Abstract
Both cis elements and host cell proteins can significantly affect HIV-1 RNA processing and viral gene expression. Previously, we determined that the exon splicing silencer (ESS3) within the terminal exon of HIV-1 not only reduces use of the adjacent 3′ splice site but also prevents Rev-induced export of the unspliced viral RNA to the cytoplasm. In this report, we demonstrate that loss of unspliced viral RNA export is correlated with the inhibition of 3′ end processing by the ESS3. Furthermore, we find that the host factor Sam68, a stimulator of HIV-1 protein expression, is able to reverse the block to viral RNA export mediated by the ESS3. The reversal is associated with a stimulation of 3′ end processing of the unspliced viral RNA. Our findings identify a novel activity for the ESS3 and Sam68 in regulating HIV-1 RNA polyadenylation. Furthermore, the observations provide an explanation for how Sam68, an exclusively nuclear protein, modulates cytoplasmic utilization of the affected RNAs. Our finding that Sam68 is also able to enhance 3′ end processing of a heterologous RNA raises the possibility that it may play a similar role in regulating host gene expression.
Keywords: 3′ end processing, Sam68, HIV-1 RNA, Rev
INTRODUCTION
Expression of the full complement of HIV-1 proteins depends on a number of post-transcriptional processes (Tang et al. 1999). Suboptimal splicing of the primary transcript generates over 30 mRNAs (Schwartz et al. 1990; Purcell and Martin 1993). Many of these mRNAs, including those that encode the viral proteins Gag, Gagpol, Vif, Vpr, Vpu, and Env, are intron containing and are therefore normally retained in the nucleus. Transport of the incompletely spliced viral RNAs is mediated by the action of the virus-encoded Rev protein. Rev interacts with the Rev Response Element (RRE) within the 9-kb and 4-kb viral RNAs and complexes with Crm1/Exportin 1 in a RanGTP-dependent manner to mediate export of these RNAs to the cytoplasm (Pollard and Malim 1998; Hope 1999).
In light of the number of RNAs that must be produced from the primary HIV-1 transcript to generate the full spectrum of viral proteins, the regulation of splicing plays a central role in the virus life cycle. This regulation is exercised at several levels. At the level of cis-acting signals, splicing is controlled by the suboptimal nature of the 3′ splice sites (Staffa and Cochrane 1994; O’Reilly et al. 1995) as well as the exon splicing enhancers (ESEs) and exon splicing silencers (ESSs) throughout the RNA, which act to promote and inhibit recognition of the adjacent splice site, respectively (Amendt et al. 1994; Staffa and Cochrane 1995; Si et al. 1998; Caputi et al. 1999; Bilodeau et al. 2001). Moreover, the ESE and ESS within the terminal exon also regulate viral RNA transport. Deletion of the ESE results in loss of Rev-induced transport of the unspliced RNA to the cytoplasm (Pongoski et al. 2002). The inhibition of Rev-induced RNA transport is ESS dependent because deletion of both the ESE and the ESS restores transport of the RNA.
In addition to viral sequences, host factors can also negatively or positively modulate viral RNA splicing and transport (Luo et al. 1994; Powell et al. 1997; Bilodeau et al. 2001; Jacquenet et al. 2001; Tange and Kjems 2001; Tange et al. 2001; Pongoski et al. 2002). Sam68 (Src associated during mitosis of 68 kDa), a member of the STAR (signal transduction and activation of RNA) family of proteins (Fumagalli et al. 1994; Taylor and Shalloway 1994) is able to increase Rev function in a number of cell lines (Reddy et al. 1999, 2000; Soros et al. 2001; Li et al. 2002a,b). Other members of the STAR protein family have activities that are indistinguishable from Sam68 (Soros et al. 2001; Reddy et al. 2002). These proteins all have a domain with homology to the KH RNA binding motif (Gibson et al. 1993; Siomi et al. 1993) located within a conserved region designated the GSG domain (Di Fruscio et al. 1999). Sam68 is a nonshuttling, nuclear protein that shows accumulation within foci in the nucleus (Huang 2000; Soros et al. 2001). In addition to its effect on HIV-1 Rev function, Sam68 can also enhance cytoplasmic utilization of RNAs and modulate alternative splicing of the CD44 gene (Matter et al. 2002; Coyle et al. 2003). A truncation mutant of Sam68 (lacking the C-terminal 100 amino acids, designated Sam68ΔC) is a potent inhibitor of Rev activity (Reddy et al. 1999; Soros et al. 2001). In contrast to Sam68, Sam68ΔC is localized predominately in the cytoplasm (Reddy et al. 1999, Soros et al. 2001) and its inhibition of HIV-1 gene expression is associated with accumulation of both the protein and viral RNA at the nuclear periphery (Soros et al. 2001).
In this report, we demonstrate that the ESS within the terminal exon (ESS3) blocks cleavage and polyadenylation of the RNA in addition to its effects on splicing. Furthermore, we show that expression of Sam68 is able to enhance 3′ end processing of unspliced viral RNA concomitant with an increase in viral protein synthesis. Stimulation of 3′ end processing is observed for all viral constructs tested as well as in a heterologous context (doublesex [dsx] gene), suggesting that Sam68 may affect multiple genes within the cell. However, the Sam68 stimulation of RNA 3′ end processing was not accompanied by significant changes in the cytoplasmic accumulation of viral RNA. This finding suggests that the Sam68-induced increase in HIV-1 gene expression must be achieved through increasing the translation efficiency of the viral RNA.
RESULTS
The ESS within the terminal exon of HIV-1 inhibits 3′ end processing of viral RNA
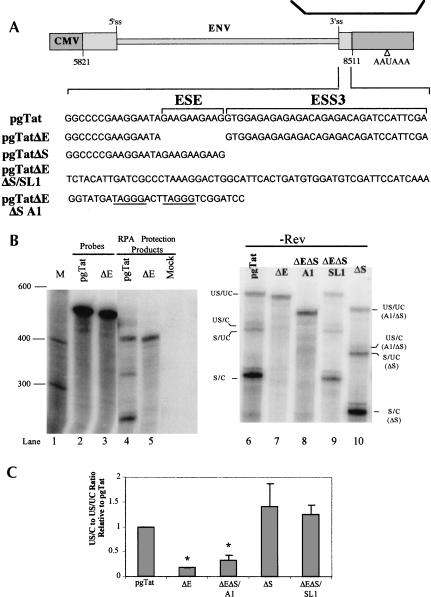
In this study, we use the previously characterized HIV-1 reporter, pgTat, derived from the env portion of HIV-1 hxb3 (Fig. 1 ▶; Malim et al. 1989). The vector contains the first coding exons of Tat and Rev and the sequence encoding gp120 but lacks the last 105 C-terminal amino acids of gp41 and much of the second coding exon of Rev. Therefore, export of the intron-containing env RNA and subsequent gp120 expression is observed only when Rev is provided in trans. With regard to RNA processing signals, the vector contains the native 5′ and 3′ splice sites (ss) and ESE and ESS3 previously identified in the terminal exon (Staffa and Cochrane 1995).
FIGURE 1.
Inhibition of env RNA 3′ end processing by ESS3. (A) Schematic of reporter constructs used. Boxes denote exon sequences, the bar the intron sequences within pgTat. Four variants of pgTat were analyzed to assay the impact of various deletions within the terminal exon on 3′ processing of the RNAs generated. Shown are the sequences of a portion of the terminal exon. The vector pgTat contains both the ESE and ESS3, pgTatΔE lacks the ESE, pgTatΔS lacks the ESS3, pgTatΔEΔS SL1 has replaced both the ESE and ESS3 with a sequence derived from the EDA exon of the fibronectin gene, and pgTatΔEΔS A1 has replaced the ESE and ESS3 with two copies of the consensus hnRNP A1-binding site (underlined sequence). The solid line above indicates the position of the RNA probe used to monitor both splicing and 3′ end processing of the generated RNAs. Due to variations in the exon sequence of the various pgTat mutants, RPA probes were generated for each construct tested. (B) Cells were transfected with pgTat, pgTatΔE (ΔE), pgTatΔS (ΔS), pgTatΔEΔS SL1 (ΔEΔS SL1) or pgTatΔEΔS A1 (ΔEΔS A1). Total RNA was isolated and used in RNA protection assays. Following treatment with ribonuclease, samples were fractionated on denaturing PAGE gels and the location of bands determined following exposure to phosphor screens. (Left) An example of the RNA protection data obtained, showing the size markers (M), input probes (Probes), and protected products (RPA Protection products) observed for untransfected cells (mock) or cells transfected with either pgTat or pgTatΔE (ΔE). (Right) An example of the protections seen for the full panel of mutants tested. Identities of the various bands observed are indicated; unspliced, uncleaved (US/UC); unspliced, cleaved (US/C); spliced, uncleaved (S/UC); spliced, cleaved (S/C). Expected sizes of protected bands for pgTat are: US/UC 441 nt, US/C 365 nt, S/UC 353 nt, and S/C 277 nt. (C) Summary of RPA data. Shown is a compilation of multiple trials indicating the changes in US/C to US/UC ratio for the various constructs tested relative to pgTat. Statistical analysis of data indicates that results for pgTatΔE and pgTatΔΔΔΔEΔS A1 (*) are different from pgTat as well as from each other (p < 0.05).
Previously, we demonstrated that the ESS3 within the terminal exon of HIV-1 not only inhibits use of the adjacent 3′ ss but also blocks Rev-mediated export of the unspliced viral RNA to the cytoplasm (Pongoski et al. 2002; Asai et al. 2003). This block to Rev-mediated export was not due solely to the inhibition of splicing because replacement of ESS3 with high affinity hnRNP A1-binding sites was found to yield a comparable reduction in splicing but did not inhibit RNA transport (Asai et al. 2003). Consequently, to identify the basis for the ESS3-induced block to unspliced RNA transport, our focus shifted to events other than splicing that are implicated in controlling RNA nuclear/cytoplasmic transport. Previous work established that inhibiting 3′ end processsing of RNA polymerase II transcripts prevents RNA transport (Custodio et al. 1999; Brodsky and Silver 2000; Hilleren and Parker 2001; Hilleren et al. 2001; Dower and Rosbash 2002). This point is particularly relevant to HIV-1, as an earlier report found that Rev-mediated export is dependent on proper 3′ end processing of the viral RNA (Huang and Carmichael 1996). Taken together, these observations suggest that the effect of ESS3 on Rev function could be mediated by alterations in the extent of viral RNA polyadenylation.
To test this hypothesis, we examined five constructs: pgTat, containing both the ESE and ESS3; pgTatΔE, which lacks the ESE; pgTatΔS, which lacks the ESS3; pgTatΔEΔS SL1, which has replaced the ESE and ESS3 with a similar size sequence from the EDA exon of the human fibronectin gene; and pgTatΔEΔS A1, in which we replaced the ESE and ESS3 with two consensus hnRNP A1 binding sites (see Fig. 1A ▶). To analyze 3′ end processing of both spliced and unspliced viral RNAs, we performed RNA protection assays (RPAs) using a probe that spanned both the 3′ ss and the polyadenylation signal. Four protection products are generated corresponding to the unspliced, uncleaved RNA (US/ UC); unspliced, 3′ -end-processed RNA (US/C); spliced, un-cleaved RNA (S/UC), and spliced, 3′ -end-processed RNA (S/C). As described earlier (Pongoski et al. 2002), deletion of the ESE causes a significant inhibition of viral RNA splicing that is restored upon removal of both the ESE and ESS3 (Fig. 1B ▶, lane 7 versus lane 9). Analysis of total RNA isolated from cells transfected with the plasmids demonstrates that the ESS3 inhibits not only splicing but also the formation of the 3′ -end-processed RNAs. Deletion of the enhancer (pgTatΔE) leads to a significant reduction in both the level of splicing and 3′ end processing (Fig. 1B ▶, lanes 5,7 vs. lanes 4,6). Replacement of the native ESE and ESS3 with two high affinity hnRNP A1-binding sites (pgTatΔEΔS A1) also inhibits splicing but yields higher levels of US/C RNA (Fig. 1B ▶, lane 8; Fig. 1C ▶). Deletion of the ESS3 alone (pgTatΔS) resulted in all four forms of viral RNA being detected but more of the S/UC and S/C RNAs accumulated (Fig. 1B ▶, lane 10). A compilation of RPA data from multiple, independent trials is provided in Figure 1C ▶. Analysis demonstrated that only pgTatΔE and pgTatΔEΔS A1 yield significant differences in the extent of unspliced env RNA 3′ end processing relative to pgTat. Furthermore, RNAs from pgTatΔEΔS A1 reproducibly have higher levels of 3′ end processing than pgTatΔE.
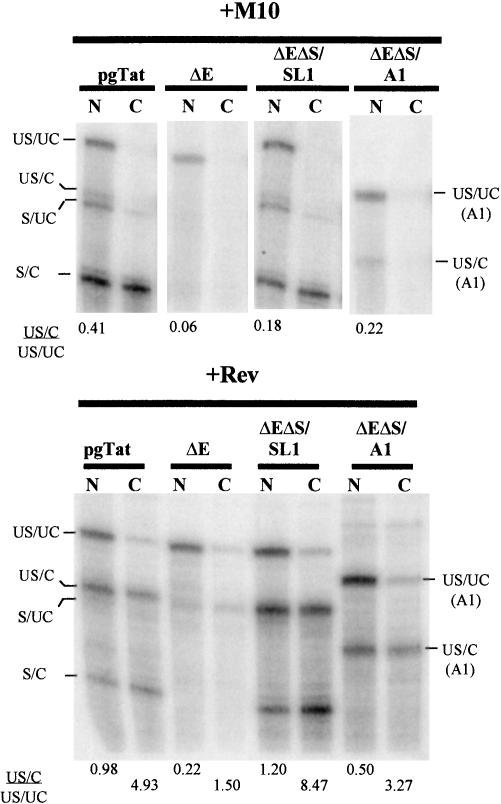
The differences in the extent of cleavage at the polyadenylation signal of the unspliced RNA among the various constructs allowed us to ask which form of the unspliced viral RNA was transported into the cytoplasm upon Rev expression. Cells were transfected with pgTat or deletion/ substitution variants thereof in the presence or absence of functional Rev, nuclear and cytoplasmic fractions isolated, and RNA analyzed. Upon comparison of nuclear and cytoplasmic fractions in the presence of Rev, we observed differences in the ratio of cleaved to uncleaved unspliced viral RNAs (US/C to US/UC ratio; Fig. 2 ▶). In the nucleus, the US/UC form is equivalent or predominant to the US/C form of the RNA. However, US/C RNA is the major form of unspliced viral RNA in the cytoplasm, indicating that Rev selectively transports this RNA. Cytoplasmic accumulation of unspliced RNA was reduced in the case of pgTatΔE.
FIGURE 2.
Selective transport by Rev of 3′ -end-processed RNA to the cytoplasm. To examine which of the different RNAs are transported to the cytoplasm, cells transfected with pgTat, pgTatΔE (ΔE), pgTatΔEΔS SL1 (ΔEΔS SL1), or pgTatΔEΔS A1 (ΔEΔS A1) in the absence (+M10) or presence (+Rev) of functional Rev were separated into nuclear and cytoplasmic fractions and RNA isolated. M10 is a variant of Rev, having several point mutations within the NES, rendering it unable to transport its target RNA to the cytoplasm. RNA was used in ribonuclease protection assays using the RNA probes described in Figure 1 ▶. Following treatment with ribonuclease, samples were fractionated on denaturing PAGE gels and the location of bands determined following exposure to phosphor screens. Indicated at right and left are the identities of the various bands observed; unspliced, uncleaved (US/UC); unspliced, cleaved (US/C); spliced, uncleaved (S/ UC); spliced, cleaved (S/C). Shown at bottom of each lane is the US/C to US/UC ratio for each cell fraction examined.
Sam68 reverses the block to env RNA expression mediated by the exon splicing silencer
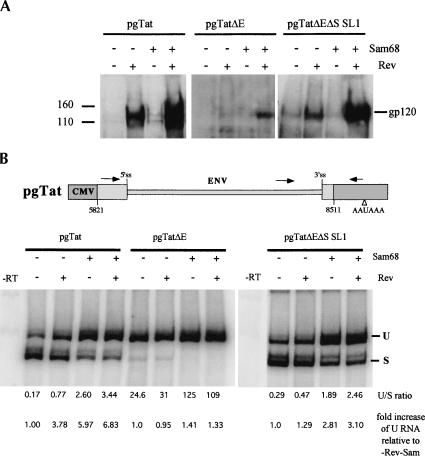
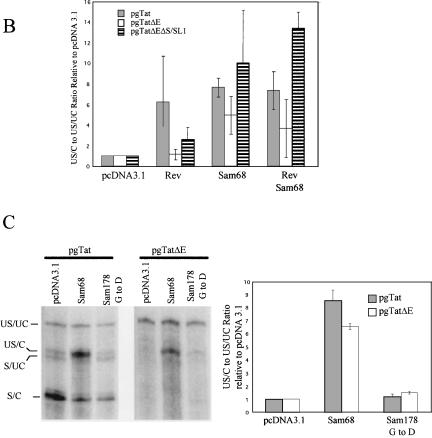
Recent studies have demonstrated that overexpression of Sam68, a host cell protein, can dramatically stimulate Rev function. As a demonstration of Sam68 function, we studied the effect of its co-expression on gp120 production from pgTat, pgTatΔE, or pgTatΔEΔS SL1. Consistent with previous work (Soros et al. 2001), Sam68 expression resulted in increased production of gp120 from pgTat and pgTatΔEΔS SL1 relative to that seen with Rev alone (Fig. 3A ▶). In the absence of Rev, little expression above background was detected. In contrast, expression of Rev alone was not sufficient to induce gp120 expression from pgTatΔE. However, coexpression of Rev and Sam68 did induce significant levels of gp120 production from pgTatΔE.
FIGURE 3.
Effect of Sam68 on HIV-1 env RNA expression and splicing. (A) Effect of Sam68 on env expression. Cells were transfected with pgTat, pgTatΔE, or pgTatΔEΔS SL1 in the presence (+) or absence (−) of Rev and/or Sam68. Two days post-transfection, cells were harvested and lysates fractionated on SDS-PAGE gels. Following transfer to PVDF membranes, blots were probed with antibody against gp120. (B) Effect of Sam68 on env RNA splicing. Shown is a schematic of the expression vector used to assay the effect of coexpression of Sam68 mutants on viral RNA splicing. (Arrows) Position of primers used to detect the unspliced (U) and spliced (S) forms of the RNA by RT-PCR. To assess whether the effects of Sam68 are dependent on the presence of either ESE or ESS3, transfections were performed with pgTat or vectors lacking only the ESE (pgTatΔE) or both the ESE and ESS3 (pgTatΔEΔS SL1) in the presence or absence of Rev and Sam68. Forty-eight hours post-transfection, total RNA was extracted and the level of unspliced and spliced RNAs determined by RT-PCR. Reactions were performed in the linear range of amplification. Unspliced to spliced RNA (U/S) ratios were determined following exposure to phosphor screens. To confirm that signals were not due to contaminating DNA, parallel reactions were run using pooled samples in the absence of RT (−RT).
In principle, Sam68 could achieve these effects by altering viral RNA processing/transport, increasing cytoplasmic utilization of the RNA by the translational apparatus of the cell, or both. However, the inability of Sam68 to shuttle between the nucleus and cytoplasm favors a model in which it affects a nuclear event such as 5′ capping, splicing, or 3′ cleavage/polyadenylation of the RNA. To assess what effect Sam68 had on viral RNA splicing, we transfected cells with pgTat, pgTatΔE, or pgTatΔEΔS SL1 in the presence or absence of Rev and/or Sam68 and determined the levels of unspliced and spliced viral RNAs by RT-PCR. Cotransfection with either Rev or Sam68 increased the ratio of unspliced to spliced RNA (U/S) for pgTat and pgTatΔEΔS SL1, with an increase in the amount of unspliced env RNA being observed (Fig. 3B ▶). Expression of Sam68 yielded the greatest increase in the level of unspliced viral RNA.
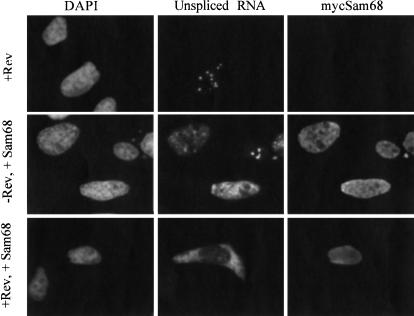
We showed previously that continued viral RNA synthesis was required for Rev to induce transport of the RNA (Iacampo and Cochrane 1996). This observation suggests that there is a window of opportunity during viral RNA synthesis or shortly thereafter when the HIV-1 RNA is accessible to Rev-mediated export. If the interaction with Rev does not occur at this time, the unspliced RNA becomes committed to nuclear sequestration and degradation. This commitment to the sequestration pathway appears to be facilitated by the ESS3 within the terminal exon, because this element is able to block both splicing and Rev-mediated export of viral RNA to the cytoplasm (Pongoski et al. 2002). The ability of Sam68 to induce gp120 expression from pgTatΔE (Fig. 3A ▶) suggests that it might enhance Rev function through counteracting the effect of the ESS3. To test this hypothesis, we transfected cells with the env expression vector pgTatΔE with plasmids expressing Rev, Sam68, or both proteins. Localization of unspliced RNA was subsequently determined by in situ hybridization. As shown in Figure 4 ▶, expression of either Rev or Sam68 alone does not result in detectable cytoplasmic accumulation of unspliced RNA from the pgTaTΔE vector. Although a low level of unspliced pgTat&dDgr;E RNA was detected in the cytoplasm in the presence of Rev alone upon fractionation (Fig. 2 ▶), this may be below the threshold of detection for the in situ assay. In contrast, coexpression of both Sam68 and Rev is found to induce the transport of the unspliced env RNA to the cytoplasm. This observation indicates that Sam68 facilitates cytoplasmic transport of RNA from this vector.
FIGURE 4.
Sam68 is able to restore Rev-induced transport of unspliced RNA from pgTatΔE. To assess whether Sam68 stimulates Rev function through inhibition of ESS3 activity, cells were transfected with pgTatΔE in the presence of Rev alone (+Rev), Sam68 alone (−Rev, +Sam68), or both Rev and Sam68 (+Rev, +Sam68). Cells were subsequently fixed and the localization of unspliced env RNA (unspliced RNA), Sam68 (mycSam68), and nuclei (DAPI) determined. Magnification was 630×.
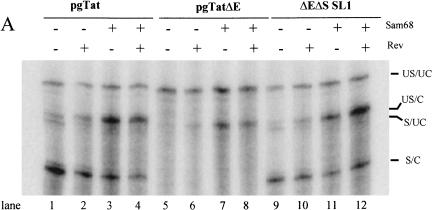
Effects of Sam68 on viral RNA 3′ end processing
The localization of unspliced RNA from pgTatΔE in discrete foci within the nucleus (Fig. 4 ▶) is very similar to observations in other systems when 3′ end processing of RNA is disrupted (Custodio et al. 1999; Brodsky and Silver 2000; Hilleren and Parker 2001; Hilleren et al. 2001; Dower and Rosbash 2002). The ability of Sam68 to reverse this localization suggests that it might play a role in viral RNA processing. To test whether Sam68 expression was affecting the extent of unspliced env RNA 3′ end processing, we looked for its effect on 3′ end cleavage of the unspliced env RNA. Cells were transfected with pgTat, pgTatΔE, or pgTatΔEΔS SL1 in the presence and absence of Sam68 and the extent of 3′ end processing of unspliced and spliced viral RNA analyzed using the probes detailed in Figure 1 ▶. Coexpression of Sam68, in the presence or absence of Rev, resulted in a marked increase in the proportion of unspliced RNA that underwent 3′ end processing for all constructs tested (Fig. 5A ▶, cf. lanes 3,4,7,8,11,12 and lanes 1,2,5,6,9,10). The effect of Sam68 on the processing of pgTatΔE RNA is particularly striking. Although very little 3′ end processing occurred for pgTatΔE RNA in the absence of Sam68 (Fig. 5A ▶, lanes 5,6), coexpression of Sam68 in the presence or absence of Rev significantly increased the level of 3′ -end-processed forms of the unspliced RNA (Fig. 5A ▶, lanes 7,8). A summary of results from multiple, independent assays is presented in Figure 5B ▶. Of particular note, Sam68 stimulation of 3′ end processing occurred without an overall increase in the level of spliced RNA for any of the constructs tested. To test whether this effect required interaction of Sam68 with RNA, we examined whether a mutant form of Sam68 (Sam 178 G to D), deficient in RNA binding (Chen et al. 1997), affected its function (Fig. 5C ▶). Although Sam68 increased the level of US/C RNA from pgTat and pgTatΔE, expression of the Sam 178 G to D mutant had little or no effect.
FIGURE 5.
Sam68 coexpression stimulates 3′ end processing of unspliced env RNA. (A) Sam68 enhances 3′ end processing of unspliced env RNA. Cells were transfected with pgTat, pgTatΔE, or pgTatΔEΔS SL1 (ΔEΔS SL1) in the presence or absence of vectors expressing Rev and Sam68. Total RNA was subsequently extracted and used in RNA protection assays to look for changes in the extent of 3′ end processing of unspliced and spliced viral RNAs. Following digestion, samples were run on denaturing PAGE gels and the position of bands determined following exposure to phosphor screens. Indicated on the right are the identities of the various bands expected; unspliced, uncleaved (US/UC); unspliced, cleaved (US/C); spliced, uncleaved (S/UC); spliced, cleaved (S/C). At the bottom of each lane is the ratio of US/C to US/UC for each of the samples analyzed. (B) Summary of RPA data. Shown is a compilation of data from multiple trials examining changes in the US/C to US/UC ratio for the indicated plasmid in response to coexpression of Rev alone, Sam68 alone, or both Rev and Sam68. Values are expressed relative to ratios observed in the absence of Rev or Sam68. (C) RNA binding is required for Sam68 function. Cells were transfected with pgTat or pgTatΔE with a control vector (pcDNA3.1), one expressing Sam68 (Sam68), or an RNA binding mutant of Sam68 (Sam 178 G to D). Two days post-transfection, total RNA was harvested and used in a RPA assay to detect the various forms of env RNA present. (Right) Compilation of RPA data from multiple trials, all values normalized to the extent of cleavage observed upon cotransfection with pcDNA3.1.
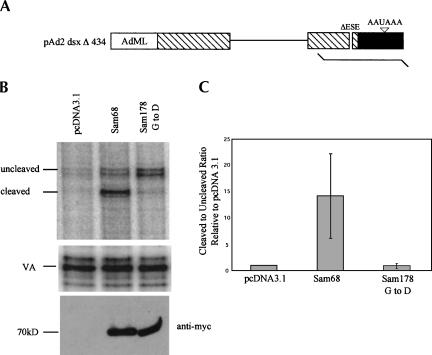
In light of above results, we used the pAd2dsxΔ434 vector derived from the Drosophila doublesex (dsx) gene to test whether the ability of Sam68 to enhance 3′ end formation was a feature unique to HIV-1 env RNA or reflected a more general enhancement of RNA 3′ end processing (Fig. 6 ▶; Szymczyna et al. 2003). This vector contains dsx intron and exon sequences driven by the adenovirus major late promoter but lacks the ESE required for efficient splicing and 3′ end formation. Upon transfection of pAd2dsxΔ434 alone, only a small fraction of the RNA undergoes 3′ end cleavage. Coexpression of Sam68 increased the extent of 3′ end processing ~10-fold. In contrast, no such increase occurred when the Sam 178 G to D mutant was expressed.
FIGURE 6.
Sam68 stimulates 3′ end processing of Drosophila doublesex (dsx) RNA. (A) Schematic of Drosophila doublesex (pAd2 dsxΔ434) expression construct used, boxes and lines denoting exon and intron sequences, respectively. The line below denotes the location of the RNA probe used in the RPA assays. (B) Effect of Sam68 on dsx RNA processing. Cells were transfected with pAd2 dsxΔ434 in the presence of control vector (pcDNA3.1), Sam68 (Sam68), or the Sam68 RNA binding mutant Sam 178 G to D (Sam 178 G to D). Two days post-transfection, total RNA was isolated and levels of uncleaved (UC) and cleaved (C) dsx RNA determined by RPA. Protected products were detected following exposure to phosphor screens. Shown are the ratios of cleaved to uncleaved RNA for each of the samples analyzed. To verify equivalent loading of RNA, parallel reactions were run to detect expression from the cotransfected VA plasmid. Similarly, Western blots were performed to confirm expression of the myc-tagged Sam68 and Sam 178 G to D mutant (anti-myc). (C) Summary of RPA data. Shown is a compilation of data from multiple trials examining changes in the cleaved to uncleaved dsx RNA upon coexpression of Sam68 or Sam 178 G to D. Results are expressed relative to values observed upon transfection with pAd2 dsxΔ434 alone.
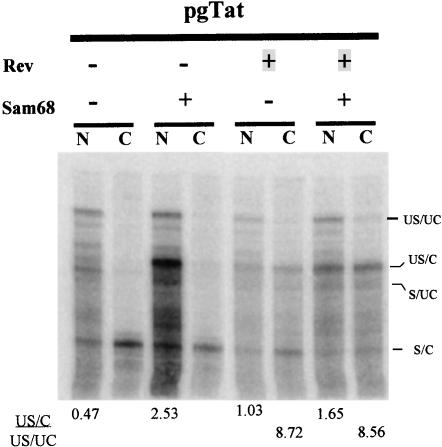
Sam68 stimulation of 3′ end processing does not lead to a correlative increase in cytoplasmic accumulation of viral RNA
To test whether the increase in 3′ end processing induced by Sam68 resulted in an enhancement of unspliced viral RNA export, we examined the nuclear and cytoplasmic fractions for the various forms of env RNA (Fig. 7 ▶). As expected, Rev increased the amount of US/C RNA in the cytoplasm. However, expression of both Rev and Sam68 did not lead to a substantial increase in the amount of US/C RNA in the cytoplasm despite higher levels of this RNA in the nucleus. This result contrasts with our finding that coexpression of Sam68 increased the amount of gp120 from the same vector (Fig. 3A ▶). The significance of this difference between the effects of Sam68 on protein expression and RNA transport is considered further in the Discussion.
FIGURE 7.
Effect of Sam68 on the nuclear/cytoplasmic distribution of HIV-1 env RNA. Cells were transfected with pgTat in the presence or absence of functional Rev and/or Sam68. Nuclear and cytoplasmic fractions were subsequently prepared and RNA extracted and used in RNA protection assays. Following fractionation on denaturing PAGE gels, the location of bands was determined by exposure to phosphor screens. Indicated at right are the identities of the various bands observed; unspliced, uncleaved (US/UC); unspliced, cleaved (US/C); spliced, uncleaved (S/UC); spliced, cleaved (S/C). Also shown is the ratio of US/C to US/UC for each of the samples analyzed.
DISCUSSION
Previous studies on cis-acting sequences affecting HIV-1 RNA processing have cataloged a number of elements that regulate usage of the adjacent 3′ ss (Amendt et al. 1994; O’Reilly et al. 1995; Staffa and Cochrane 1995; Si et al. 1998; Caputi et al. 1999; Bilodeau et al. 2001; Jacquenet et al. 2001). This report is the first demonstration that the ESS3 within the terminal exon of HIV-1 blocks RNA transport by inhibiting the cleavage and subsequent polyadenylation of the unspliced RNA. Based on the extent of cleavage of RNA containing ESS3 versus consensus hnRNP A1-binding sites, the effect on 3′ end processing is not solely attributable to an inhibition of splicing. That is, both the ESS3 and consensus hnRNP A1 binding sites result in a similar inhibition of env RNA splicing (Fig. 1 ▶; Asai et al. 2003), but only RNAs containing ESS3 are blocked in cleavage and transport to the cytoplasm (Fig. 2 ▶). The inhibition of 3′ end processing is ESS3 dependent because its substitution with other sequences (pgTatΔEΔS SL1) restores processing to levels seen for pgTat. These observations suggest that the complex assembled on ESS3 consists of more than the multiple hnRNP A1 molecules that have been shown to bind to this region (Tange et al. 2001; Zhu et al. 2001; Damgaard et al. 2002; Marchand et al. 2002).
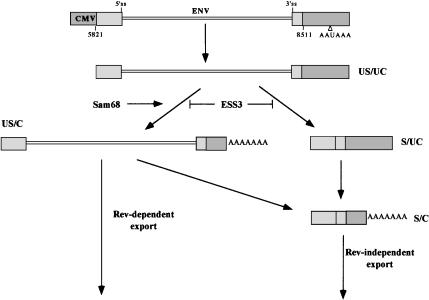
Analysis of the data in Figure 1 ▶ shows that the most abundant form of env RNA is the S/C, indicating that once the RNA is either spliced or cleaved, it is generally committed to becoming fully processed (i.e., spliced and cleaved). These results are consistent with recent observations that splicing and 3′ end cleavage are mutually stimulatory (Bentley 2002; Millevoi et al. 2002; Neugebauer 2002; Proudfoot et al. 2002). The detection of both US/C and S/UC species indicates that splicing and cleavage can occur independently but that the unspliced or uncleaved forms are either rapidly processed to S/C RNA or degraded. This poses a problem for HIV-1 because Rev-dependent transport requires generation of the US/C form of the RNA as shown in Figure 2 ▶ and as demonstrated previously (Huang and Carmichael 1996). The similar extent of viral RNA processing and transport seen for pgTat and pgTatΔEΔS SL1 (Figs. 1 ▶, 2 ▶) suggests that the ESE is required for RNA splicing and 3′ end processing only to counter the effect of the ESS3. These data, combined with previous work showing that Rev-transported RNA is a labile species within the nucleus (Iacampo and Cochrane 1996), suggest a model for the fate of viral RNA following transcription (Fig. 8 ▶). If splicing occurs first, the S/UC RNA is processed to the S/C form and subsequently transported to the cytoplasm in a Rev-independent fashion. However, if 3′ end processing occurs first, unspliced RNA has two potential fates. Either it is bound by Rev and is transported to the cytoplasm or it undergoes splicing to generate the S/C form of the RNA. However, some env RNA generated does not follow any of the above pathways. If the ESS3-directed complex forms, the RNA does not commit to either splicing or 3′ cleavage. As a result, this ESS3-env RNA complex remains trapped within the nucleus and is most likely degraded. The balance between the productive versus the dead end fate of the unspliced env RNA appears to be determined by the relative activities of the ESE and ESS3.
FIGURE 8.
Outline of HIV-1 env RNA metabolism. Following synthesis, the US/UC form of env RNA has two potential fates: (1) undergoing cleavage and polyadenylation to generate the US/C form or (2) being spliced to produce S/UC RNA. The ESS3 acts at this stage to block both processing events. In contrast, Sam68 selectively promotes the formation of the US/C form of env RNA. Once generated, the S/UC RNA can be further processed by 3′ end cleavage/ polyadenylation to generate S/C RNA that is subsequently transported to the cytoplasm in a Rev-independent fashion. The US/C RNA has two possible fates: (1) undergoing splicing to generate S/C RNA or (2) interacting with Rev and being transported into the cytoplasm through the Crm-1 pathway. Indicated are the identities of the various RNAs observed; unspliced, uncleaved (US/UC); unspliced, cleaved (US/C); spliced, uncleaved (S/UC); spliced, cleaved (S/C).
In light of the above model, factors that inhibit or stimulate Rev activity could do so by modulating the extent of cleavage/polyadenylation of the unspliced RNA. One factor that augments Rev activity is Sam68 (Reddy et al. 1999, 2000; Soros et al. 2001; Li et al. 2002a). As shown here, Sam68 not only increased gp120 production from pgTat but also rescued Rev-dependent expression of gp120 from pgTatΔE (Fig. 3A ▶), an effect correlated with the stimulation of 3′ end cleavage of the RNA. The finding that Sam68 is also able to stimulate gp120 production and 3′ end processing of unspliced RNA from pgTatΔEΔS SL1 suggests that Sam68 functions by affecting the cleavage process directly and not by antagonizing ESS3 activity. The ability of Sam68 to elicit a similar enhancement of dsxΔ434 3′ end formation further supports the hypothesis that it is acting at a more general level to alter RNA processing. In light of previous observations regarding the coupled nature of RNA splicing and 3′ end processing (Bentley 2002; Millevoi et al. 2002; Neugebauer 2002; Proudfoot et al. 2002), it is of note that Sam68 stimulation of 3′ end processing occurs without an increase in the level of spliced viral RNA. Cotransfection of Sam68 results in increased levels of unspliced RNA and decreased amounts of the spliced form in the presence or absence of Rev (Fig. 3B ▶). Although the increased levels of unspliced env RNA could be attributed to a stabilization of the RNA due to the presence of a poly(A) tail, the reduction in levels of the spliced form suggests an uncoupling of 3′ end processing and splicing. Otherwise, the stimulation of 3′ end processing would be expected to increase the level of cleaved, spliced viral RNA.
The mechanism by which Sam68 achieves the stimulation of 3′ end processing of unspliced viral RNA is unclear, although it has been recently demonstrated that Sam68 can function much like a SR protein in the stimulation of alternative splicing within the CD44 system (Matter et al. 2002). SR and SR-related proteins have been demonstrated to affect both splicing and RNA 3′ end processing (Zahler et al. 1992; Lou et al. 1996; Manley and Tacke 1996; Blencowe et al. 1999; McCracken et al. 2002). The absence of an equivalent protein–protein interaction domain (the RS repeats) within Sam68 raises the question as to how it may be mediating equivalent interactions. Although recent work has documented the interaction of Sam68 with both hnRNP K and the transcriptional cofactor CBP (Hong et al. 2002; Yang et al. 2002), the nature of these interactions does not immediately suggest a model for the effect of Sam68 on RNA processing. However, analysis of Sam68 high affinity binding sites within RNA has determined a preference for UAAA that forms part of the AAUAAA consensus sequence of the eukaryotic polyadenylation signal (Lin et al. 1997; Itoh et al. 2002). This fact raises the possibility that Sam68 may interact directly with the polyadenylation signal to recruit the necessary machinery for RNA cleavage and polyadenylation. In support of this hypothesis, a mutation within the RNA-binding domain of Sam68 (Sam 178 G to D), which results in loss of RNA binding, was unable to stimulate 3′ end processing of either env or dsxΔ434 RNA. Further analysis of the domains of Sam68 required for stimulation of Rev activity and the testing for interactions with components of the polyadenylation machinery could provide insights into its mechanism of action. However, the observation that Sam68 is able to modulate RNA 3′ end processing reveals a novel activity for this factor and suggests that it may play a similar role in the case of host genes whose polyadenylation signals may be suboptimal.
Although we have found a correlation between the ability of Sam68 to stimulate RNA 3′ end formation and the production of HIV-1 structural protein (gp120), the increase in protein synthesis is not a result of a comparable increase in cytoplasmic accumulation of the unspliced viral RNA (Fig. 7 ▶). This is despite the increased level of nuclear US/C RNA, the favored substrate for Rev export. However, these findings are similar to recent work examining the effect of Sam68 in a comparable system. Sam68 increased protein production of HIV-1 Gag expressed using the CTE transport element without affecting the cytoplasmic accumulation of the RNA, suggesting that it acts to enhance the translation of the RNA (Coyle et al. 2003). However, the question remains how this nuclear, nonshuttling protein (Soros et al. 2001) can alter the cytoplasmic fate of the RNA. One suggestion is that Sam68 deposits a mark on the RNA that promotes its utilization by the translational machinery (Coyle et al. 2003) comparable to the exon junction complex left after an RNA is spliced (Kataoka et al. 2000; Le Hir et al. 2000a,b). Recent work has demonstrated that splicing of an RNA can enhance synthesis of the encoded protein in a manner that is not directly correlated with changes in RNA abundance and without affecting the cytoplasmic accumulation of the affected RNA (Lu and Cullen 2003; Nott et al. 2003). In light of this precedent, it is possible that Sam68, in the course of promoting 3′ end formation, induces formation of an RNP complex that affects the translation efficiency of the RNA. Given the role of the poly(A) tail in promoting translation through the binding of PABP and interaction with eIF4G (Gallie 1998; Wells et al. 1998), complexes that facilitate or inhibit these interactions would be anticipated to have significant effects on the translation efficiency of a particular mRNA. Further studies will be necessary to test these predictions.
MATERIALS AND METHODS
Expression constructs
The following plasmids have been previously described: Bl SVhygro, SVH6Rev (ΔOlsen et al. 1990), pgTat (Malim et al. 1989), Bl env-HindIII (Seguin et al. 1998), pgTatΔE, pgTatΔEΔS SL1, pgTatΔEΔS A1 (Asai et al. 2003), pcDNA3.1 (Invitrogen), and pcDNA3.1-based myc-tagged Sam68 (Soros et al. 2001). The pAd2dsxΔ434 was provided by Dr. B. Blencowe (University of Toronto, Canada) and has been described previously (Szymczyna et al. 2003).
Cell lines and transfections
HeLa and 293 cells were maintained in Is-cove’s modified Dulbecco’s media (IMDM) supplemented with 10% fetal bovine serum (FBS), 50 μg/mL gentamycin sulfate, and 2.5 μg/mL amphotericin B. For transient expression studies, vectors were introduced by calcium phosphate transfection (Kriegler 1990). Two days post-transfection, cells were harvested for analysis.
For the analysis of the effect of Sam68 on gp120 production from pgTat or mutants thereof, transfected cells were harvested two days post-transfection in 2× dissociation buffer (0.125 M Tris-HCl at pH 6.8, 4% SDS, 20% glycerol, 0.2% bromophenol blue), boiled, and fractionated on 7% SDS PAGE gels. Following transfer to PVDF membrane, blots were probed with monoclonal mouse anti-gp120 antibody (kindly provided by H. Schaal, Heinrich-Heine-University of Duesseldorf, Germany) and developed using horseradish peroxidase conjugated donkey anti-mouse antibody and the Western Lightning kit (Perkin-Elmer).
In situ hybridization
Transfections for in situ hybridization were performed on 2 × 105 HeLa cells as follows: 1.25 μg pgTatΔE, 0.25 μg BlSVhygro or BlSVH6Rev, and 5 μg pcDNA3.1 or the myc-tagged Sam68 as described in Results. DNA was equalized to 6.5 μg per transfection. In situ hybridization was performed as previously described (Seguin et al. 1998). Digoxi-genin-labeled Env-HindIII probe, antisense to HIV-1 env mRNA, was used to probe unspliced HIV RNA. The probes were synthesized using the digoxigenin RNA labeling kit (Roche) and XhoI-digested Bl-env-HindIII template. Cotransfected myc-tagged proteins were detected with the monoclonal anti-myc antibody (Invitrogen Inc.). FITC-conjugated sheep anti-digoxigenin antibody (Boehringer Mannheim) and Texas-red-labeled anti-mouse were used to detect the antisense probes and myc-tagged proteins, respectively.
RNA analysis
In most instances, Hela/293 cells (1.5 × 106 cells) were transfected with 2 μg of pgTat, pgTatΔE, pgTatΔEΔS SL1, or pgTatΔEΔS A1, 0.4 μg SVhygro or SV Rev, and 8 μg of pcDNA3.1 or pc mycSam68. Forty-eight hours post-transfection, total RNA was harvested using the protocol of Chomczynski and Sacchi (1987). RT-PCR analysis was performed as described previously (Pongoski et al. 2002) with the modification that CR-Tat (5′ -AGTGGTGGGCCTAGTTGCAGTA-3′ ) was used as the reverse primer. Briefly, cDNA was generated from 3 μg of total RNA using DR12 (5′ -AGGGGTGGACAG-3′ ) and M-MLV reverse transcriptase according to the manufacturer’s protocol (GIBCO). PCR was subsequently performed with a 32P end-labeled reverse primer and two forward primers that specifically detect cDNA representing spliced and unspliced RNA. Using Taq polymerase (Boehringer-Mannheim), 30 cycles of amplification were performed as follows: 1 min 94°C, 1 min 56°C, 2 min 72°C.
To monitor the status of the 3′ end of both spliced and unspliced env RNA, RNA probes containing the last 87 nt of the intron and extending 90 nt 3′ of the consensus AAUAAA polyadenylation signal for each construct tested were used in RNA protection assays. In the case of Ad2dsxΔ434, analysis was performed as described previously (Szymczyna et al. 2003). Probe was incubated with 10 or 30 μg of total RNA in 80% formamide, 40 mM PIPES at pH 6.4, 1 mM EDTA, 0.4 M NaCl for 12–16 h at 50°C. Following hybridization, samples were digested according to manufacturer’s suggestions (Ambion). Following ethanol precipitation, samples were re-suspended in 90% formamide, 2 mM EDTA and fractionated on 4% or 6% polyacrylamide, 8 M urea gels. Position of bands was determined following exposure to phosphor screens and scanning using a PhosphorImager. To test for the selective transport of particular env RNA forms, nuclear and cytoplasmic fractions were prepared as previously described (McCracken et al. 1998) with the following amendments: Cells were harvested in 1× TBS, 2 mM EDTA and EDTA was added to the hypotonic lysis buffer to a final concentration of 10 mM to remove any remaining polysomal RNA from the nuclear fraction (Weil et al. 2000). RNA was isolated and analyzed from the individual fractions as indicated above.
Acknowledgments
K.A. is supported by an MD/Ph.D studentship from CIHR. M.M. holds a studentship and A.C. is the recipient of a Scientist award from the Ontario HIV Treatment Network (OHTN). We would like to thank P. Sadowski, M. Shulman, B. Blencowe, and K. Boris-Lawrie for comments regarding this article. Work was funded by grants from the OHTN and the Canadian Institutes of Heath Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5263904.
REFERENCES
- Amendt, B.A., Hesslein, D., Chang, L.-J., and Stoltzfus, C.M. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of Human Immuno-deficiency Virus Type 1. Mol. Cell. Biol. 14: 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, K., Platt, C., and Cochrane, A. 2003. Control of HIV-1 env RNA splicing and transport: Investigating the role of hnRNP A1 in exon splicing silencer (ESS3a) function. Virology 314: 229–242. [DOI] [PubMed] [Google Scholar]
- Bentley, D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336–342. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., Domsic, J.K., Mayeda, A., Krainer, A.R., and Stoltzfus, C.M. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 75: 8487–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J., Bowman, J., McCracken, S., and Rosonina, E. 1999. SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol. 77: 277–291. [PubMed] [Google Scholar]
- Brodsky, A. and Silver, P.A. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Mayeda, A., Krainer, A., and Zahler, A. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18: 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Boisvert, F.M., Bazett-Jones, D.P., and Richard, S. 1997. Self-association of the single-KH-doamin family members Sam68, GRP33, GLD-1 and Qk-1: Role of the KH domain. Mol. Cell. Biol. 17: 5707–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Coyle, J.H., Guzik, B.W., Bor, Y.C., Jin, L., Eisner-Smerage, L., Taylor, S.J., Rekosh, D., and Hammarskjold, M.L. 2003. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell. Biol. 23: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio, N., Carmo-Fonseca, M., Geraghty, F., Pereira, H., Grosveld, F., and Antoniou, M. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18: 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard, C.K., Tange, T.O., and Kjems, J. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8: 1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fruscio, M., Chen, T., and Richard, S. 1999. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc. Natl. Acad. Sci. 96: 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower, K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli, S., Totty, N., Hsuan, J., and Courtneidge, S. 1994. A target for Src in mitosis. Nature 368: 871–874. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R. 1998. A tale of two termini: A functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216: 1–11. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., Thompson, J.D., and Heringa, J. 1993. The KH domain occurs in a diverse set of RNA-binding proteins. FEBS Lett. 324: 361–366. [DOI] [PubMed] [Google Scholar]
- Hilleren, P. and Parker, R. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenyltion during 3′ -end formation of nascent transcripts. RNA 7: 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T. 2001. Quality control of mRNA 3′ -end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Hong, W., Resnick, R., Rakowski, C., Shalloway, D., Taylor, S., and Blobel, G. 2002. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol. Cancer Res. 1: 48–55. [PubMed] [Google Scholar]
- Hope, T.J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365: 186–191. [DOI] [PubMed] [Google Scholar]
- Huang, S. 2000. Review: Perinucleolar structures. J. Struct. Biol. 129: 233–240. [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacampo, S. and Cochrane, A. 1996. Human Immunodeficiency Virus Type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 70: 8332–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Haga, I., Li, Q., and Fujisawa, J. 2002. Identification of cellular mRNA targets for RNA-binding protein Sam68. Nucleic Acids Res. 30: 5452–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquenet, S., Mereau, A., Bilodeau, P.S., Damier, L., Stoltzfus, C., and Branlant, C. 2001. A second exon splicing silencer within the Human Immunodeficiency Virus Type 1 tat exon 2 represses splicing of tat mRNA and binds protein hnRNP H. J. Biol. Chem. 276: 40464–40475. [DOI] [PubMed] [Google Scholar]
- Kataoka, N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673–682. [DOI] [PubMed] [Google Scholar]
- Kriegler, M. 1990. Gene transfer and expression: A laboratory manual, 1st ed. Stockton Press, New York.
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000a. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Moore, M.J., and Maquat, L.E. 2000b. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon–exon junctions. Genes & Dev. 14: 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li, J., Liu, Y., Kim, B.O., and He, J.J. 2002a. Direct participation of Sam68, the 68-kilodalton Src-associated protein in mitosis, in the CRM1-mediated Rev nuclear export pathway. J. Virol. 76: 8374–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Liu, Y., Park, I.W., and He, J.J. 2002b. Expression of exogenous Sam68, the 68-kilodalton SRC-associated protein in mitosis, is able to alleviate impaired Rev function in astrocytes. J. Virol. 76: 4526–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q., Taylor, S., and Shalloway, D. 1997. Specificity and determinants of Sam68 RNA binding: Implications for the biological function of K homology domains. J. Biol. Chem. 272: 27274–27280. [DOI] [PubMed] [Google Scholar]
- Lou, H., Gagel, R.F., and Berget, S.M. 1996. An intron enhancer recognized by splicing factors activates polyadenylation. Genes & Dev. 10: 208–219. [DOI] [PubMed] [Google Scholar]
- Lu, S. and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y., Yu, H., and Peterlin, B.M. 1994. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J. Virol. 68: 3850–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim, M.H., Hauber, J., Le, S.-Y., Maizel, J.V., and Cullen, B.R. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254–257. [DOI] [PubMed] [Google Scholar]
- Manley, J.L. and Tacke, R. 1996. SR proteins and splicing control. Genes & Dev. 10: 1569–1579. [DOI] [PubMed] [Google Scholar]
- Marchand, V., Mereau, A., Jacquenet, S., Thomas, D., Mougin, A., Gattoni, R., Stevenin, J., and Branlant, C. 2002. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323: 629–652. [DOI] [PubMed] [Google Scholar]
- Matter, N., Herrlich, P., and Konig, H. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695. [DOI] [PubMed] [Google Scholar]
- McCracken, S., Rosonina, E., Fong, N., Sikes, M., Beyer, A., O’Hare, K., Shuman, S., and Bentley, D.L. 1998. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harbor Symp. Quant. Biol. 63: 301–309. [DOI] [PubMed] [Google Scholar]
- McCracken, S., Lambermon, M., and Blencowe, B.J. 2002. SRm160 splicing coactivator promotes transcript 3′ -end cleavage. Mol. Cell. Biol. 22: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi, S., Geraghty, F., Idowu, B., Tam, J., Antoniou, M., and Vagner, S. 2002. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′ -end processing. EMBO Rep. 3: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Nott, A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, H.S., Cochrane, A.W., Dillon, P.J., Nalin, C.M., and Rosen, C.A. 1990. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent upon multimer formation mediated through a basic stretch of amino acids. Genes & Dev. 4: 1357–1364. [DOI] [PubMed] [Google Scholar]
- O’Reilly, M.M., McNally, M.T., and Beemon, K.L. 1995. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology 213: 373–385. [DOI] [PubMed] [Google Scholar]
- Pollard, V. and Malim, M. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52: 491–532. [DOI] [PubMed] [Google Scholar]
- Pongoski, J., Asai, K., and Cochrane, A. 2002. Positive and negative modulation of Human Immunodeficiency Virus Type 1 Rev function by cis and trans regulators of viral RNA splicing. J. Virol. 76: 5108–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, D., Amaral, M., Wu, J., Maniatis, T., and Greene, W. 1997. HIV Rev-dependent binding of SF2/ASF to the Rev response element: Possible role in Rev-mediated inhibition of HIV RNA splicing. Proc. Natl. Acad. Sci. 94: 973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Purcell, D. and Martin, M.A. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67: 6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, T., Xu, W., Mau, J., Goodwin, C., Suhasini, M., Tang, H., Frimpong, K., Rose, D., and Wong-Staal, F. 1999. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat. Med. 5: 635–642. [DOI] [PubMed] [Google Scholar]
- Reddy, T.R., Xu, W., and Wong-Staal, F. 2000. General effect of Sam68 on Rev/Rex regulated expression of complex retroviruses. Oncogene 19: 4071–4074. [DOI] [PubMed] [Google Scholar]
- Reddy, T.R., Suhasini, M., Xu, W., Yeh, L.Y., Yang, J.P., Wu, J., Artzt, K., and Wong-Staal, F. 2002. A role for KH domain proteins (Sam68-like mammalian proteins and quaking proteins) in the post-transcriptional regulation of HIV replication. J. Biol. Chem. 277: 5778–5784. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Felber, B.K., Benko, D.M., Fenyo, E.-M., and Pavlakis, G.N. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64: 2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin, B., Staffa, A., and Cochrane, A. 1998. Control of HIV-1 RNA metabolism: The role of splice sites and intron sequences in unspliced viral RNA subcellular distribution. J. Virol. 72: 9503–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si, Z.-H., Rauch, D., and Stoltzfus, M. 1998. The exon splicing silencer in the Human Immunodeficiency Virus Type 1 tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 18: 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., Matunis, M.J., Michael, W.M., and Dreyfuss, G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros, V., Valderrarama Carvajal, H., Richard, S., and Cochrane, A. 2001. Inhibition of Human Immunodeficiency Virus Type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J. Virol. 75: 8203–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffa, A. and Cochrane, A. 1994. The tat/rev intron of Human Immunodeficiency Virus Type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 68: 3071–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15: 4597–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyna, B.R., Bowman, J., McCracken, S., Pineda-Lucena, A., Lu, Y., Cox, B., Lambermon, M., Graveley, B.R., Arrowsmith, C.H., and Blencowe, B.J. 2003. Structure and function of the PWI motif: A novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes & Dev. 17: 461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H., Kuhen, K.L., and Wong-Staal, F. 1999. Lentivirus replication and regulation. Annu. Rev. Genet. 33: 133–170. [DOI] [PubMed] [Google Scholar]
- Tange, T.O. and Kjems, J. 2001. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J. Mol. Biol. 312: 649–662. [DOI] [PubMed] [Google Scholar]
- Tange, T.O., Damgaard, C.K., Guth, S., Valcarcel, J., and Kjems, J. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20: 5748–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. and Shalloway, D. 1994. An RNA-binding protein associated with src through its SH2 and SH3 domains in mitosis. Nature 368: 867–871. [DOI] [PubMed] [Google Scholar]
- Weil, D., Boutain, S., Audibert, A., and Dautry, F. 2000. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA 6: 962–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, S.E., Hillner, P.E., Vale, R.D., and Sachs, A.B. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Yang, J.-P., Reddy, T., Truong, K., Suhasini, M., and Wong-Staal, F. 2002. Functional interaction of Sam68 and heterogenous nuclear ribonucleoprotein K. Oncogene 21: 7187–7194. [DOI] [PubMed] [Google Scholar]
- Zahler, A.M., Lane, W.S., Stolk, J.A., and Roth, M.B. 1992. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 6: 837–847. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Mayeda, A., and Krainer, A. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8: 1351–1361. [DOI] [PubMed] [Google Scholar]