Abstract
Telomerase is a reverse transcriptase that synthesizes telomeric DNA repeats at the ends of eukaryotic chromosomes. Although it is minimally composed of a conserved catalytic protein subunit (TERT) and an RNA component, additional accessory factors present in the holoenzyme play crucial roles in the biogenesis and function of the enzyme complex. Telomerase from the ciliate Tetrahymena can be reconstituted in active form in vitro. Using this system, we show that p43, a telomerase-specific La-motif protein from the ciliate Euplotes, stimulates activity and increases repeat addition processivity of telomerase. Activity enhancement by p43 requires its incorporation into a TERT•RNA•p43 ternary complex but is independent of other dissociable protein factors functioning in telomerase complex assembly. Stimulation is enhanced at elevated temperatures, supporting a role for p43 in structural stabilization of a critical region of the RNA subunit. To our knowledge, this represents the first demonstration that an authentic telomerase accessory protein can directly affect the enzymatic activity of the core enzyme in vitro.
Keywords: ciliates, Euplotes, La motif, p43, telomerase
INTRODUCTION
The linear chromosomes of most eukaryotes end in tandem arrays of short DNA repeats that are essential for genome integrity. They nucleate the formation of specialized nucleoprotein complexes, called telomeres, that distinguish these natural DNA termini from double-strand breaks and protect them from nucleolytic degradation (for review, see Cervantes and Lundblad 2002). Because of the inability of the conventional DNA replication machinery to completely replicate linear DNA molecules (Watson 1972; Olovnikov 1973), however, these telomeric sequences must be replenished periodically. This is most commonly accomplished by a specialized reverse transcriptase, the ribonucleoprotein (RNP) enzyme telomerase.
Telomerase activity was first detected and characterized in the ciliated protozoon Tetrahymena thermophila (Greider and Blackburn 1985), and it remains one of the best understood. Telomerases from all organisms contain an RNA component, a portion of which serves to specify the DNA sequence added to telomeres. As Figure 1 ▶ shows, most structural elements within the ciliate RNA subunit are highly conserved (Romero and Blackburn 1991; ten Dam et al. 1991; Lingner et al. 1994; McCormick-Graham and Romero 1995). Some of them have been implicated in contributing to several aspects of enzymatic function, such as telomerase activity and processivity (Sperger and Cech 2001; Lai et al. 2003; Mason et al. 2003), so it is now becoming evident that the RNA plays much more of a role than simply providing the template for DNA synthesis. Repeat addition is catalyzed by a catalytic protein subunit, the telomerase reverse transcriptase (TERT). This protein, first identified in Euplotes aediculatus, contains amino acid motifs common to all reverse transcriptases (Lingner and Cech 1996; Lingner et al. 1997b) and is tethered to the RNA component via its TERT-specific N-terminal domain (Bryan et al. 2000b; Lai et al. 2001).
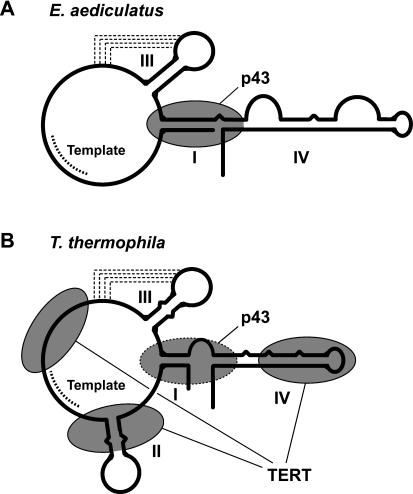
FIGURE 1.
Secondary structures of telomerase RNAs from (A) Euplotes aediculatus (Lingner et al. 1994) and (B) Tetrahymena thermophila (Romero and Blackburn 1991; ten Dam et al. 1991; McCormick-Graham and Romero 1995). Experimentally determined sites of binding of p43 (Aigner et al. 2003) and TERT (Licht and Collins 1999; Lai et al. 2003) are outlined with solid ovals, and the inferred binding site of p43 in B is depicted by an oval outlined by a dashed line.
Telomerases from most organisms show at least some degree of processive repeat addition in vitro; that is, the enzyme is capable of remaining bound to a telomeric oligonucleotide primer while extending it by multiple repeats Fig. 2 ▶). In Tetrahymena, for example, the template RNA sequence 3′-AACCCCAAC-5′ is reverse transcribed into the telomeric DNA sequence 5′-TTGGGG-3′ Fig. 2 ▶). After nucleotide polymerization reaches the 5′-end of the RNA template region, the active site of telomerase can translocate relative to the extended primer such that, upon realignment of the partially extended primer with the template 3′ end, nucleotide addition can resume (Step C in Fig. 2 ▶). In Tetrahymena, processivity has been shown to be influenced by features within both the RNA and TERT. The stem III pseudoknot and stem–loop IV of the telomerase RNA are both required for normal levels of repeat addition processivity (Lai et al. 2003; Mason et al. 2003). The identity of a specific amino acid within a reverse transcriptase motif of TERT also affects repeat addition processivity (Bryan et al. 2000a). Although there is generally no correlation between the mean telomere length of a given organism and the in vitro processivity of its telomerase, evidence suggests that processivity may be relevant for telo-mere maintenance in vivo. Exposure of human cells to a drug that predominantly reduces telomerase repeat addition processivity in vitro causes telomere shortening in vivo (Pascolo et al. 2002). Moreover, structurally distinct telomerase complexes with differing biochemical properties, including processivity, can be isolated during the different developmental stages of Euplotes crassus (Greene and Shippen 1998).
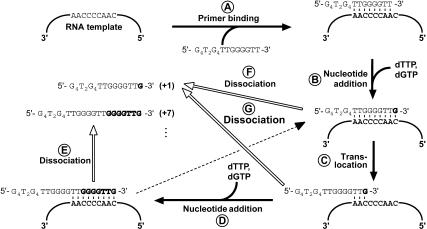
FIGURE 2.
A model for Tetrahymena telomerase action. Binding of a telomeric oligonucleotide, for instance, 5′-(G4T2)3-3′, to the enzyme involves base pairing of the 3′ end of the primer with the template region of telomerase RNA (A). Upon addition of substrate dNTPs, telom-erase catalyzes the RNA-templated extension of the primer to the 5′ end of the template region (B). At this point, the extended primer may translocate to the alignment region at the beginning of the template (C) to facilitate a new round of nucleotide addition (D), thus processively extending the primer until it dissociates from the enzyme. Product dissociation (indicated by open arrows) predominantly occurs when synthesis has reached the end of the template, either prior (E,F) or subsequent (G) to translocation. This mechanism of telomerase action results in a characteristic ladder of extension products that differ in size by 6 nt.
Upon translation in rabbit reticulocyte lysate (RRL), Tetrahymena TERT can assemble with telomerase RNA into an active telomerase RNP (Collins and Gandhi 1998; Bryan et al. 2000a) in a manner that is likely to be dependent on the action of protein chaperones (Holt et al. 1999; Forsythe et al. 2001). Indicative of a high degree of structural and functional complexity in telomerase, the in vitro reconstituted complex does not approach the degree of processivity of the enzyme isolated from cells (Collins and Gandhi 1998; Bryan et al. 2000a). Nevertheless, its activity resembles that of the endogenous enzyme in most other respects, making this system ideal for studying the function of telomerase accessory proteins.
Although the conserved core of the telomerase complex consists of TERT and the RNA subunit, all telomerase RNPs examined to date contain one or more additional protein factor(s). These accessory proteins play roles in telomerase maturation and function and are often found in other RNP complexes in the cell. For example, vertebrate telomerase RNAs contain a conserved box H/ACA small nucleolar (sno) RNA domain (Mitchell et al. 1999a; Chen et al. 2000), which recruits snoRNA-specific proteins that are critical for telomerase RNA accumulation and RNP assembly (Mitchell et al. 1999b; Dragon et al. 2000; Pogacic et al. 2000). Likewise, the budding yeast telomerase RNP is associated with the same set of Sm proteins that otherwise functions in the maturation of small nuclear (sn) RNPs involved in pre-mRNA splicing (Seto et al. 1999). In contrast, only three telomerase proteins (besides TERT), namely yeast Est1p and Est3p and Euplotes p43, appear to be telomerase specific. Est1p and Est3p, identified by a genetic approach (Lundblad and Szostak 1989; Lendvay et al. 1996), are dispensable for in vitro telomerase activity (Lingner et al. 1997a) but essential for telomere maintenance in vivo (Lendvay et al. 1996). Est1p plays a role in recruitment or activation of telomerase at the telomere (Evans and Lundblad 1999; Pennock et al. 2001; Taggart et al. 2002), but the function of Est3p is unknown.
The La-motif protein p43 was found by biochemical purification of active telomerase from Euplotes aediculatus (Lingner and Cech 1996; Aigner et al. 2000). Suggestive of a critical role in telomerase function, it is the only factor to copurify with TERT and the RNA in apparently stoichiometric amounts (Lingner and Cech 1996). We have previously shown that this protein is a bona fide telomerase-specific component that recognizes a conserved region of ciliate telomerase RNA Fig. 1 ▶) and may contribute to its proper folding (Aigner et al. 2003). We now report that p43 is an activator of ciliate telomerase that stimulates both nucleotide addition activity and processivity of the enzyme in vitro, presumably by stabilizing the RNA subunit.
RESULTS
Euplotes p43 forms a stable ternary complex with Tetrahymena TERT and telomerase RNA
Two lines of evidence from previous studies indicated that Euplotes p43 might be able to bind the Tetrahymena telom-erase RNA component and, together with Tetrahymena TERT, form a functional ternary complex. First, under conditions optimized for p43 binding to Euplotes telomerase RNA, p43 binds the Tetrahymena RNA with a dissociation constant (Kd) of around 12 nM (Aigner et al. 2003). This stability of the heterologous complex is only about fourfold less than that of the cognate interaction. The ability of the Tetrahymena RNA to substitute for the Euplotes RNA is consistent with the close relationship of their secondary structures Fig. 1 ▶). Second, the sites of TERT binding on the RNA, determined in Tetrahymena (Licht and Collins 1999; Lai et al. 2003), are distinct from the putative sites of p43 binding, as inferred from footprinting of the Euplotes p43•RNA complex (Aigner et al. 2003; Fig. 1 ▶). Based on these two observations, formation of a TERT•RNA•p43 ternary complex was not expected to be sterically hindered. However, to prevent aggregation, free p43 requires high concentrations of monovalent salt and nonionic detergent (Aigner et al. 2003)—conditions that are incompatible with both in vitro translation and telomerase function (data not shown).
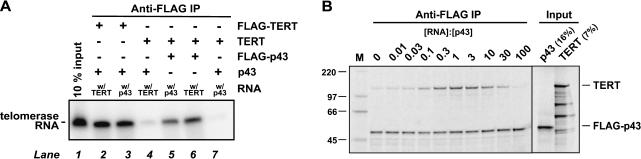
We began to address whether Euplotes p43 and Tetrahymena TERT bind Tetrahymena telomerase RNA under conditions used for activity assays (described below) by assaying coimmunoprecipitation of the RNA when either p43 or TERT was immunoprecipitated (Fig. 3A ▶). The 32P-labeled input RNA was coimmunoprecipitated on FLAG beads when either FLAG-tagged TERT (lanes 2,3) or FLAG-tagged p43 (lanes 5,6) was incubated with the untagged second protein component. RNA binding was protein dependent because only spurious amounts of RNA were coimmunoprecipitated when the protein components were untagged (lanes 4,7). RNA was coimmunoprecipitated both when p43 was incubated with preformed TERT•RNA complexes (lanes 2,6) and when TERT was incubated with preformed p43•RNA complexes (lanes 3,5).
FIGURE 3.
Euplotes p43 forms a stable ternary complex with TERT and telomerase RNA from Tetrahymena. (A) FLAG-tagged TERT (lanes 2,3) or untagged TERT (lanes 4–7) and FLAG-tagged p43 (lanes 5,6) or untagged p43 (lanes 2–4, 7) were separately translated in RRLs in the presence or absence of 32P-labeled telomerase RNA, as indicated. TERT- and p43-containing RRLs were mixed, incubated to allow binding, and subjected to anti-FLAG immunoprecipitation. Bound material was released from the beads, separated on an SDS gel, and the RNA visualized with a PhosphorImager. (Lane 1) 10% of the RNA used for immunoprecipitation. (B) RRLs containing 35S-labeled TERT and FLAG-tagged p43 were combined and unlabeled telomerase RNA was added at the relative concentrations given above the gel, whereas the concentrations of full-length p43 and TERT were held constant at ~12 nM each. The Input column shows the indicated fractions of the material used for immunoprecipitation. p43 is actually a 51-kDa protein (Aigner et al. 2000). Full-length Tetrahymena TERT is a 133-kDa protein but in vitro translation produces significant amounts of shorter fragments that do not bind RNA. (M) 14C-labeled protein markers of the indicated molecular masses (in kDa).
We next tested whether p43 forms a ternary complex with TERT and telomerase RNA from Tetrahymena under these conditions (Fig. 3B ▶). FLAG-tagged p43 and untagged TERT were translated in the presence of [35S]methionine in separate RRL reactions, and the lysates were combined so that the two proteins were present at approximately equal concentrations, estimated to be ~12 nM (Bryan et al. 2003). Next, telomerase RNA was added to achieve final RNA concentrations between 0.12 nM and 1.2 μM (that is, ranging from 0.01-fold to 100-fold RNA relative to p43/TERT), or it was omitted. After incubation to allow complex formation, samples were subjected to immunoprecipitation on anti-FLAG beads as described above, and bound material was analyzed by SDS-PAGE. PhosphorImager quantitation showed that in the absence of RNA, 2.2% ± 0.3% (mean and range of two experiments) as much TERT as p43 was coimmunoprecipitated, whereas no TERT was recovered when p43 was not present (data not shown). With increasing levels of RNA, however, the amounts of TERT that were coimmunoprecipitated increased considerably, reaching a maximum of 41% ± 6% when the concentration of the RNA was equal to that of the protein components. (The band intensities were corrected for the different methionine contents of the two proteins: 8 in p43, 22 in TERT, so the roughly equal band intensities in the lane labeled “1” mean ~2.5-fold as much p43 as TERT.) When RNA concentrations were raised further, TERT-coimmunoprecipitation dropped because formation of p43•RNA and TERT•RNA binary complexes was increasingly favored over the ternary complex.
These results indicate that a substantial fraction of RRL-expressed Euplotes p43 forms a ternary complex with Tetrahymena TERT and telomerase RNA under conditions used for activity assays (described below). Because binding of both protein subunits is largely dependent on their interactions with the RNA component, rather than based primarily on protein–protein interactions, the RNA is likely to serve as the element that connects p43 and TERT in the complex. Alternatively, the RNA may serve to stabilize a p43•TERT interaction. However, the low but significant level of interaction between Tetrahymena TERT and Euplotes p43 observed in the absence of RNA may be specific, and we cannot rule out the possibility that in a telomerase ternary complex composed entirely of Euplotes components, p43 may make contacts to both the RNA subunit and TERT that are similarly important for complex stability.
p43 enhances telomerase activity and repeat addition processivity
To elucidate whether p43 has a role in core telomerase action, we examined its effect on the activity of telomerase reconstituted in the RRL in vitro translation system (Collins and Gandhi 1998; Bryan et al. 2000a). Tetrahymena TERT expressed in RRL was mixed with preformed complexes containing telomerase RNA and purified recombinant p43 expressed in insect cells (approximate final concentrations: TERT and RNA, 20 nM each; p43, 300 nM). Total radio-labeled telomerase products formed in a standard assay increased compared to samples that only contained TERT and RNA (Fig. 4A ▶). Quantitation (Fig. 4A ▶) revealed that in this experiment, stimulation of telomerase activity was about 37%, and in repeat measurements, an increase in overall telomerase activity of 70% ± 51% (mean and standard deviation from six experiments) was observed. Although modest under these assay conditions, this stimulation was statistically significant (p = 0.0073, Student’s t test). Moreover, similar results were obtained when ternary complexes were immunopurified or when p43 translated in vitro was used instead of recombinant protein purified from insect cells (see below). In contrast, no effect was seen when heat-denatured p43 was used (data not shown). These observations further support a specific mechanism for p43 and rule out the possibility of artifactual stimulation of telomerase by buffer components or impurities in the p43 preparations. We therefore conclude that p43 acts as an activator of in vitro reconstituted telomerase.
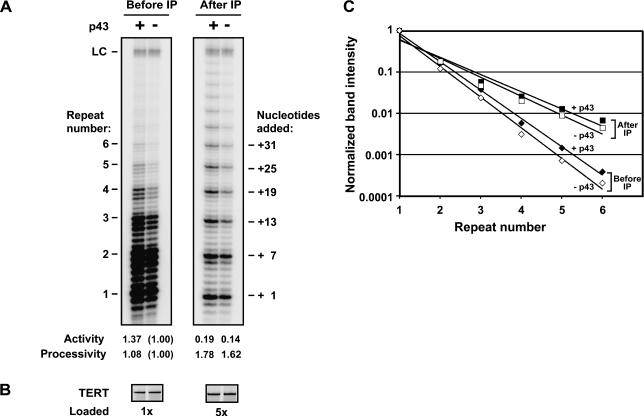
FIGURE 4.
p43 enhances both overall telomerase activity and repeat addition processivity. Purified recombinant p43 expressed in insect cells (lanes +) or p43 buffer as a control (lanes -) was incubated with telomerase RNA, followed by addition of RRL containing 35S-labeled T7-tagged TERT. Relative levels of (A) telomerase activity and (B) TERT protein in the samples were determined prior to (left panels) or after (right panels) immunopurification on T7-antibody beads. In A, the number of telomeric repeats added to the primer are indicated on the left, and the total number of nucleotides added is indicated on the right. (LC) Loading control; a radiolabeled 100-nt DNA added prior to the telomerase assay sample work-up to account for variability in recovery and gel loading. Quantitation of telomerase activity and processivity are summarized below the gel in A. For determination of the overall nucleotide incorporation activity of telomerase (“Activity”), total radioactivity in each telomerase assay lane (panel A) was quantitated, corrected for TERT levels (panel B) and for gel loading, and normalized to the activity seen in the second lane. Repeat addition processivity (“Processivity”) was quantitated as follows. The intensities of the major repeat bands (indicated in A) were normalized to the intensity of the first repeat, adjusted for specific activity, and plotted against the repeat number. Fitting of the data to a curve of the form y = C × e−x yields a log-linear relationship, and processivity is expressed as the inverse of the exponent x of the curve fit (a measure of the slope of the curve), normalized to the value of x−1 from the second lane. (C) Graphic representation of telomerase processivity. Data were plotted as described for A. (Solid and open diamonds) Data from samples in the presence and absence of p43, respectively, before immunoprecipitation. (Solid and open squares) Data from samples in the presence and absence of p43, respectively, after immunoprecipitation.
In addition to its effect on overall telomerase activity, the presence of p43 changed the length distribution of telom-erase products. As shown in Figure 4A ▶, the fraction of higher molecular weight telomerase products increased when p43 was part of the telomerase complex, indicative of a stimulatory effect on repeat addition processivity. Processivity can be quantified separately from overall nucleotide incorporation activity by measuring the intensities of corresponding repeat bands in each lane (Fig. 4A ▶, denoted by +7, +13, +19, and so forth) and normalizing them to the intensity of the first (+1) bands. Plotting the band intensities against the repeat number and fitting the data to an exponential equation yields a log-linear relationship whose slope is inversely related to processivity (Hammond and Cech 1997; Bryan et al. 2000a). When quantified in this manner, the +p43 samples reproducibly showed a modest stimulation of processivity, as judged from the decrease in the slope of the curve (Fig. 4A,C ▶).
Previous analysis of reconstituted Tetrahymena telomerase had shown that when the telomerase complex is immunopurified, the enzyme’s overall activity decreases, whereas its processivity increases (Bryan et al. 2000a). The gain in processivity upon immunopurification is probably due to the loss of an inhibitor of processivity present in the RRL. Similarly, the decrease in overall activity of the immunopurified complex is due to the loss of protein components in the RRL, including chaperones, that stimulate telomerase activity (Holt et al. 1999; Bryan et al. 2000a; Forsythe et al. 2001). In the light of these observations, it was important to test whether p43’s effect was independent of the presence of such dissociable factors. Enhancement of telomerase activity by p43 was observed both before (Fig. 4A ▶, left panel) and after isolation and extensive washing of the complex on antibody beads (right panel). Quantitation of processivity and activity (Fig. 4A,C ▶) normalized for TERT levels (Fig. 4B ▶) confirmed that the extent of p43-dependent processivity and activity stimulation was similar both before and after immunopurification.
In summary, these results indicate that p43 modestly enhances telomerase activity and also has a small but significant positive influence on repeat addition processivity. Both effects are independent of dissociable RRL components, suggesting that the protein functions in a different pathway to stimulate enzymatic activity. Similarly, p43-dependent stimulation of telomerase processivity apparently is not achieved by mere suppression of the inhibitory activity present in the RRL but must instead employ a distinct mechanism.
The extent of telomerase stimulation correlates with the fraction of p43-containing telomerase complexes formed
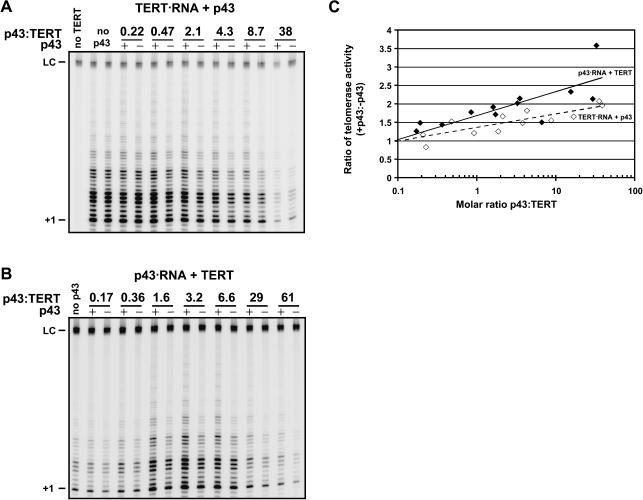
If p43 exerts its effect on telomerase by binding to the telomerase complex, the degree of stimulation of telomerase activity by p43 is expected to be dependent on the concentration of p43 during ternary complex assembly. Using RRL-expressed 35S-labeled proteins, p43 concentration can be varied by mixing the RRLs containing the TERT•RNA complex and p43 at different volume ratios. Thus a range of molar ratios of p43:TERT spanning 2.5 orders of magnitude can be achieved, as determined from SDS gels by Phosphor-Imager quantitation. As shown in Figure 5 ▶, substoichiometric amounts of p43 had limited effect on activity, whereas 10–50-fold excess of p43 over TERT approximately doubled telomerase activity.
FIGURE 5.
The extent of telomerase stimulation by p43 depends on the p43 concentration and the order of ternary complex assembly. (A) RRLs containing p43 (lanes +) or control RRL (lanes -) were mixed with RRLs containing preformed complexes consisting of telomerase RNA and RRL-expressed TERT at the indicated molar ratios of p43:TERT and assayed for telomerase activity. The concentration of TERT decreased at high ratios of p43:TERT, resulting in decreased overall activity. (B) RRLs containing TERT and RRLs containing preformed complexes consisting of telomerase RNA and RRL-expressed p43 (lanes +) or telomerase RNA only (lanes -) were mixed at the indicated molar ratios of p43:TERT and assayed for telomerase activity. At low p43:TERT, RNA is limiting and at very high p43:TERT, TERT is limiting, resulting in decreased activity at the high and low ends. (C) Quantitation of two such p43 titrations. For each titration point, the level of telomerase activity in the presence of p43 relative to that in the absence of p43 was determined and plotted against the molar ratio of p43:TERT. (Open diamonds) p43 added to preformed complexes of TERT and telomerase RNA (as described in A); (solid diamonds) TERT added to preformed complexes of p43 and telomerase RNA (as described in B). (Dashed and solid lines) Log-linear fits of the datapoints denoted by the open and solid symbols, respectively.
The protein concentrations in these samples ranged from ~2 nM p43 and ~10 nM TERT for the first titration point (molar ratio of p43:TERT = 0.22) to ~40 nM p43 and ~1 nM TERT for the last titration point (molar ratio of p43:TERT = 38). These values are close to the Kd observed for the p43•RNA complex under conditions optimized for this interaction (Aigner et al. 2003) but the Kd for the current buffer conditions may be higher and/or the fraction of p43 molecules competent for RNA specific binding may be lower. The increase of telomerase activity stimulation by p43 with increasing p43 concentration was statistically highly significant (data not shown) and therefore likely due to an increase in the fraction of ternary complexes formed.
In the experiment described in Figure 5A ▶, p43 was added to preformed TERT•RNA complexes. Upon changing the order of addition of the protein components such that TERT is added to preformed p43•RNA complexes, p43-dependent stimulation was again seen (Fig. 5B ▶). However, as is apparent upon quantitation of the titration data (Fig. 5C ▶), here the levels of stimulation were approximately twofold higher. The difference in the slopes of the lines fitted to the two data sets (TERT•RNA + p43, Fig. 5A ▶; and p43•RNA + TERT, Fig. 5B ▶) was statistically significant at the 80% confidence level. Moreover, the R2 value, a measure of the “goodness” of the fit, suggests that the two data sets are distinct because fits to the individual data sets result in higher R2 values than when the data sets are combined (R2 = 0.74 for TERT•RNA + p43 and R2 = 0.67 for p43•RNA + TERT, but R2 = 0.54 for the combined data sets). Based on these two criteria, we believe the observed difference in stimulation is modestly significant. This apparent dependence on the order of ternary complex is intriguing and deserves further investigation because it suggests that TERT may have a higher affinity to telomerase RNA that is prebound by p43. This may be due to the fact that p43 alters the conformation of the RNA to allow more efficient TERT binding and is suggestive of such an order of complex assembly in vivo.
p43 does not merely enhance the nuclease resistance of telomerase RNA
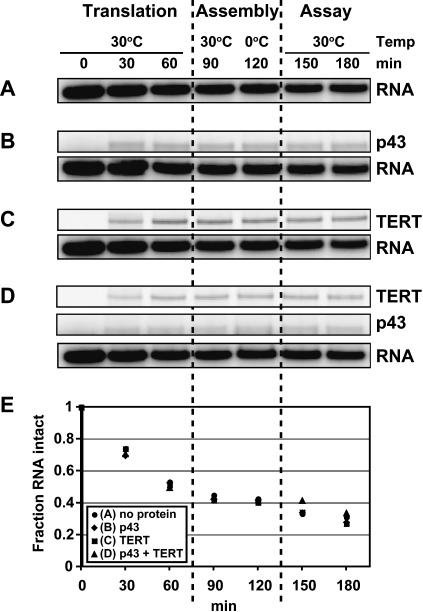
Under standard telomerase reconstitution conditions, the RNA subunit is added prior to protein translation at a concentration of 20 nM, roughly equal to that of TERT in the RRL after translation is completed (Bryan et al. 2000a, 2003). If p43 rendered the RNA component of telomerase less susceptible to nucleolytic degradation—via inhibition of nuclease activity or by blocking nuclease access to the RNA—then it would simply increase the fraction of active telomerase complexes. However, this scenario appeared unlikely because decreasing the concentration of telomerase RNA in the RRL by 50-fold (to 0.4 nM) or increasing it by 40-fold (to 800 nM) did not significantly change the degree of p43 stimulation (data not shown), suggesting that nuclease resistance of the RNA was not affected by p43 binding. To test this directly, we assayed the level of RNA during a timecourse that included translation of TERT and/or p43, complex assembly, and telomerase assay Fig. 6 ▶). Although RNA levels decreased considerably throughout the time-course (and the RNA subunit was expected to become limiting after protein translation was completed), no significant differences were seen between the fractions of intact RNA remaining in RRLs expressing no protein (Fig. 6A ▶), p43 only (Fig. 6B ▶), TERT only (Fig. 6C ▶), or coexpressing p43 and TERT (Fig. 6D ▶), as shown by Phophorimager quantitation (Fig. 6E ▶). Because similar results were obtained when recombinant p43 from insect cells was used and when the telomerase assay temperature was raised to 35°C (see below), we conclude that protection of telomerase RNA from nucleolytic degradation can be discounted as the basis of p43’s stimulation of activity.
FIGURE 6.
Binding of p43 does not protect telomerase RNA against degradation during the assay. Trace-32P-labeled telomerase RNA at the concentration used in a standard telomerase reaction was incubated in RRLs expressing no protein (A), p43 (B), TERT (C), or in RRL coexpressing p43 and TERT (D). After translation for 1 h, the RRLs were incubated at 30°C and on ice for 30 min each to allow complex formation, followed by a mock assay for telomerase activity for 1 h at 30°C. Aliquots were removed at 30-min intervals, subjected to gel electrophoresis, and analyzed using a PhosphorImager. (E) Quantitation of the data in A–D. The amount of RNA present at each timepoint was normalized to the amount of RNA at time = 0 min and plotted against the time of incubation.
p43 stimulation is enhanced at elevated temperatures
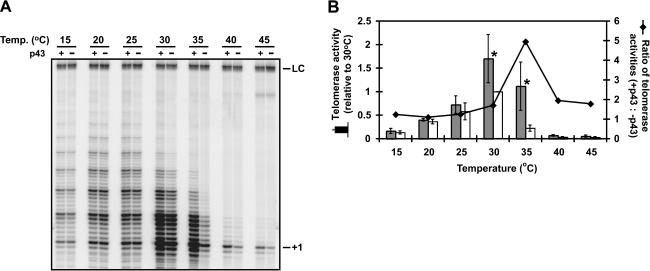
If increased RNase resistance can be ruled out, what alternative mechanism might be responsible for the observed increase in enzyme activity when p43 is bound to telomerase RNA? One likely possibility is that p43 contributes to structural stabilization of the RNA, as suggested both by direct RNA structure mapping (Aigner et al. 2003) and by the fact that p43 belongs to a class of proteins previously implicated as RNA chaperones (Aigner et al. 2000; Wolin and Cedervall 2002). We therefore reasoned that if productive folding of the RNA subunit is indeed promoted by p43, then conditions that destabilize RNA structure—for example, elevated temperatures—should enhance telomerase’s dependence on p43. Figure 7 ▶ shows that this may indeed be the case. Increasing the assay temperature by just 5°C (from 30°C to 35°C) resulted in an almost fivefold drop in telomerase activity when p43 was not included in the reactions. Strikingly, p43 was able to counteract this temperature-induced loss in activity, providing a fivefold stimulation of telomerase at 35°C compared to samples lacking p43. No statistically significant effect of p43 was seen above 35°C, perhaps because at these temperatures, p43 itself becomes thermally destabilized and is no longer able to bind the RNA. Control experiments confirmed that the observed peak in activity enhancement at 35°C was due to the temperature increase itself, and not due to the temperature-dependent change in the pH of the Tris buffer employed (data not shown). Our observations are therefore consistent with the idea that p43 contributes to telomerase function by stabilizing its RNA subunit in a manner that promotes ribonucleoprotein assembly.
FIGURE 7.
The stimulatory effect of p43 on telomerase activity is most pronounced at slightly elevated temperature. (A) Telomerase complexes were assembled with and without p43 as described in the legend to Figure 4 ▶ and assayed at the indicated temperatures. (B) Quantitation of the data shown in A. Mean telomerase activities (± standard deviations) in the presence (gray columns) and absence (white columns) of p43 were determined at the indicated temperatures in three independent experiments and are expressed relative to the mean telomerase activity measured at 30°C in the absence of p43. (Asterisks) Statistically significant (p < 0.01, Student’s t test) differences in activity. The line represents the ratios of telomerase activities with p43 relative to those without p43 at the indicated temperatures.
DISCUSSION
Previous results established Euplotes p43 as a bona fide telomerase-specific subunit that recognizes the RNA component and may influence its structure (Aigner et al. 2003). Now we have studied p43’s role in telomerase function, using the well-characterized Tetrahymena in vitro reconstitution system (Collins and Gandhi 1998; Bryan et al. 2000a). Our observations lend credence to the notion that this p43-induced structural stabilization of the RNA may play a role in the core enzymatic function of telomerase. In addition, the fact that Euplotes p43 is able to act, in a heterologous fashion, on Tetrahymena telomerase strongly suggests the existence of functional p43 homologs in other—and perhaps all—ciliates. Indeed, preliminary sequence data obtained from The Institute for Genomic Research Web site at http://www.tigr.org shows that the Tetrahymena genome contains an ORF predicting a 65-kDa protein product containing a La motif with 36% amino acid identity (51% similarity) to the La motif of Euplotes p43 (our unpublished results). This ORF may therefore encode the Tetrahymena ortholog of p43 (which is in fact a 51-kDa protein; Aigner et al. 2000).
p43-induced stabilization of stem IV, a conserved interrupted stem–loop element of ciliate telomerase RNA, has been suggested by the fact that this region shows enhanced susceptibility toward digestion by the double-strand-specific RNase V1 when the RNA is bound by p43 (Aigner et al. 2003). Stem IV is critically involved both in the correct folding of other portions of the RNA and, even though distant from the template region in the RNA secondary structure, in enzyme activity and processivity (Sperger and Cech 2001; Lai et al. 2003; Mason et al. 2003). Our data are therefore consistent with a model in which this critical RNA element undergoes structural “fine-tuning” by p43 under normal physiological conditions, but relies more heavily on p43-induced stabilization under conditions that weaken RNA interactions, such as elevated temperature Fig. 7 ▶). These p43-induced changes in RNA structure may increase the affinity of TERT to bind to the RNA, as is suggested by the dependence of the degree of p43 activation on the order of ternary complex assembly Fig. 5 ▶). Alternatively, p43 may directly affect the conformation of the enzyme’s active site.
The modest overall magnitude of telomerase activation by p43 in our experiments may be due to dissociation of p43 from the ternary complex. In this case, the fraction of telomerase present as a TERT•RNA•p43 ternary complex would decrease during the course of the telomerase assay and thus, the contribution of p43-containing telomerase particles within the large pool of TERT•RNA complexes would be considerably underestimated. This scenario is supported by the fact that at higher molar ratios of p43:TERT, more activation is observed Fig. 5 ▶); approaches the effect of p43 saturation in Figure 5C ▶ (note semi-log scale). In addition, a subset of p43 molecules may be mis-folded and therefore incompetent for RNA binding, whereas other aberrantly folded p43 molecules might retain RNA-binding activity yet fail to stimulate activity. Thus, the stimulation reported here provides a minimum estimate of the effect of p43 on telomerase activity and processivity.
Although not yet observed in other organisms, binding of accessory protein factors to the RNA subunit as a means to directly regulate one or more aspects of telomerase enzyme activity may have a precedent in ciliates. In Euplotes crassus, a dramatic increase in the size of the telomerase particle is observed during macronuclear development (Greene and Shippen 1998), a stage in the ciliate life cycle in which gene-sized chromosomes are generated by excision from regular chromosomes and de novo telomere addition (for review, see Prescott 1994). This so-called macronuclear development is accompanied by a shift in the expression profile of this organism’s three TERT genes (Karamysheva et al. 2003) and correlates with an increase in telomerase processivity and relaxation of primer specificity in vitro (Bednenko et al. 1997; Greene and Shippen 1998). Because protection from nuclease digestion in stem I and adjacent regions of telomerase RNA is observed in telomerase isolated from developing macronuclei, these changes in the biochemical properties of telomerase have been attributed to an as-yet unidentified dissociable protein factor, dubbed the chromosome healing factor (CHF), that is proposed to bind to the RNA (Bednenko et al. 1997; Greene and Shippen 1998). Given that p43 recognizes this same region of the RNA in vegetatively growing (nondeveloping) Euplotes aediculatus cells, stem I and its vicinity may serve as a dedicated binding site for proteins that modulate telomerase function in ciliates.
A very different way for a protein to alter telomerase activity was recently described by Lin and Blackburn (2004). The yeast homolog of human PinX1 (Zhou and Lu 2001), yPinX1p (also known as Gno1p), is a nucleolar protein that competes with TERT for binding to telomerase RNA and thereby sequesters the catalytic subunit in a nonfunctional complex that lacks the RNA subunit (Lin and Blackburn 2004).
The roles we observe for p43 in telomerase enzymatic action do not exclude the possibility that p43 may also contribute to telomerase RNP maturation. Initially in evolution, p43 might have exclusively shepherded the biogenesis of the telomerase complex, but then the RNP might have evolved to partially rely on p43 binding for structural stabilization as well. Indeed, such a dual role for p43 as a telomerase-specific La-motif protein is suggested by the La protein itself. Although primarily functioning in the biogenesis of all pol III precursor transcripts (Wolin and Cedervall 2002), La is required by a subset of these RNAs, namely some tRNAs, for stabilization of their mature forms, thus facilitating efficient aminoacylation (Yoo and Wolin 1997; Chakshusmathi et al. 2003). As may be the case for p43’s effect on the conformation of telomerase RNA, the dependence of these yeast tRNAs on structural stabilization by La is limited in wild-type cells under normal growth conditions but is very pronounced when RNA structure is disturbed by mutation (Yoo and Wolin 1997) or changes in growth temperature (Chakshusmathi et al. 2003).
Ideally, the roles of p43 in telomerase, especially with respect to RNP biogenesis, would be tested by compromising p43’s function in vivo. The recent advances in RNAi methodology in ciliated protozoa (Galvani and Sperling 2002; Mollenbeck et al. 2003) may make this approach more feasible, and using this approach, evidence has been presented that p43 appears to anchor telomerase in the Euplotes macronucleus (Mollenbeck et al. 2003). In addition, given that Tetrahymena is much more genetically tractable than Euplotes, the finding of a p43 ortholog in Tetrahymena, if confirmed, would allow further elucidation of the function of ciliate La-motif proteins in telomerase biogenesis and catalysis.
MATERIALS AND METHODS
Plasmids, protein expression, and RNA transcription
Plasmid pET28a-TERT encoding N-terminally T7-tagged Tetrahymena TERT has been described (Bryan et al. 2000b). Plasmid pFLAG-TERT-His encoding N-terminally FLAG-tagged and C-terminally His-tagged TERT was a generous gift from T. Bryan. It was modified from plasmid pFLAG-TERT (Bryan et al. 2003) by insertion of oligonucleotides to remove the stop codon at the C terminus of TERT. Plasmid pET15b-p43-FLAG encoding N-terminally FLAG-tagged Euplotes p43 was constructed by subcloning a synthetic DNA duplex encoding the FLAG epitope flanked by NcoI and NdeI restriction sites into the respective sites of plasmid pET15b-p43 (Aigner et al. 2000) TERT and p43 were produced from these plasmids in the TnT rabbit reticulocyte lysate (RRL) transcription/translation system according to the manufacturer’s instructions (Promega). In cases where [35S]methionine labeling of proteins was not required, unlabeled methionine at 20 μM final concentration was used in place of [35S]methionine. Alternatively, purified N-terminally FLAG-tagged p43 expressed in insect cells was used where indicated.
In vitro transcription and purification of telomerase RNA was done as described (Zaug and Cech 1995; Bryan et al. 2000a).
Telomerase reconstitution
Telomerase was reconstituted from Tetrahymena TERT and telomerase RNA as described (Bryan et al. 1998, 2000a). To allow formation of the ternary complex consisting of in vitro transcribed telomerase RNA and RRL-expressed TERT and p43, the proteins were expressed in separate RRL reactions (containing 20 nM telomerase RNA where indicated), mixed, and incubated for 30 min at 30°C, followed by 30 min on ice. Aliquots of the 35S-labeled proteins were run on SDS gels to allow quantitation of protein levels by PhosphorImager analysis as described (Bryan et al. 2003).
Where indicated, p43 (15 μM) from insect cells was incubated with telomerase RNA (1 μM) in p43 binding buffer (Aigner et al. 2003) for 1 h on ice. The preformed complex was then diluted 1:50 into RRL containing TERT and incubated as above to allow ternary complex formation.
Immunoprecipitation and telomerase assay
Immunoprecipitation on T7-antibody or FLAG-antibody beads and telomerase assays were done as described (Bryan et al. 2000a). Briefly, samples were incubated with blocked antibody beads, followed by extensive washing of the beads at elevated salt concentration (300 mM potassium glutamate) and resuspension in 10 μL of buffer TMG (10 mM Tris-acetate at pH 8.0, 1 mM MgCl2, 10% glycerol).
Telomerase activity assays were done as described (Bryan et al. 2000a). Briefly, 10-μL samples of RRL or a 1:1 slurry of antibody beads in TMG were incubated for 1 h at 30°C in a final reaction volume of 20 μL in the presence of 50 mM Tris-Cl (pH 8.3), 1.25 mM MgCl2, 5 mM DTT, 1 μM primer 5′-(G4T2)3-3′, 100 μM dTTP, and 10 μM radiolabeled dGTP (80 Ci/mmole). Assay products were extracted in phenol and chloroform, recovered by ethanol precipitation, and separated on denaturing 10% polyacrylamide gels.
NOTE ADDED IN PROOF
The 65-kDa La-motif protein predicted from our database searches of the Tetrahymena genome has now been identified in telomerase purified from Tetrahymena extracts by an epitope-tag approach (Witkin and Collins 2004).
Acknowledgments
We thank Tracy Bryan for generous gifts of plasmids and help with quantitating protein levels, and members of the Cech lab for fruitful discussions. This work was supported by grant GM28039 from the NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7400704.
REFERENCES
- Aigner, S., Lingner, J., Goodrich, K.J., Grosshans, C.A., Shevchenko, A., Mann, M., and Cech, T.R. 2000. Euplotes telomerase contains a La motif protein produced by apparent translational frameshifting. EMBO J. 19: 6230–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner, S., Postberg, J., Lipps, H.J., and Cech, T.R. 2003. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42: 5736–5747. [DOI] [PubMed] [Google Scholar]
- Bednenko, J., Melek, M., Greene, E.C., and Shippen, D.E. 1997. Developmentally regulated initiation of DNA synthesis by telomerase: Evidence for factor-assisted de novo telomere formation. EMBO J. 16: 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, T.M., Sperger, J.M., Chapman, K.B., and Cech, T.R. 1998. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. 95: 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, T.M., Goodrich, K.J., and Cech, T.R. 2000a. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 275: 24199–24207. [DOI] [PubMed] [Google Scholar]
- ———. 2000b. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 6: 493–499. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Tetrahymena telomerase is active as a monomer. Mol. Biol. Cell 14: 4794–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes, R.B. and Lundblad, V. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14: 351–356. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi, G., Kim, S.D., Rubinson, D.A., and Wolin, S.L. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J. 22: 6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.-L., Blasco, M.A., and Greider, C.W. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514. [DOI] [PubMed] [Google Scholar]
- Collins, K. and Gandhi, L. 1998. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. 95: 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., Pogacic, V., and Filipowicz, W. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20: 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, S.K. and Lundblad, V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120. [DOI] [PubMed] [Google Scholar]
- Forsythe, H.L., Jarvis, J.L., Turner, J.W., Elmore, L.W., and Holt, S.E. 2001. Stable association of hsp90 and p23, but not hsp70, with active human telomerase. J. Biol. Chem. 276: 15571–15574. [DOI] [PubMed] [Google Scholar]
- Galvani, A. and Sperling, L. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18: 11–12. [DOI] [PubMed] [Google Scholar]
- Greene, E.C. and Shippen, D.E. 1998. Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes & Dev. 12: 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C.W. and Blackburn, E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Hammond, P.W. and Cech, T.R. 1997. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 25: 3698–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, S.E., Aisner, D.L., Baur, J., Tesmer, V.M., Dy, M., Ouellette, M., Trager, J.B., Morin, G.B., Toft, D.O., Shay, J.W., et al. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes & Dev. 13: 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva, Z., Wang, L., Shrode, T., Bednenko, J., Hurley, L.A., and Shippen, D.E. 2003. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell 113: 565–576. [DOI] [PubMed] [Google Scholar]
- Lai, C.K., Mitchell, J.R., and Collins, K. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.K., Miller, M.C., and Collins, K. 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 11: 1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay, T.S., Morris, D.K., Sah, J., Balasubramanian, B., and Lundblad, V. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht, J.D. and Collins, K. 1999. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & Dev. 13: 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. and Blackburn, E.H. 2004. Nucleolar protein PinX1p regulates telomerase by sequestering its protein catalytic subunit in an inactive complex lacking telomerase RNA. Genes & Dev. 18: 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J. and Cech, T.R. 1996. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. 93: 10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Hendrick, L.L., and Cech, T.R. 1994. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 8: 1984–1998. [DOI] [PubMed] [Google Scholar]
- Lingner, J., Cech, T.R., Hughes, T.R., and Lundblad, V. 1997a. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. 94: 11190–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Hughes, T.R., Shevchenko, A., Mann, M., Lundblad, V., and Cech, T.R. 1997b. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567. [DOI] [PubMed] [Google Scholar]
- Lundblad, V. and Szostak, J.W. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Mason, D.X., Goneska, E., and Greider, C.W. 2003. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 23: 5606–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Graham, M. and Romero, D.P. 1995. Ciliate telomerase RNA structural features. Nucleic Acids Res. 23: 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., Cheng, J., and Collins, K. 1999a. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., Wood, E., and Collins, K. 1999b. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555. [DOI] [PubMed] [Google Scholar]
- Mollenbeck, M., Postberg, J., Paeschke, K., Rossbach, M., Jonsson, F., and Lipps, H.J. 2003. The telomerase-associated protein p43 is involved in anchoring telomerase in the nucleus. J. Cell Sci. 116: 1757–1761. [DOI] [PubMed] [Google Scholar]
- Olovnikov, A.M. 1973. A theory of marginotomy. J. Theor. Biol. 41: 181–190. [DOI] [PubMed] [Google Scholar]
- Pascolo, E., Wenz, C., Lingner, J., Hauel, N., Priepke, H., Kauffmann, I., Garin-Chesa, P., Rettig, W.J., Damm, K., and Schnapp, A. 2002. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 277: 15566–15572. [DOI] [PubMed] [Google Scholar]
- Pennock, E., Buckley, K., and Lundblad, V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396. [DOI] [PubMed] [Google Scholar]
- Pogacic, V., Dragon, F., and Filipowicz, W. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20: 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, D.M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58: 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, D.P. and Blackburn, E.H. 1991. A conserved secondary structure for telomerase RNA. Cell 67: 343–353. [DOI] [PubMed] [Google Scholar]
- Seto, A.G., Zaug, A.J., Sobel, S.G., Wolin, S.L., and Cech, T.R. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribo-nucleoprotein particle. Nature 401: 177–180. [DOI] [PubMed] [Google Scholar]
- Sperger, J.M. and Cech, T.R. 2001. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40: 7005–7016. [DOI] [PubMed] [Google Scholar]
- Taggart, A.K., Teng, S.C., and Zakian, V.A. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026. [DOI] [PubMed] [Google Scholar]
- ten Dam, E., van Belkum, A., and Pleij, K. 1991. A conserved pseu-doknot in telomerase RNA. Nucleic Acids Res. 19: 6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, J.D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239: 197–201. [DOI] [PubMed] [Google Scholar]
- Witkin, K.L. and Collins, K. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes & Dev. 18: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S.L. and Cedervall, T. 2002. The La protein. Annu. Rev. Biochem. 71: 375–403. [DOI] [PubMed] [Google Scholar]
- Yoo, C.J. and Wolin, S.L. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402. [DOI] [PubMed] [Google Scholar]
- Zaug, A.J. and Cech, T.R. 1995. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1: 363–374. [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.Z. and Lu, K.P. 2001. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107: 347–359. [DOI] [PubMed] [Google Scholar]