Abstract
Synthesis of eukaryotic ribosomal RNAs (rRNAs) includes methylation of scores of nucleotides at the 2′-O-ribose position (Nm) by small nucleolar RNP complexes (snoRNPs). Sequence specificity is provided by the snoRNA component through base-pairing of a guide sequence with rRNA. Here, we report that methylation snoRNPs can be targeted to many new sites in yeast rRNA, by providing the snoRNA with a novel guide sequence, and that in some cases growth and translation activity are strongly impaired. Novel snoRNAs can be expressed individually or by a unique library strategy that yields guide sequences specific for a large target region. Interference effects were observed for sites in both the small and large subunits, including the reaction center region. Targeting guide RNAs to nucleotides flanking the sensitive sites caused little or no defect, indicating that methylation is responsible for the interference rather than a simple antisense effect or misguided chaperone function. To our knowledge, this is the only approach that has been used to mutagenize the backbone of rRNA in vivo.
Keywords: snoRNA, snoRNP, snoRNA gene library, rRNA methylation, rRNA mutagenesis
INTRODUCTION
There are two major modified nucleotides (nt) in all ribosomal RNAs, namely, 2′-O-methylated nucleotides (Nms) and pseudouridines (Ψs), which are formed during or shortly after transcription (Maden 1990). Although the composition and level of modification vary among organisms and between kingdoms in a general way (Ofengand 2002), many modifications are heavily clustered in regions of the ribosome known to be functionally important (Maden 1990; Decatur and Fournier 2002; Hansen et al. 2002). This distribution suggests that modifications play beneficial roles in the structure and function of the ribosome. Consistent with this view, blocking modification globally causes severe growth defects in yeast (Tollervey et al. 1993; Zebarjadian et al. 1999), whereas disrupting modification of individual sites has only slight or no apparent effect (Lowe and Eddy 1999; Samarsky and Fournier 1999; Badis et al. 2003; Bonnerot et al. 2003; King et al. 2003). Importantly, evaluating effects of individual modifications has been limited in most cases to screening for major growth defects only.
In eukaryotes, modification of cytoplasmic rRNA occurs in the nucleolus, mediated by two large families of snoRNPs, that is, the box C/D and box H/ACA snoRNPs, which are specific for Nms and Ψs, respectively (Decatur and Fournier 2003). The family designations are based on the names of distinguishing sequence elements in the snoRNA components. The methylating snoRNPs are the subject of this study. In yeast and other eukaryotes, the methylating snoRNPs contain one site-specific guide snoRNA and a set of four core proteins common to all C/D snoRNPs. Each snoRNA contains one box C and one box D, located near the 5′ and 3′ ends of the RNA, respectively. These elements are part of a kink-turn motif involved in binding of the core proteins, including the methylase (Fatica and Tollervey 2003). Many C/D guide snoRNAs have a second set of related elements (boxes C′ and D′), located in the interior of the molecule, and these have similar but not identical structures and functions as boxes C and D. Methylation is targeted by a long (10–21 nt) guide sequence in the snoRNA located upstream of box D/D′, and the reaction occurs within the region of complementarity, at a substrate site 5 nt upstream of box D or D′. Where two targeting domains exist, these may be for the same or different rRNA molecules.
The focus of this report is characterizing the effects of introducing Nms into yeast rRNA at novel locations. By expressing a C/D snoRNA with a new guide sequence, a point mutation can be made in rRNA in a site-specific way by using natural machinery. We reasoned that strong interference effects would be observed at some sites due to altered properties of the rRNA caused by methylation (Davis 1998). The ability to target Nm to new sites with snoRNAs was first demonstrated during discovery and characterization of the guide function of the C/D snoRNAs (Cavaille et al. 1996; Kiss-Laszlo et al. 1996; Ni 1998). For one novel yeast site, the cell growth rate was slightly impaired (Kiss-Laszlo et al. 1996). We have extended this type of experimentation by targeting methylation to preselected sites in the yeast ribosome in regions known or reasonably predicted to be functionally important, and to all nucleotides in a segment of the large subunit (LSU) that encompasses the peptidyl transferase center (PTC). In the latter thrust, we developed a novel strategy for generating a library of snoRNA genes with guide sequences specific for the desired target region. Severe and strong growth defects were identified in both approaches. Preliminary results from these experiments were reported in a methods paper that describes the procedures we used to create novel guide snoRNAs (Liu et al. 2001).
We also address the question of whether modifying snoRNPs have other effects on their RNA substrates beyond creating a site-specific modification, such as chaperone-like effects that influence pre-rRNA folding or assembly of rRNP complexes (Bachellerie et al. 1995; Maxwell and Fournier 1995; Steitz and Tycowski 1995). If chaperone-like functions exist, it is reasonable to expect these reactions involve the antisense guide sequences through which the snoRNP binds to its RNA substrate. To determine if the growth defects are due to an antisense effect or misguided chaperone function, we examined the consequences of outfitting interfering snoRNAs with guide sequences that target methylation to adjacent or other nearby nucleotides. Our results argue against an antisense phenomenon of the type likely to occur in a chaperone function. The tight clustering and close spacing of natural snoRNP-mediated modifications in yeast rRNA in several rRNA segments also argue against each snoRNP being involved in an rRNA folding event. Taken together, these results suggest that targeting modification is the only role for the modifying snoRNPs examined, and that this situation quite likely applies to other modifying snoRNPs as well.
RESULTS
Strategy for probing rRNA in vivo with novel methylating snoRNPs
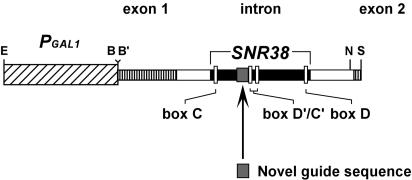
In our approach, novel guide snoRNAs are expressed conditionally from a plasmid-encoded transcription unit regulated by a galactose-inducible promoter (Fig. 1 ▶). The experimental transcription unit was derived from a chromosomal segment that encodes mRNA for the translation elongation factor TEF4 (Kinzy et al. 1994). A single intron encodes a snoRNA (snR38) that targets methylation to a site in the LSU of the ribosome (G2811; Ni 1998). The experimental snoRNAs contain a 20- or 21-nt deletion between box D′ and the predicted box C′ and accumulate and function normally (see below). Genes for the new snoRNAs were created either on an individual basis by a PCR strategy or as a library with overlapping guide elements that span a region of interest generated by replacing the guide element in the parental snoRNA with a new 13- or 14-nt sequence. The growth rates of cells expressing the new guide RNAs were examined on solid medium containing galactose.
FIGURE 1.
Structure of the parental gene used to express novel guide snoRNAs. New guide RNAs were derived from a modified coding sequence for a natural snoRNA (snR38), which is embedded within the intron of a gene for translation elongation factor 4 (TEF4). The transcription unit contains exon 1, the intron, and 36 bp of exon 2 and is expressed from the galactose-inducible GAL1 promoter. The coding sequence of the variant snR38 gene is 21 nt shorter than the natural gene and was used as the parental gene for the novel guide snoRNAs. New guide sequences were inserted at the appropriate site by a PCR-based method, to replace the natural 13-nt guide. Several restriction enzyme sites used for DNA manipulation are indicated. E indicates EcoRI; B, BamHI; B′, BglII; N, NruI; and S, SacI.
Strong growth defects can be caused by novel Nm guide snoRNAs
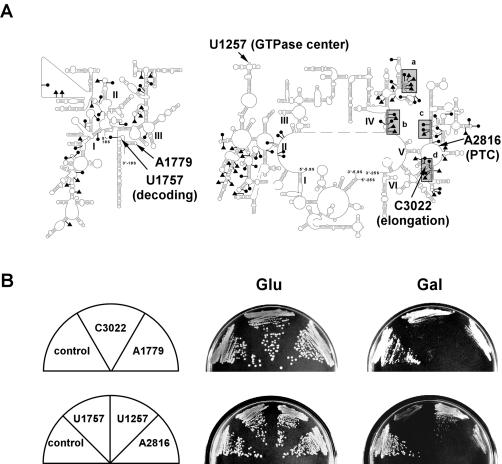
The feasibility of using methylating snoRNPs for interference mapping was evaluated initially by targeting rRNA regions known to be important for ribosome function in Escherichia coli (Fig. 2A ▶). In each case, conditional expression of the novel guide RNA caused a slow-growth phenotype (Fig. 2B ▶). Methylation was demonstrated by a primer extension assay procedure (Maden et al. 1995) at sites A1779 (E. coli: A1518), U1257 (U1083), and A2816 (A2451), but not at sites U1757(U1495), and C3022 (C2658; data not shown). Evidence shown below indicates that methylation is the basis for the growth defects observed, rather than a simple antisense effect caused by the 13-nt guide element in the novel snoRNAs.
FIGURE 2.
Expression of novel methylation snoRNAs can impair growth. (A) Five nucleotides (arrows) located in rRNA domains required for translation were targeted for methylation. Two sites are in 18S rRNA regions involved in decoding and tRNA binding (A1779 and U1757). The other three are linked to the GTPase center (U1257), peptidyl transferase center (A2816), and elongation (C3022). The corresponding E. coli sites are A1779 (A1518), U1757 (U1495), U1257 (U1083), A2816 (A2451), and C3022 (C2658). Natural methylated nucleotides (solid circles) and pseudouridines (solid triangles) are indicated, with four highly modified segments of 25S rRNA highlighted. (B) Yeast cells expressing these novel snoRNAs (indicated by the targets) have defects in growth. The parental snR38, which targets G2811 (G2446) of 25S rRNA was used as control. Pictures were taken after growth on selective plates containing glucose (Glu) or galactose (Gal) for 2 and 3 d, respectively.
A library strategy for probing specific segments of rRNA
With a view to eventually constructing interference maps for specific structural domains, we developed a procedure for creating a library of snoRNAs that can, in principle, methylate every site (except those at the ends) in a preselected RNA region of modest size; details of the methodology are provided elsewhere (Liu et al. 2001). The library was tailored to a region that encompasses the PTC. The key steps involved in creating the library are as follows: (1) generating random fragments with DNase I; (2) fusing the random fragments to a linker oligonucleotide that contains cleavage sites for both type I and type II restriction enzymes; (3) producing a pool of 14-bp fragments with the type I enzyme BpmI; and (4) following cleavage with appropriate restriction enzymes, cloning the new guide elements into matching sites in a snoRNA expression plasmid. Because the double-stranded rDNA fragments will be incorporated in both orientations in the library, half of the cloned inserts will be proper antisense guide sequences, and half will have the same sequence as the rRNA. Sequencing of 46 independent isolates showed the content of appropriate guide sequences to be 33% (Liu et al. 2001). The data show the library construction strategy is a good one.
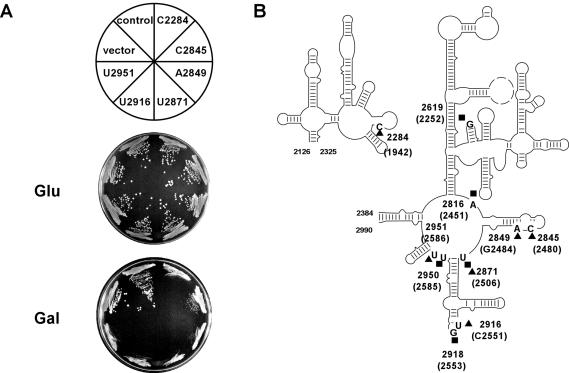
The potential of the library strategy to identify important sites was evaluated by screening the growth properties of yeast transformants. Screening for slow and lethal growth phenotypes identified six sites (Fig. 3 ▶). Strikingly, five of these are at or adjacent to sites in domain V that are already known to be important in E. coli rRNA for tRNA binding (Garrett and Rodriguez-Fonseca 1996; Green and Noller 1997; Nissen et al. 2000; Yusupov et al. 2001). The sixth site C2284 (C1942) occurs in a helix in domain IV, which in E. coli is involved in subunit association (Merryman et al. 1999). These results add strong support to our proposition that methylating snoRNPs can be used to identify functionally important rRNA sites, and that probing with libraries of guide RNAs can be effective.
FIGURE 3.
Nucleotides (nts) identified by a snoRNA gene library targeted to a region in the LSU rRNA that includes domain V. (A) Growth inhibition in yeast cells expressing the snoRNAs encoded in the snoRNA gene library, as indicated by the target sites. (B) The locations of target nucleotides identified are indicated (solid triangles). Five sites in E. coli rRNA involved in P-site or A-site binding by tRNA are shown by solid squares. Of the six yeast nucleotides identified, five are at or within 1–2 nt of sites implicated in tRNA binding in E. coli. The sixth, C2284 (C1942), occurs in a segment important for ribosomal subunit association.
The nucleotide specificity of 17 snoRNAs that did not cause strong growth defects was also determined. These snoRNAs were specific for the target region, consistent with the library sequence data, and the targeted sites are well distributed in that region. The sites correspond to nucleotides C2989, A2323, C2329, A2372, A2386, C2469, G2533, A2568, C2576, G2605, G2644, A2743, U2755, G2820, U2976, C2981, and C3081. The fact that only a minority of the guide RNAs caused growth defects supports the view that the interference effects are site specific in nature, and that most sites are probably not sensitive. We show below that several additional PTC sites are sensitive, indicating that a saturation condition was not achieved in this initial probing analysis.
Growth defects correlate with global translation activity
To determine if the slow-growth defects observed with the novel guide RNAs reflect loss of protein synthesis activity, the rate of amino acid incorporation was analyzed in vivo for the six sites in the PTC region identified from screening of the snoRNA gene library. A dramatic decrease in incorporation rate was observed for each strain. Compared with a vector-only control, the calculated rates of global translation (expressed as cpm/OD600 per min) were 24% (C2284), 14% (C2845), 30% (A2849), 44% (U2871), 14% (U2916), and 44% (U2951). In work to be presented elsewhere, translation activity was also examined for eight other interfering snoRNAs targeted to the central loop region of the PTC. The impact of these latter snoRNAs on cell growth included no effect, moderate effect, and severe or lethal effects. The corresponding amino acid incorporation activities for cells targeted by these snoRNAs ranged downward from control cell levels to 10% of the control activity and, in one case, no activity. Taken together, the results show that the extents of growth impairment correlate with loss of translational activity, as expected.
The methylation elements in the snoRNA are required for the growth defects
Methylation was not detected in all cases described (see Discussion), so the growth defects could also be due to an antisense phenomenon, such as interference with rRNA folding or protein binding, rather than to modification. With a view to distinguishing among these possibilities, we analyzed the effects of (1) mutating the box C′ /D′ motif and adjoining guide sequence that are essential for methylation activity, and (2) directing methylation to other nucleotides near the sensitive sites, to characterize the size of the sensitive rRNA region. The first such analyses were carried out with the interfering snoRNA that targets nucleotide U1757 (U1495) in 18S rRNA.
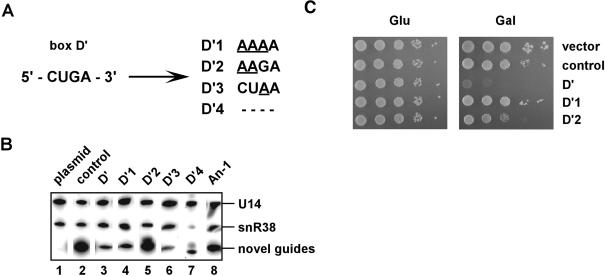
Mutational analysis has shown that the C′ and D′ elements are essential for methylation activity with an internal guide sequence, but are not required for snoRNA accumulation (Kiss-Laszlo et al. 1998). Four mutant variants of box D′ were analyzed, including three with nucleotide substitutions and one that lacked all of box D′ (D′1–4 in Fig. 4A ▶). The growth block was relieved in three of the four cases and the mutant snoRNAs accumulated in each case, although to different extents (Fig. 4B ▶, lanes 4–7). A point mutation at the third position (CUGA →CUAA; D′3) did not nullify the growth defect (data not shown), whereas alterations of two or three nucleotides (CUGA →AAGA or AAAA) yielded near–wild-type or wild-type growth (Fig. 4C ▶, D′2 and D′1). Similarly, complete deletion of box D′ (D′4) restored normal growth (data not shown). The mutational effects show that the box D′ motif is required for the interference effects, suggesting that the defects are due to methylation. The effects observed with the box D′mutations are also consistent with phylogenetic and genetic results that show the sequence of box D′ is more variable than that of the canonical box D (Kiss-Laszlo et al. 1996, 1998; Nicoloso et al. 1996).
FIGURE 4.
Mutations in snoRNA elements required for methylation block the snoRNA interference effect. (A) Sites of mutation introduced into the novel U1757 (U1495) snoRNA. The wild-type box D′ motif (left) is shown, and mutations in this element are underlined (right). Deletion of box D′ is indicated with dashes. The set of mutant snoRNAs also included a variant with a point mutation in the antisense guide sequence, located 4 nt upstream of box D′ (An-1, data not shown). (B) Steady-state levels of the mutant snoRNAs. Northern blots were carried out on total RNA isolated from cells containing the following plasmids: empty plasmid (lane 1, plasmid); the parental control snoRNA (snR38), which was plasmid encoded (lane 2, control); the initial U1757 (U1495) guide snoRNA (lane 3, D′); and the experimental mutant variants of the U1757 (U1495) snoRNA (lanes 4–8). Natural U14 and snR38 snoRNAs derived from the genome served as internal controls. RNAs were separated on a urea-8% polyacrylamide gel, and blots were probed with a mixture of 5′-labeled oligonucleotides specific for snR38 and U14 snoRNAs. The snoRNA genes are identified at the top of the panel by the mutated elements. (C) Growth properties of cells with the mutant guide RNA genes. Transformants were diluted serially 1:10 and spotted on solid selective medium containing glucose (Glu) or galactose (Gal).
A point mutation in the U1757 (U1495) guide sequence at the fourth nucleotide upstream of box D′ completely abolished the growth defect caused by this snoRNA (data not shown). Release of the growth block in this case is presumably due to loss of continuous complementarity between the guide sequence and the rRNA target segment. No effect on snoRNA accumulation was apparent (Fig. 4B ▶, lane 8). Taken together, these mutational results show that elements required for methylation from an internal guide sequence are also required for the growth defect. We conclude that methylation potential is essential for the slow-growth defect observed.
Interference by the U1757 guide RNA is not due to an antisense effect
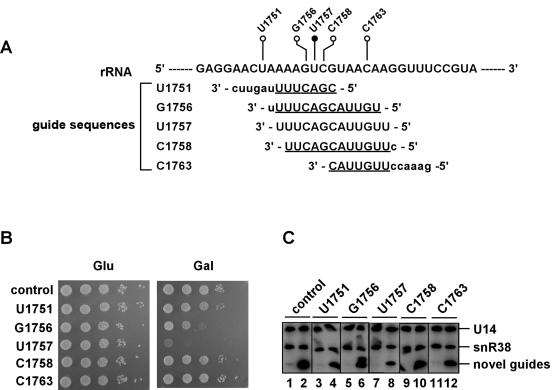
To investigate the possibility that growth interference by the U1757 (U1495) guide RNA is due to an antisense phenomenon rather than methylation, we tested the ability of guide RNAs targeted to nearby sites to impair growth. If an antisense effect is the basis of the impaired growth, we reasoned that targeting neighboring nucleotides should also cause interference. To this end, we targeted the two nucleotides that flank U1757 and two other nearby sites (Fig. 5A ▶). The new guide elements were incorporated into the same parental snoRNA. The guide sequences for these sites, G1756 (G1494) and C1758 (C1496), differ by only one nucleotide from that of the U1757 guide snoRNA, at the 3′ and 5′ ends, respectively.
FIGURE 5.
The growth defects do not extend to neighboring rRNA sites. (A) Alignment of the guide sequences tested. The guide sequences of the interfering U1757 (U1495) snoRNA and those corresponding to neighboring sites are aligned with the complementary rRNA substrate. The nucleotides of the U1757 (U1495) guide sequence are in uppercase. Identical nucleotides in the other guide sequences are also in uppercase, and the uncommon nucleotides are shown in lowercase. The predicted sites of methylation are shown for U1757 (U1495; solid circle) and for the neighboring sites examined (open circles). (B) Growth phenotypes of cells with overlapping guide sequences. Cells expressing the control snoRNA (snR38) or the experimental guide snoRNAs were diluted and spotted on plates containing glucose (Glu) or galactose (Gal) and incubated for 2 or 3 d, respectively, and the growth rates compared. The names of transformants correspond to the sites targeted for modification. (C) Expression data for the novel guide RNAs. Northern blots were performed on total RNA isolated from cells containing the experimental snoRNAs. RNA patterns are shown for cells grown in glucose (odd-numbered lanes) or galactose medium (even-numbered lanes). The specificities of the guide RNAs are shown above each pair of lanes.
Importantly, the guide snoRNAs targeted to C1758 (C1496) had no observable deleterious effect on cell growth, and that targeted to G1756 (G1494) impaired growth much less than the initial U1757 (U1495) snoRNA (Fig. 5B ▶). Guide RNAs were also targeted to sites six nucleotides up-and downstream, and neither of these had an adverse effect on growth (Fig. 5B ▶). All four of the new guide RNAs accumulated at levels comparable to the parental snR38 control; interestingly, accumulation exceeded that of the toxic U1757 (U1495) snoRNA (Fig. 5C ▶, lanes 2,4,6,8,10,12; and see below). Identical growth was observed for all experimental cells on glucose (Fig. 5B ▶), where expression of the test snoRNAs was repressed (Fig. 5C ▶, lanes 1,3,5,7,9,11). From these results, we conclude that the growth defect observed for U1757 (U1495) guide snoRNA is not due to a simple antisense effect.
Interference effects correlate with the importance of the targeted nucleotide
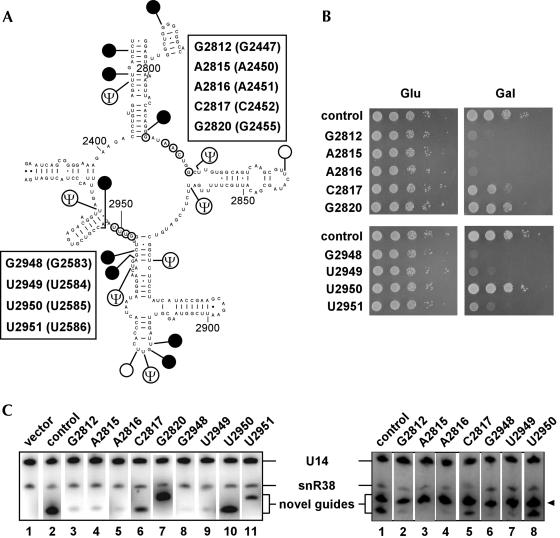
Because of the importance of understanding the basis of the interference effects, we extended our study to other interfering snoRNAs. Two more groups of nucleotides in the loop region of domain V of 25S rRNA were investigated. One group is localized in the upper part of the loop region (Fig. 6A ▶) and includes five sites known or predicted to be important. The sites and corresponding positions in E. coli are: G2812 (G2447), A2815 (A2450), A2816 (A2451), C2817 (C2452), and G2820 (G2455). Three of the sites, G2812 (G2447), A2815 (A2450), A2816 (A2451), together with G2402 (G2061) were predicted to form the active site of the PTase center (Nissen et al. 2000), based on the crystal structure of the 50S subunit from Haloarcula marismortui (Ban et al. 2000). They have also been implicated in the PTase reaction and/or tRNA binding from both genetic and biochemical evidence (Garrett and Rodriguez-Fonseca 1996; Green and Noller 1997).
FIGURE 6.
Interference analysis of two sets of neighboring nucleotides in the peptidyl transferase center (PTC). (A) Location of two groups of targeted sites in the PTC. The sites selected include nucleotides linked to snoRNA-induced growth defects and/or implicated in PTC function. The targeted nucleotides are circled, and each set of sites is listed in a box, with the corresponding Escherichia coli sites in parentheses. Naturally methylated nucleotides in the PTC region are indicated by solid circles (2′-O-methylations, Nms) or open circles (base methylations), and pseudouridines (Ψs) are also shown. (B) Effects on growth of targeting the neighboring sites. Growth phenotypes of cells expressing the snoRNAs in each group were compared on selective plates containing glucose (Glu) or galactose (Gal) after incubation for 2 or 3 d, respectively. The test strains are identified by the corresponding target sites. (C) Underaccumulation of inhibitory snoRNAs. Northern blot results are shown for cells expressing individual snoRNAs (left) or coexpressing (right) an inhibitory snoRNA (indicated by the targets on top of the panel) and a nontoxic snoRNA (G2820 guide). Probing was with a mix of oligonucleotides specific for snR38 and U14, and bands corresponding to the novel snoRNAs, endogenous snR38 and U14 are labeled. The G2820 guide snoRNA is also indicated by an arrowhead.
Less information is available about the importance of the C2817 (C2452) site. In archaeal rRNAs, point mutations at the equivalent position have been shown to create resistance to drugs that inhibit PTase activity, such as chloramphenicol (Aagaard et al. 1994) and sparsomycin (Tan et al. 1996). This situation suggests that C2817 (C2452) is required for the binding or action of the antibiotics. In addition, results from chemical probing of ribosomes with or without a PTase drug also argue that this site is involved in binding of antibiotic inhibitors (Rodriguez-Fonseca et al. 1995). Consistent with having an influence on the reaction center, E. coli ribosomes with a point mutation at this position (C2452U) have twofold to threefold lower PTase activity (Semrad and Green 2002). Interestingly, the reactivity of this nucleotides to dimethyl sulfate (DMS) has been shown to be pH dependent (Muth et al. 2001), suggesting that C2817 (C2452) could undergo structural rearrangement during the PTase reaction. This proposal is supported by a recent observation from the crystal structure of the large ribosomal subunit of Deinococcus radiodurans (Bashan et al. 2003). A conformational change at the equivalent position in the LSU occurs on binding of a PTase inhibitor, sparsomycin. Finally, no information is available about the functional role of G2820 (G2455)—it was selected solely on the basis of its close proximity to the reaction center.
Consistent with the known important roles of the G2812 (G2447), A2815 (A2450), and A2816 (A2451) sites, methylation targeted to these nucleotides resulted in lethal growth phenotypes (Fig. 6B ▶). In the context of a possible antisense effect, it is striking that in two cases guide RNAs targeted to adjacent nucleotides gave very disparate results. A slight slow growth phenotype was observed for C2817 (C2452) cells, and wild-type growth was seen for G2820 (G2455) cells, in strong contrast to the lethal growth effect observed for A2816 (A2451). Similarly, growth was strongly impaired in cells targeted for G2812 (G2447), which is adjacent to the position targeted in the control cells; these cells express the parental version of the snR38 used in our study that guides a natural Nm at G2811 (G2446). Northern data indicate that the novel guide snoRNAs were produced in each case, and that the inhibitory snoRNAs underaccumulated (Fig. 6C ▶, left panel, lanes 2–7; and see below). Because the guide RNAs differ by only one nucleotide at the ends of the guide sequences, these results also argue that the growth defect is not caused by a simple antisense interference effect. More likely, the results reflect the importance of the target nucleotide.
The second group of sites examined is localized in the lower part of the loop region (Fig. 6A ▶), and includes nucleotides G2948 (G2583), U2949 (U2584), U2950 (U2585), and U2951 (U2586). Results from genetic and biochemical studies suggest these nucleotides play important roles in tRNA binding and/or PTase activity. Mutations at sites G2948 (G2583), U2949 (U2584), and U2950 (U2585) cause dominant lethal growth defects, and large ribosomal subunits or ribosomes with mutations at these sites have reduced or extremely depressed PTase and protein synthesis activity (Porse et al. 1996; Green et al. 1997; Saarma et al. 1998; Green and Noller 1999; Polacek et al. 2003). The U2949 (U2584) and U2950 (U2585) nucleotides are among those in the large ribosomal subunits that are either protected from chemical probing by tRNA or tRNA fragments bound to the P site or involved in P site tRNA binding based on results from footprinting experiments with PTase-specific antibiotics (Garrett and Rodriguez-Fonseca 1996; Noller 1999). In addition, site U2950 (U2585) has been shown to be important for P site tRNA binding in damage selection experiments (Bocchetta et al. 1998). Finally, U2951 (U2586) is involved in a tertiary interaction between domain V and domain IV in the large ribosomal rRNA (Larsen 1992) and has also been implicated in the interaction between the sequence containing U2949 (U2584) and U2951 (U2586) with the P loop (Green et al. 1997).
Severe growth defects were obtained when methylation was targeted to G2948 (G2583), U2949 (U2584), or U2951 (U2586; Fig. 6B ▶), consistent with biochemical and genetic evidence showing that these nucleotides have important roles in tRNA binding at the P site and in the PTase reaction. Remarkably, targeting U2950 (U2585) had no effect on growth, whereas strong growth defects were observed for cells in which the adjacent sites U2949 (U2584) or U2951 (U2586) were targeted. The normal growth phenotype for the U2950 (U2585) cells is in contrast to effects observed for base mutations at this site, which presumably reflects different effects on rRNA structure of the Nm and base mutations. As in the similar cases presented above, the anti-sense sequences of these snoRNAs differed from that of the U2950 (U2585) guide RNA by only one nucleotide (in the case of the U2949 guide) or two (in the case of the U2951 guide) at the end(s).
Northern blot analysis showed that the near-normal and normal growth phenotypes observed for C2817 (C2452) cells (first group) and U2950 (U2585) cells (second group), were not due to lack of production of the corresponding guide RNA (Fig. 6C ▶, left panel, lanes 6,10). However, the data do reveal the interesting phenomenon, seen earlier for the U1757 (U1495) guide snoRNA (Fig. 5C ▶): The most deleterious snoRNAs occur at considerably lower levels than the others (Fig. 6C ▶, left panel, cf. lanes 2,6,7,10 and lanes 3,4,5,8,9). In principle, the reduced yield could reflect a defect in snoRNA or snoRNP production resulting from inhibition of transcription of the GAL1 promoter or, more likely, an increased turnover rate for the toxic snoRNPs. To investigate these possibilities, we coexpressed a nontoxic snoRNA—the G2820 guide RNA, with several other snoRNAs with or without deleterious effects. We reasoned that if the reduced levels of the toxic snoRNAs observed were due to impaired transcription of the GAL1 promoter, then coexpression of the G2820 guide RNA would also be inhibited. Northern blot analysis indicates that the expression of the G2820 guide RNA in all cases tested was identical to that in the control cells and was not affected by the presence of the toxic snoRNAs (Fig. 6C ▶, right panel, arrowhead). In contrast, the toxic snoRNAs underaccumulated as before (Fig. 6C ▶, cf. the corresponding snoRNAs in the right and left panels). These results indicate that underaccumulation of the toxic snoRNAs resulted from increased turnover rate rather than a defect in synthesis. Taken together, the results obtained with the set of PTC nucleotides also argue strongly that methylation, rather than an antisense effect, is responsible for the growth defects observed. We conclude that mutation of a functionally important nucleotide is the basis of the interfering effects.
DISCUSSION
The results provide valuable insights into the basis of interference effects caused by methylation snoRNPs with new guide sequences. They also enhance our understanding of how natural modifications are created and shed light on the interesting and important question of whether modifying snoRNPs function as chaperones in rRNA folding. From the perspective of using methylation snoRNPs for functional mapping, the results show that the interference effects are caused not by a simple antisense phenomenon but by modification of specific sites that are functionally important. Supporting evidence comes from demonstrations showing that (1) altering snoRNA elements required for methylation abolished the growth defects caused by an interfering guide snoRNA, and (2) shifting the specificity of an interfering guide RNA to adjacent or other nearby nucleotides yielded normal growth or growth that was much less impaired. In work to be presented elsewhere, we show that the growth defects correlate with two types of ribosome defects: impaired production of ribosomes and reduced translational activity.
A key question about the modifying snoRNPs is whether they have an active chaperone function; that is, do they mediate rRNA folding or rRNP assembly? That issue is relevant to the interference effects of interest here, and our results provide new insights into this matter as well. The possibility that snoRNPs function as chaperones in rRNA folding and ribosome assembly was raised when sequences complementary to rRNA were first discovered, initially in the U3 snoRNA and subsequently for several other C/D snoRNAs that we now know are methylation guide RNAs (Bachellerie et al. 1995; Maxwell and Fournier 1995; Steitz and Tycowski 1995). If a methylating snoRNP functions as a chaperone of rRNA, it would most likely act in a site-specific manner using the same sequence that guides methylation. Interactions between snoRNPs and ribosomal proteins do not appear to be essential for the occurrence of methylation, as methylation can be targeted to mini-rRNA substrates in vivo (Cavaille et al. 1996; Ganot et al. 1999). This situation suggests that protein–protein interactions between the experimental snoRNPs and the rRNP substrates are not necessary for the observed growth defects.
The possibility that the interfering effects are caused by a misguided chaperone seems very unlikely, because of the limited site-specific nature of the effects. If the effects are due to an aberrant chaperone, it is reasonable to expect that misfolding could also occur when the snoRNP is targeted to adjacent and other neighboring nucleotides. This was not the case, rather the effects were highly selective within several different rRNA segments.
Strong specificity was observed for the U1757 (U1495) site in the small subunit (Fig. 5 ▶) and for several nucleotides in the PTC region of the LSU (Fig. 6 ▶). Targeting the flanking sites involved shifting the 13-nt guide sequences up- or downstream by only one position. Although an antisense effect that exhibits a sharp sequence dependence of the sort seen here seems possible, this situation would be very unusual and not likely to occur for all of the sites examined. Based on current knowledge of how a methylation guide snoRNA works, it is more reasonable to conclude that the effects are nucleotide specific as expected, and that modification of an important nucleotide is the basis of the interference effects.
Of the 15 guide snoRNAs that cause strong growth defects (Figs. 2 ▶, 3 ▶, 6 ▶), methylation was detected in six cases only, that is, A1779 (A1518), A2815 (A2450), A2816 (A2451), A2849 (G2484), U2871 (U2506), and U2951 (U2586; data not shown). Possible reasons for the negative results could be a low level of rRNA containing the novel methylation, which results from poor modification efficiency or rapid turnover of rRNA containing the unusual modification and/or corresponding snoRNP. In this last regard, toxic snoRNAs were shown to underaccumulate, probably because of an increased turnover rate (Fig. 6C ▶), perhaps within pre-rRNP complexes. Another possible reason for the negative methylation results could be limitations of the modification assay, which does not detect all modifications known to exist, apparently due to sequence context effects (Maden et al. 1995; Maden 2001). It seems likely this could apply to a few cases only.
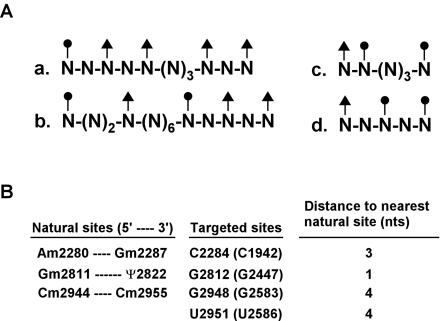
Additional evidence that not all modifying snoRNPs are chaperones comes from examining the distribution of natural modifications, relative to each other and to the novel methylation sites. In several segments of rRNA Nm and Ψ modifications are separated by only one or a few nucleotides (Fig. 2 ▶), and it seems improbable that each cognate snoRNP chaperones a folding event for such a short substrate region. The case is most persuasive for rRNA segments that possess multiple, tightly clustered modifications, such as the unusually dense subset highlighted in Figure 2A ▶ (shaded blocks) and Figure 7A ▶. This set includes one segment of 5 nt with three modifications (Fig. 7A ▶, d) and another segment of 11 nt with five modifications (Fig. 7A ▶, a). The sequences include both Nm and Ψ modifications and occur in both helical and loop regions of the secondary structure.
FIGURE 7.
Close spacing of natural modifications in rRNA suggests that at least some snoRNPs do not interfere with each other. (A) Modification patterns in the four highlighted regions (see Fig. 2 ▶). Individual nucleotides are designated “N,” and the sites of modification are identified with solid circles (Nm) or solid triangles (Ψ). (B) Some interfering guide RNAs target rRNA nucleotides that are close to sites of natural modification. The shortest distances between several targeted sites and nearby sites that are naturally modified are shown. Natural modifications are listed in the left column, and four targeted sites are listed in the middle column. The distances between the targeted sites and nearest naturally modified nucleotides are shown in the right column.
In addition, some of the sites targeted by the novel interfering snoRNPs are only 1 to 4 nt from a site(s) of natural modification, as shown for three target regions (Fig. 7B ▶). Because natural modifications can have similar spacings, presumably the close proximity of the test sites to other modification sites does not preclude methylation at the new sites. Although some of the snoRNPs that modify these segments could be chaperones, the likelihood that all are seems very remote. In this context, we define a chaperone as an active machine that mediates a folding event. Distinct from this, it seems quite possible, even likely, that rRNA folding can be influenced in a more passive way during the modification process, as a consequence of snoRNP binding and release. With regard to the Nm and Ψ modifications themselves, it is well established that these alterations affect RNA folding and conformational dynamics (Davis 1998). Our conclusion that the interference effects are caused by modification is supported by a final line of evidence, which is also indirect, but interesting and relevant. Screening the PTC region with a library of methylation guide snoRNAs identified only six different guide RNAs that caused strong growth defects (Fig. 3 ▶), many fewer than would be expected for a simple antisense effect.
Taken together, the results from these different types of experimentation demonstrate the validity of using snoRNA-guided modification for functional mapping of ribosomal RNA, and presumably other RNAs that are substrates for snoRNPs. The attraction of this approach, of course, is that point mutations can be created in a target RNA in vivo by harnessing natural cellular machinery. These same results also argue that the parental methylating snoRNP used in our study is not a chaperone, which casts doubt on the notion that all methylating snoRNPs are chaperones.
MATERIALS AND METHODS
General procedures
The yeast strain used was YS625 (Liang and Fournier 1995). Media, culturing (Kaiser et al. 1994), and transformation conditions are as cited (Gietz et al. 1992). Methods for preparing yeast total RNA (Balakin et al. 1993) and Northern blot analysis of snoRNAs (Chanfreau et al. 1998) are also as described.
Plasmid constructs
A plasmid-encoded variant of a wild-type snoRNA (snR38) was used as a control for all other snoRNA gene constructs. This plasmid (pBL152) was generated by insertion of a 1.2-kb EcoRI–SacI snoRNA expression cassette (Fig. 1 ▶) of pJN32 (Ni 1998) into plasmid pRS314 (Sikorski and Hieter 1989). The corresponding snR38 guide RNA has a 21-nt deletion between box D′ and the predicted box C′ , to allow the experimental snoRNAs to be readily distinguished from wild-type snR38. The deletion eliminated an NdeI site in the coding sequence.
The U1757 guide snoRNA gene was made by a two-step PCR-based method (Chen and Przybyla 1994), using pJN32 (Ni 1998) as template; pJN32 contains the same coding sequence of the recombinant snR38 gene as pBL152. In brief, a first round of amplification was performed with primers BLO-38 and BU, which are complementary to exon 1 of the TEF4 gene and the coding region of the snR38 gene in pJN32, respectively. In the second round, the PCR product of the first-round reaction was used as a mega primer, together with BLO-39, to generate a 0.5-kb fragment. Primer BLO-39 is complementary to the intron of the TEF4 gene, and the resulting 0.5 kb fragment contains mutant snR38 with the natural 13-nt guide sequence replaced with a new guide specific for U1757. This fragment was digested with BglII and NruI restriction enzymes and used to replace the BglII/NruI fragment of pJN32, resulting in plasmid pBL118. The A1779 and C3022 guide snoRNA genes were constructed as pBL118, and the corresponding plasmids are pBL115 and pBL140, respectively. The parental plasmid (pBL134) of pBL150 was generated as described for pBL118, except that the NdeI site was restored. pBL150 was generated by insertion of an EcoRI/SacI fragment of pBL134 into pRS314 and was used to establish a library of snoRNA genes (Liu et al. 2001).
An EcoRI/SacI fragment of pBL118 was inserted into pRS314, generating pBL143. Guide snoRNA genes for targets U1751, G1756, C1758, and C1763 were generated by one-step PCR using 5′ primer BLO-38 and 3′ primers BU-6b, BU-1b, BU+1b, and BU+6b, respectively, with pBL143 as template. The corresponding PCR fragments containing new methylation guide sequences were inserted into the BglII and NdeI sites of pBL150, resulting in plasmids pBL158 (G1756), pBL159 (U1751), pBL160 (C1758), and pBL161 (C1763), with the targets in the parentheses. Plasmids pBL162 (U1257), pBL227 (G2812), pBL225 (A2815), pBL163 (A2816), pBL230 (G2948), pBL231 (U2949), pBL232 (U2950), and pBL226 (C2817) were generated by the same strategy. Plasmids Y-17 (U2951) and Y-29 (G2820) were isolated from a snoRNA library (Liu et al. 2001). Plasmid Y-29u was produced by insertion of an EcoRI/SacI fragment of Y-29 into pRS316 (Sikorski and Hieter 1989) and was used for coexpression of G2820 guide RNA with several other guide snoRNAs analyzed in Figure 6C ▶.
The snoRNA genes, which were derived from the U1757 guide RNA and had mutations in the box elements or guide sequences, were constructed by either a one- or a two-step PCR strategy as described above. The plasmids that contain snoRNA genes with mutations in the box D′ element include pBL147 (CUGA →CUAA), pBL151 (deletion), pBL156 (CUGA →AAAA), and pBL171 (CUGA →AAGA), with the mutations indicated in parentheses. All of these snoRNA genes have the same guide sequences and sequence context as that of pBL143. Plasmid pBL148 is identical to pBL143 except for a point mutation (C →U) at the fourth nucleotide of the antisense sequence upstream of box D′.
In vivo labeling of proteins with [35S]methionine
The procedure for in vivo incorporation of [35S]methionine was based on a previously described method (Carr-Schmid et al. 1999). YS625 cells harboring different experimental snoRNA genes were grown in 200 mL minimum medium containing galactose at 30°C to OD600 of 0.4 to 0.8. Ten milliliters of cell culture was removed and labeled with 10 μCi of [35S]methionine ( 1175 Ci/mmole, NEN) for 1, 2, 3, 4, and 5 min, respectively. Samples of 0.5 mL labeled cells were taken at each time point and mixed immediately with 0.2 mL ice-cold stop solution containing 70 μL unlabeled 60 mM methionine, 1μL of 10 mg/mL cycloheximide, and 129 μL of 50% TCA. After incubation on ice for 20 min, cells were heated for 20 min at 65°C and filtered through GF/C filters (Whatman). Filters were washed 3× with 5 mL of 5% TCA and then 3× with 5 mL of 95% ethanol. After drying at 65°C, radioactivity was analyzed by scintillation counting. Triplicate samples were analyzed for each strain at each time point, and average values were used for each time point.
DNA oligonucleotides
Probes used for Northern analyses of snoRNAs were as follows: snR38, 5′-TCAGAAATACAAATATCAACATAT-3′; U14, 5′-CGA TGGGTTCGTAAGCGTACTCCTACCGTG-3′.
Sequences of primers used for construction of the novel guide snoRNA genes will be provided on request.
Acknowledgments
We are grateful to Wayne Decatur for help with the figures. This study was supported by National Institutes of Health grant GM19351.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7190104.
REFERENCES
- Aagaard, C., Phan, H., Trevisanato, S., and Garrett, R.A. 1994. A spontaneous point mutation in the single 23S rRNA gene of the thermophilic arachaeon Sulfolobus acidocaldarius confers multiple drug resistance. J. Bacteriol. 176: 7744–7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie, J.P., Michot, B., Nicoloso, M., Balakin, A., Ni, J., and Fournier, M.J. 1995. Antisense snoRNAs: A family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci. 20: 261–264. [DOI] [PubMed] [Google Scholar]
- Badis, G., Fromont-Racine, M., and Jacquier, A. 2003. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin, A.G., Schneider, G.S., Corbett, M.S., Ni, J., and Fournier, M.J. 1993. SnR31, snR32, and snR33: Three novel, non-essential snRNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 21: 5391–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Agmon, I., Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H.A., et al. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11: 91–102. [DOI] [PubMed] [Google Scholar]
- Bocchetta, M., Xiong, L., and Mankin, A.S. 1998. 23S rRNA positions essential for tRNA binding in ribosomal functional sites. Proc. Natl. Acad. Sci. 95: 3525–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot, C., Pintard, L., and Lutfalla, G. 2003. Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol. Cell 12: 1309–1315. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid, A., Valente, L., Loik, V.I., Williams, T., Starita, L.M., and Kinzy, T.G. 1999. Mutations in elongation factor 1β, a guanine nucleotide exchange factor, enhance translational fidelity. Mol. Cell. Biol. 19: 5257–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J., Nicoloso, M., and Bachellerie, J.P. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383: 732–735. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17: 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. and Przybyla, A.E. 1994. An efficient site-directed mutagenesis method based on PCR. Biotechniques 17: 657–659. [PubMed] [Google Scholar]
- Davis, D.R. 1998. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies). In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 85–102. AMS Press, Washington, DC.
- Decatur, W.A. and Fournier, M.J. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27: 344–351. [DOI] [PubMed] [Google Scholar]
- ———. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Fatica, A. and Tollervey, D. 2003. Insights into the structure and function of a guide RNP. Nat. Struct. Biol. 10: 237–239. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Jady, B.E., Bortolin, M.L., Darzacq, X., and Kiss, T. 1999. Nucleolar factors direct the 2′-O-ribose methylation and pseudo-uridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19: 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, R.A. and Rodriguez-Fonseca, C. 1996. The peptidyl transferase center. In Ribosomal RNA: Structure, evolution, processing, and function in protein biosynthesis (eds. R.A. Zimmermann and A.E. Dahlberg), pp. 327–355. CRC, Boca Raton, FL.
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R. and Noller, H.F. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66: 679–716. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry 38: 1772–1779. [DOI] [PubMed] [Google Scholar]
- Green, R., Samaha, R.R., and Noller, H.F. 1997. Mutations at nucleotides G2251 and U2585 of 23 S rRNA perturb the peptidyl transferase center of the ribosome. J. Mol. Biol. 266: 40–50. [DOI] [PubMed] [Google Scholar]
- Hansen, M.A., Kirpekar, F., Ritterbusch, W., and Vester, B. 2002. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA 8: 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, C., and Mitchell, A. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- King, T.H., Liu, B., McCully, R.R., and Fournier, M.J. 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11: 425–435. [DOI] [PubMed] [Google Scholar]
- Kinzy, T.G., Ripmaster, T.L., and Woolford Jr., J.L. 1994. Multiple genes encode the translation elongation factor EF-1 γ in Saccharomyces cerevisiae. Nucleic Acids Res. 22: 2703–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., and Kiss, T. 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., and Kiss, T. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, N. 1992. Higher order interactions in 23S rRNA. Proc. Natl. Acad. Sci. 89: 5044–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, W.Q. and Fournier, M.J. 1995. U14 base-pairs with 18S rRNA: A novel snoRNA interaction required for rRNA processing. Genes & Dev. 9: 2433–2443. [DOI] [PubMed] [Google Scholar]
- Liu, B., Ni, J., and Fournier, M.J. 2001. Probing RNA in vivo with methylation guide small nucleolar RNAs. Methods 23: 276–286. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Maden, B.E. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 39: 241–303. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 25: 374–382. [DOI] [PubMed] [Google Scholar]
- Maden, B.E., Corbett, M.E., Heeney, P.A., Pugh, K., and Ajuh, P.M. 1995. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77: 22–29. [DOI] [PubMed] [Google Scholar]
- Maxwell, E.S. and Fournier, M.J. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64: 897–934. [DOI] [PubMed] [Google Scholar]
- Merryman, C., Moazed, D., Daubresse, G., and Noller, H.F. 1999. Nucleotides in 23S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285: 107–113. [DOI] [PubMed] [Google Scholar]
- Muth, G.W., Chen, L., Kosek, A.B., and Strobel, S.A. 2001. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA 7: 1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Ni, J. 1998. The major function of the small nucleolar RNAs is nucleotide modification in ribosomal RNA. University of Massachusetts at Amherst, Amherst, MA.
- Nicoloso, M., Qu, L.H., Michot, B., and Bachellerie, J.P. 1996. Intronencoded, antisense small nucleolar RNAs: The characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol. 260: 178–195. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Noller, H.F. 1999. On the origin of the ribosome: Coevolution of subdomains of tRNA and rRNA. In The RNA world, 2nd ed. (eds. R.F. Gesteland et al.), pp. 197–219. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- Ofengand, J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514: 17–25. [DOI] [PubMed] [Google Scholar]
- Polacek, N., Gomez, M.J., Ito, K., Xiong, L., Nakamura, Y., and Mankin, A. 2003. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell 11: 103–112. [DOI] [PubMed] [Google Scholar]
- Porse, B.T., Thi-Ngoc, H.P., and Garrett, R.A. 1996. The donor substrate site within the peptidyl transferase loop of 23S rRNA and its putative interactions with the CCA-end of N-blocked aminoacyl-tRNA(Phe). J. Mol. Biol. 264: 472–483. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fonseca, C., Amils, R., and Garrett, R.A. 1995. Fine structure of the peptidyl transferase centre on 23S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J. Mol. Biol. 247: 224–235. [DOI] [PubMed] [Google Scholar]
- Saarma, U., Spahn, C.M., Nierhaus, K.H., and Remme, J. 1998. Mutational analysis of the donor substrate binding site of the ribosomal peptidyltransferase center. RNA 4: 189–194. [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A. and Fournier, M.J. 1999. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 27: 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrad, K. and Green, R. 2002. Osmolytes stimulate the reconstitution of functional 50S ribosomes from in vitro transcripts of Escherichia coli 23S rRNA. RNA 8: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, J.A. and Tycowski, K.T. 1995. Small RNA chaperones for ribosome biogenesis. Science 270: 1626–1627. [DOI] [PubMed] [Google Scholar]
- Tan, G.T., DeBlasio, A., and Mankin, A.S. 1996. Mutations in the peptidyl transferase center of 23S rRNA reveal the site of action of sparsomycin, a universal inhibitor of translation. J. Mol. Biol. 261: 222–230. [DOI] [PubMed] [Google Scholar]
- Tollervey, D., Lehtonen, H., Jansen, R., Kern, H., and Hurt, E.C. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72: 443–457. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Zebarjadian, Y., King, T., Fournier, M.J., Clarke, L., and Carbon, J. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19: 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]