stereochemistry (Laskowski et al. 1993), with 90.3% of the residues in the most favored, 9.3% in the additionally allowed, 0.3% in the generously allowed, and none in the disallowed regions of the Ramachandran plot. Except for the sequence alignment, all figures were prepared with RIBBONS (Carson 1997), and they depict TruD molecule B.
Atomic coordinates
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank (accession code 1SB7).
Acknowledgments
We thank J. Bolduc, T. Edwards, T. Hamma, J. Pitt, A. Roll-Mecak, B. Shen, J. Simon, B. Stoddard, A. Weiner, and Y. Yu for discussions. Supported by grants from the National Institutes of Health (GM63576 and RR15943) and the Rita Allen Foundation. A.R.F. is a Rita Allen Foundation Scholar and a W.M. Keck Foundation Distinguished Young Scholar in Medical Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7240504.
REFERENCES
- Aravind, L. and Koonin, E.V. 1999. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 48: 291–302. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Urban, A., Ma, X., Yu, Y.T., Motorin, Y., and Branlant, C. 2003. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Ψ-synthase also acting on tRNAs. RNA 9: 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., et al. 1998. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr. D 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1997. Ribbons. Meth. Enzymol. 277: 493–505. [PubMed] [Google Scholar]
- Del Campo, M., Ofengand, J., and Malhotra, A. 2004. Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA 10: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié, S. 1997. Preparation of selenomethionyl proteins for phase determination. Meth. Enzymol. 276: 523–530. [PubMed] [Google Scholar]
- Ericsson, U.B., Andersson, M.E., Engvall, B., Nordlund, P., and Hall-berg, B.M. 2004. Expression, purification, crystallization and preliminary diffraction studies of the tRNA pseudouridine synthase TruD from Escherichia coli. Acta Crystallogr. D 60: 775–776. [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré, A.R. 2003. RNA-modifying enzymes. Curr. Opin. Struct. Biol. 13: 49–55. [DOI] [PubMed] [Google Scholar]
- Foster, P.G., Huang, L., Santi, D., and Stroud, R.M. 2000. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat. Struct. Biol. 7: 23–27. [DOI] [PubMed] [Google Scholar]
- Hoang, C. and Ferré-D’Amaré, A.R. 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 71: 929–939. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. and Kim, S.-H. 1991. Sparse matrix sampling: A screening method for crystallization of proteins. J. Appl. Cryst. 24: 409–411. [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Jung, J. and Lee, B. 2001. Circularly permuted proteins in the protein structure database. Protein Sci. 10: 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya, Y. and Ofengand, J. 2003. A novel unanticipated type of pseudourdine synthase with homologs in bacteria archaea and eukarya. RNA 9: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya, Y., Del Campo, M., Ofengand, J., and Malhotra, A. 2004. Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold. J. Biol. Chem. 279: 18107–18110. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 24: 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.J., Macarthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–290. [Google Scholar]
- Lemke, C.T., Smith, G.D., and Howell, P.L. 2002. S-SAD, Se-SAD and S/Se-SIRAS using Cu Kα radiation: Why wait for synchrotron time? Acta Crystallogr. D 58: 2096–2101. [DOI] [PubMed] [Google Scholar]
- Lindqvist, Y. and Schneider, G. 1997. Circular permutations of natural protein sequences: Structural evidence. Curr. Opin. Struct. Biol. 7: 422–427. [DOI] [PubMed] [Google Scholar]
- Ma, X., Zhao, X., and Yu, Y.-T. 2003. Pseudouridylation of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, E.G. 2002. Chips off the old block. Nat. Struct. Biol. 9: 320–322. [DOI] [PubMed] [Google Scholar]
- Newby, M.I. and Greenbaum, N.L. 2002. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 9: 958–965. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pan, H., Agarwalla, S., Moustakas, D.T., Finer-Moore, J., and Stroud, R.M. 2003. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. 100: 12648–12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phannachet, K. and Huang, R.H. 2004. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. Nucleic Acids Res. 32: 1422–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, C. and Schneider, R. 1991. Database of homology-derived protein structures and structural meaning of sequence alignment. Proteins 9: 56–68. [DOI] [PubMed] [Google Scholar]
- Sivaraman, J., Sauve, V., Larocque, R., Stura, E.A., Schrag, J.D., Cygler, M., and Matte, A. 2002. Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP. Nat. Struct. Biol. 9: 353–358. [DOI] [PubMed] [Google Scholar]
- Sivaraman, J., Iannuzzi, P., Cygler, M., and Matte, A. 2004. Crystal structure of the RluD pseudouridine synthase catalytic module, an enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli. J. Mol. Biol. 335: 87–101. [DOI] [PubMed] [Google Scholar]
- Spedaliere, C.J. and Mueller, E.G. 2004. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA 10: 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, A.E., Orengo, C.A., and Thornton, J.M. 2002. Plasticity of enzyme active sites. Trends Biochem. Sci. 27: 419–426. [DOI] [PubMed] [Google Scholar]
- Uliel, S., Fliess, A., and Unger, R. 2001. Naturally occurring circular permutations in proteins. Protein Eng. 14: 533–542. [DOI] [PubMed] [Google Scholar]
- Yu, Y.-T., Shu, M.-D., and Steitz, J.A. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. and Yu, Y.-T. 2004. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10: 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]