Abstract
GP64, the major envelope glycoprotein of budded virions of the baculovirus Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV), is involved in viral attachment, mediates membrane fusion during virus entry, and is required for efficient virion budding. Thus, GP64 is essential for viral propagation in cell culture and in animals. Recent genome sequences from a number of baculoviruses show that only a subset of closely related baculoviruses have gp64 genes, while other baculoviruses have a recently discovered unrelated envelope protein named F. F proteins from Lymantria dispar MNPV (LdMNPV) and Spodoptera exigua MNPV (SeMNPV) mediate membrane fusion and are therefore thought to serve roles similar to that of GP64. To determine whether F proteins are functionally analogous to GP64 proteins, we deleted the gp64 gene from an AcMNPV bacmid and inserted F protein genes from three different baculoviruses. In addition, we also inserted envelope protein genes from vesicular stomatitis virus (VSV) and Thogoto virus. Transfection of the gp64-null bacmid DNA into Sf9 cells does not generate infectious particles, but this defect was rescued by introducing either the F protein gene from LdMNPV or SeMNPV or the G protein gene from VSV. These results demonstrate that baculovirus F proteins are functionally analogous to GP64. Because baculovirus F proteins appear to be more widespread within the family and are much more divergent than GP64 proteins, gp64 may represent the acquisition of an envelope protein gene by an ancestral baculovirus. The AcMNPV pseudotyping system provides an efficient and powerful method for examining the functions and compatibilities of analogous or orthologous viral envelope proteins, and it could have important biotechnological applications.
The baculoviruses are a large family of enveloped DNA viruses that are pathogenic to invertebrates, infecting primarily insects in the order Lepidoptera, but also certain members of the Diptera and Hymenoptera (3, 9). Baculoviruses have supercoiled, double-stranded DNA genomes of approximately 80 to 180 kbp. Baculoviruses are classified into two genera, the nucleopolyhedroviruses (NPVs) and the granuloviruses (GVs), and the NPVs can be further subdivided into groups I and II based on phylogenetic studies of many genes (5, 11, 13).
GP64 is the major envelope protein of the budded-virus (BV) form of group I NPVs such as Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) and Orgyia pseudotsugata MNPV (OpMNPV). GP64 is involved in viral attachment and is required for low-pH-mediated membrane fusion during virus entry (4, 12). GP64 is also required for efficient budding of the BV (24). Thus, GP64 is required for propagation of infection in cell culture and in Trichoplusia ni larvae (22). Although GP64 proteins have been identified in other group I NPVs, recent data from the complete genomic sequences of a number of baculoviruses suggest that gp64 genes are not present in group II NPVs or GVs. An envelope protein gene that is unrelated to gp64 was recently identified in Lymantria dispar MNPV (LdMNPV) (27) and Spodoptera exigua MNPV (SeMNPV) (15), and homologs of this gene have been identified in other group II NPVs and GVs. This protein has been named the F (fusion) protein (33). F proteins from LdMNPV (ORF 130, or Ld130) and SeMNPV (ORF 8, or Se8) are present in the virion membrane and mediate pH-triggered membrane fusion (15, 27). Like envelope proteins from some other virus groups, the baculovirus F proteins are cleaved internally by cellular proprotein convertases such as furin, and cleavage is necessary for fusion activity (33). These observations suggest that F proteins may represent a functional analog of GP64 in some (perhaps all) baculoviruses that do not have a gp64 gene. To experimentally address this possibility, we deleted the gp64 locus of a bacterial artificial chromosome containing an AcMNPV genome (bacmid) (17) and inserted F protein genes from two group II NPVs and a GV. We show that the F protein genes from LdMNPV and SeMNPV can rescue the gp64 knockout in AcMNPV. In addition, we show that the predicted furin cleavage site present in the SeMNPV F protein is required for processing of the SeMNPV F protein and rescue of the gp64-null virus. We also examined the functions of orthomyxovirus and rhabdovirus envelope proteins in the gp64-null pseudotyping system.
MATERIALS AND METHODS
gp64-null AcMNPV bacmid.
The gp64 gene of an AcMNPV bacmid (bMON14272; Invitrogen) was deleted by using a modification of the method reported by Bideshi and Federici (2). A chloramphenicol resistance gene (cat) cassette was amplified by PCR with primers P5′Spe(ChlR) (5′-ACTAGTGCTGGATCGGGCCCTAAATACCTG-3′) and P3′(ChlR)Bgl-II (5′-AGATCTTTACGCCCCGCCCTGCCACTCATC-3′) and with plasmid pRE112 as a template (7). The PCR product was cloned into the pCR-II-Blunt vector (Invitrogen) to generate plasmid pChlR-CRIIblunt. The insert containing the cat cassette was excised from pChlR-CRIIblunt by SpeI and BglII digestion and was used to replace the SpeI-BglII fragment (the gp64 gene) in pAcEcoHΔSma, a plasmid containing the AcMNPV gp64 locus and flanking sequences (22). The resulting plasmid, pAcEcoHΔSma,gp64(ChlR), was digested with EcoRI and HindIII; 60 ng of the gel-purified EcoRI-HindIII fragment containing the cat cassette was cotransformed with 1 μg of bMON14272 into electrocompetent BJ5183 cells (Stratagene); and colonies resistant to both kanamycin and chloramphenicol were selected.
Donor plasmids containing envelope protein genes.
Genes (Ld130, Se8, Px26, VSV-G, and gp75) encoding heterologous viral envelope proteins and AcMNPV gp64 (as a positive control) were amplified by PCR using Vent DNA polymerase (New England Biolabs) and cloned downstream of the AcMNPV gp64 promoter on a modified pBacGus5 plasmid (Novagen), pmBG5. pmBG5 was modified by digesting pBacGus5 with NcoI and SmaI to remove sequences encoding His tags and protease cleavage sites. The 5′ overhang was filled in with Klenow fragment and religated, so that the AcMNPV gp64 early-plus-late promoter was immediately upstream of a multiple cloning site and a β-glucuronidase gene (GUS) reporter driven by a p6.9 late viral promoter. As an example, the AcMNPV gp64 open reading frame (ORF) was PCR amplified with primers P5′Bam2327Acgp64 (5′-GGATCCAAGATGGTAAGCGCTATTGTTTT-3′) and P3′AcEcoHdstopE (5′-GAATTCTTAATATTGTCTATTACGGTTTCTA-3′) by using pAcEcoHΔSma as a template, digested with BamHI and EcoRI, and cloned into similarly digested pmBG5. The gp64 promoter and ORF were subsequently PCR amplified and subcloned into pΔFBgus(R), a donor plasmid of pFastBac1 (Invitrogen) from which the polyhedrin promoter had been removed and replaced with the GUS cassette from pBacGUS5. Genes for heterologous envelope proteins were similarly amplified, cloned into pmBG5, and subcloned into pΔFBgus(R). In some cases, genes (Ld130, Px26, and gp75) were placed under the control of the stronger polyhedrin promoter to increase expression. For the Px26, VSV-G, gp75, and Ld130 constructs under the control of the gp64 promoter, the p6.9 promoter-GUS cassette was inserted in the orientation opposite that shown in Fig. 1 (with the GUS transcription oriented toward Tn7R). All clones containing PCR-derived sequences were confirmed by sequencing. To confirm expression of Px26, sequences encoding a c-myc epitope tag were fused to the 3′ end of the Px26 ORF. Primers P5′Px996 (5′-GACCCTCGATTTCATGACGCTGTG-3′) and P3′Px26cmycEcoRI (5′-GTGAATTCCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCCAGCGGTTTAAGTATCG-3′) were used to amplify the 3′ end of Px26 and to add a c-myc epitope tag by PCR using pΔFBgusPx26 as a template. The PCR product was cloned into the pCR-4-Blunt vector (Invitrogen), released by SacI and EcoRI, and used to replace the 3′ end of the Px26 gene in pΔFBgusPx26, generating plasmid pΔFBgusPx26c-myc.
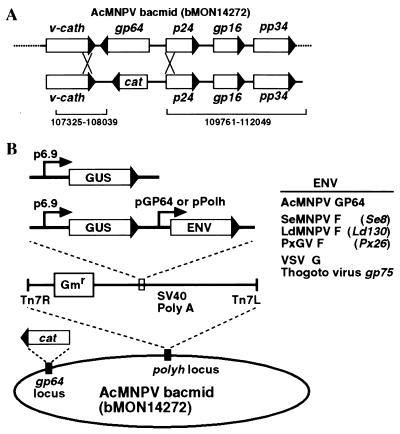
FIG. 1.
(A) Strategy for generation of a gp64-null AcMNPV bacmid by homologous recombination in E. coli. The gp64 locus of the AcMNPV bacmid (bMON14272) is shown above the EcoRI-HindIII fragment that was used as a transfer vector to replace the gp64 locus with a chloramphenicol resistance gene (cat). Sequences included for homologous recombination (107325 to 108039 and 109761 to 112049) are indicated (1). (B) Strategy for insertion of envelope protein gene constructs into the polyhedrin locus of the gp64-null AcMNPV bacmid. Inserts include a p6.9 promoter-GUS reporter and no envelope protein gene (top) and a p6.9 promoter-GUS reporter plus envelope protein genes (ENV) under the control of either the gp64 promoter (pGP64) or the polyhedrin promoter (pPolh) (center). Envelope protein gene cassettes were inserted into the att b site (indicated by right and left insertion sites, Tn7R and Tn7L) in the polyhedrin locus by Tn 7-based transposition. Envelope proteins and genes (italicized) are listed on the right.
Transpositions of inserts from donor plasmids into the gp64-null bacmid were confirmed by gentamicin resistance, blue-white screening according to the BAC-to-BAC manual (Invitrogen), and PCR using a primer corresponding to sequences in the GUS reporter gene in the donor plasmid (P-gus-jm [5′-GTG AAGAGTATCAGTGTGCATG-3′]) in combination with a primer corresponding to bacmid sequences adjacent to the transposition site (either the M13/pUC forward primer [5′-CCCAGTCACGACGTTGTAAAACG-3′] or the M13/pUC reverse primer [5′-AGCGGATAACAATTTCACACAGG-3′]).
Transfection-infection assay.
DNAs were prepared from 1.5-ml cultures of 2 to 3 independent colonies carrying the bacmid with the inserted heterologous envelope protein gene according to the BAC-to-BAC manual (Invitrogen) and were analyzed in parallel. For transfections, 12.5% of each DNA preparation was used to transfect 9 × 105 Sf9 or Sf9Op1D (28) cells in a 6-well plate by calcium phosphate precipitation. At 72 h (protocol 1) or 10 days (protocol 2) posttransfection, the supernatant (passage 1) was centrifuged for 10 min at 2,200 × g to remove debris, and this clarified supernatant (50 or 500 μl) was used to infect 9 × 105 Sf9 cells for 48 h (protocol 1) or 72 h (protocol 2). Both transfected and infected cells were stained for GUS activity (according to the BAC-to-BAC manual [Invitrogen]) to monitor transfection efficiency and to detect infection by virions generated in transfected cells. Supernatants from infected Sf9 cells (passage 2) were stored at 4°C. In all cases where rescue was not observed by using protocol 1, no rescue was observed by using protocol 2. As a control to confirm that no lethal mutation had been acquired during propagation of the AcMNPV bacmid in Escherichia coli, all bacmid DNAs were also used to transfect Sf9Op1D cells (which express a GP64 protein) (28) and the supernatants from those transfections were used to infect Sf9 cells.
Transfections into CHO cells were performed with Lipofectin (Invitrogen), and 3 μg of DNA was used to transfect 70% confluent cells in 60-mm plates according to the manufacturer's instructions.
Virus amplification and preparation.
Viruses carrying envelope protein genes that rescued the gp64-null phenotype were amplified by infecting one T150 flask containing 107 Sf9 cells with 500 μl of passage 2 supernatant, and cells were split every 3 to 5 days for 10 to 13 days. Viruses carrying envelope proteins that did not rescue the gp64 deletion were amplified in a similar manner but with Sf9Op1D cells and Sf9Op1D cell-derived supernatant. Amplified pseudotyped viruses were titered on Sf9Op1D cells by a TCID50 (50% tissue culture infective dose) assay (25) and scored for infection by examining cells for GUS expression. Budded virions were purified by centrifugation through a 25% sucrose pad, and Western blot analysis was performed as described previously (20).
RESULTS
Disruption of the gp64 gene in an AcMNPV bacmid.
The critical roles that GP64 plays in AcMNPV budding and propagation of infection were demonstrated by early physical and functional studies (4, 12, 14, 16, 32) and by studies of gp64-null viruses that were generated by homologous recombination in insect cells (22, 24). To determine whether the recently identified baculovirus F proteins serve as functional analogs of GP64 in baculoviruses with no gp64 gene, we modified an AcMNPV bacmid (17) by first deleting the gp64 gene and then inserting heterologous envelope protein genes. The gp64 gene of the AcMNPV bacmid bMON14272 was deleted by homologous recombination in recA+ E. coli BJ5183 cells by using a linear EcoRI-HindIII fragment of DNA containing a cat cassette flanked by 2,288 bp of sequences upstream, and 714 bp of sequences downstream, of the gp64 locus (Fig. 1A). Three colonies resistant to kanamycin and chloramphenicol were selected and confirmed by PCR to contain the desired recombinant gp64-null bacmid (data not shown). All three gp64-null bacmids failed to propagate an infection after transfection into Sf9 cells, while in parallel experiments, the original bacmid was capable of initiating a spreading infection (data not shown). gp64-null bacmid DNA was then transformed into DH10B cells containing a helper plasmid (pMON7124) that encodes a Tn7 transposase (17). The resulting E. coli strain (gp64− bacmid-DH10B+pMON7124) was used to insert reporter and envelope protein genes into the gp64-null bacmid for subsequent studies. To examine the abilities of several heterologous envelope proteins to rescue the gp64-null AcMNPV, each heterologous envelope protein gene was inserted into the polyhedrin locus of the above gp64-null bacmid by using a donor plasmid and Tn7-based transposition (17). Each donor plasmid contained a GUS reporter gene plus the heterologous envelope protein gene under the control of the AcMNPV gp64 promoter. In some cases, the envelope protein gene was also placed under the control of the much stronger polyhedrin promoter (Fig. 1B). As a positive control, the Ac MNPV gp64 gene was inserted into a donor plasmid. A donor plasmid containing no envelope protein gene served as a negative control. The resulting bacmids were named vAcgp64− (no gp64 gene), vAcgp64−/Acgp64 (gp64 reinserted), and vAcgp64−/env (“env” stands for the heterologous envelope protein gene that was inserted) (Fig. 1B).
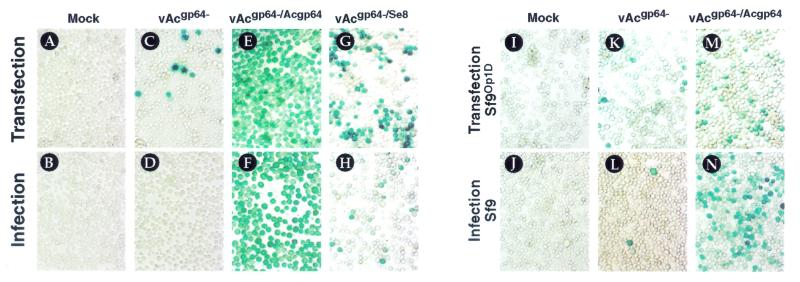
Following the insertion of each heterologous envelope protein gene, bacmid DNAs from at least two independent colonies were used to separately transfect Sf9 cells, and cells were scored for the spread of infection and generation of infectious virions by a transfection-infection assay (see Materials and Methods). After transfection of Sf9 cells with each bacmid construct, culture supernatants were harvested from transfected cells at 72 h (or, alternatively, at 10 days) posttransfection and used to infect a second group of Sf9 cells, which were incubated for 48 h (or, alternatively, for 72 h). If infectious virions are produced from the transfected cells, infection should be detected in the second group of cells. Both transfected and infected cells were stained for GUS reporter activity to detect entry and infection by the virus. Our results show that although the gp64-null (vAcgp64−) bacmid entered cells by transfection and the late promoter was activated, infectious virions were not produced and infection did not spread (Fig. 2C and D). In parallel experiments using the gp64-null bacmid that was repaired by reinsertion of gp64 (vAcgp64−/Acgp64), infection spread in both transfected and infected cells (Fig. 2E and F). Furthermore, transfection of the gp64-null bacmid into Sf9Op1D cells (which express GP64) (28) also resulted in generation of infectious virions (Fig. 2K and L), indicating that the defect can be rescued by supplying the GP64 protein in trans. Thus, the inability of the gp64-null bacmid to propagate an infection is due to the lack of GP64 and not to possible second-site mutations.
FIG. 2.
Transfection-infection assay for viral propagation. Sf9 cells were transfected with the indicated bacmids, incubated for 72 h, and stained for GUS activity (A, C, E, and G). Supernatants from the transfected cells (top panels) were used to infect Sf9 cells (B, D, F, and H), which were incubated for 48 h and then stained for GUS activity. Staining of cells in the lower panels (infected cells) indicates that infectious virions were generated in the transfected cells. Bacmids that fail to propagate an infection in Sf9 cells can propagate an infection in cells expressing OpMNPV GP64 (Sf9Op1D). Sf9Op1D cells were transfected with the indicated bacmids (I, K, and M) and incubated for 72 h; then supernatants were transferred to Sf9 cells and stained for GUS activity after 48 h (J, L, and N).
Envelope proteins from group II NPVs rescue an AcMNPV gp64-null bacmid.
To determine whether F proteins from heterologous baculoviruses could rescue the gp64-null bacmid, we placed three F protein genes under the control of the AcMNPV gp64 promoter and inserted each construct into the gp64-null bacmid (Fig. 1B and Table 1). Transfection of Sf9 cells with gp64-null bacmids carrying the F protein gene from SeMNPV or LdMNPV resulted in rescue of the gp64-null AcMNPV and production of infectious virions (Fig. 2G and H; Table 1, vAcgp64−/Se8 and vAcgp64−/Ld130). In contrast, the gp64-null bacmid carrying the F protein gene from the Plutella xylostella GV (PxGV) (Table 1, vAcgp64−/Px26) (10) did not produce infectious virions. To ensure that the PxGV F protein can be expressed in Sf9 cells, a bacmid containing sequences encoding the PxGV F protein with a c-myc epitope tag (vAcgp64−/Px26c-myc) was generated. After transfection of the vAcgp64−/Px26c-myc bacmid into Sf9 cells, the c-myc-tagged PxGV F protein was detected by Western blot analysis of infected cell lysates (data not shown), indicating that the PxGV F protein could be expressed. However, neither the wild-type nor the c-myc-tagged PxGV F protein rescued gp64-null AcMNPV, suggesting that PxGV F was unable to substitute for GP64 function. To determine whether a stronger promoter driving expression of the PxGV F protein could result in rescue of the gp64 deletion, we tested a gp64-null bacmid containing the Px26 gene under the control of the polyhedrin promoter (vAcgp64−/pPolh-Px26). Although a parallel LdMNPV F gene construct (under polyhedrin promoter control) complemented GP64 function, the PxGV F gene construct did not (Table 1). When both bacmids encoding Px26 were transfected into Sf9Op1D cells, infectious virions were produced (Table 1), indicating that the lack of virion production in Sf9 cells was due to lack of a functional envelope protein and not to a second-site mutation. Thus, our results show that the F protein genes from LdMNPV and SeMNPV rescued a gp64-null bacmid, but the F protein gene from PxGV did not.
TABLE I.
Pseudotyped gp64-null bacmids and viruses
| gp64-null bacmid | Envelope protein | Promoter | Rescue | Titer (IU/ml)a in:
|
|
|---|---|---|---|---|---|
| Sf9 cells | Sf9Op1D cells | ||||
| Controls | |||||
| vAcgp64− | None | − | NA | 8.3 × 106 | |
| vAcgp64−/Acgp64 | AcMNPV GP64 | gp64 | ++ | 2.9 × 108 | ND |
| Baculovirus F proteins | |||||
| vAcgp64−/Se8 | SeMNPV F | gp64 | + | 2.5 × 107 | ND |
| vAcgp64−/Se8K149 | SeMNPV FK149 | gp64 | − | NA | 2.2 × 107 |
| vAcgp64−/Ld130 | LdMNPV F | gp64 | + | 5.0 × 106 | 8.3 × 106 |
| vAcgp64−/pPolh-Ld130 | LdMNPV F | polyh | + | 8.3 × 104 | 6.9 × 104 |
| vAcgp64−/Px26 | PxGV F | gp64 | − | NA | 1.9 × 106 |
| vAcgp64−/pPolh-Px26 | PxGV F | polyh | − | NA | 1.4 × 107 |
| Other viral Env proteins | |||||
| vAcgp64−/VSV-G | VSV G | gp64 | + | 5.9 × 107 | ND |
| vAcgp64−/Thogp75 | Thogoto GP75 | gp64 | − | NA | 2.8 × 106 |
| vAcgp64−/pPolh-Thogp75 | Thogoto GP75 | polyh | − | NA | ND |
NA, not applicable; ND, not determined.
Thogoto virus GP75 is incorporated into AcMNPV virions but does not rescue a gp64-null bacmid.
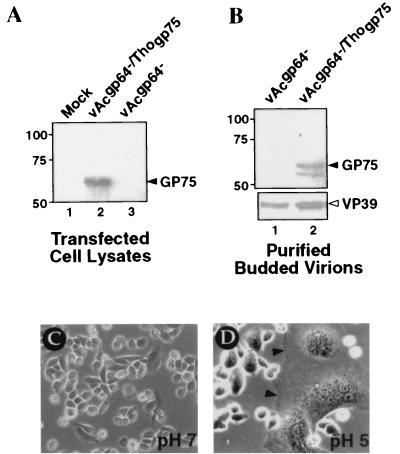
Two envelope proteins from viruses unrelated to baculoviruses were also examined in these studies: vesicular stomatitis virus (VSV) G protein and Thogoto virus GP75. VSV-G shows no sequence similarity to either GP64 or F proteins but was included as a positive control, since a previous study showed that it was able to rescue a gp64 deletion (20). In agreement with the findings of that study, the VSV G gene rescued the gp64 deletion in AcMNPV (Table 1). Although the Thogoto virus is a single-stranded RNA virus (orthomyxovirus), the Thogoto virus GP75 envelope protein shares 28% amino acid sequence identity with the AcMNPV GP64 protein and therefore appears to be derived from a common ancestor (23). We therefore asked whether GP75 could rescue a gp64-null AcMNPV bacmid. The Thogoto virus gp75 gene, under the control of either the gp64 promoter or the polyhedrin promoter, failed to rescue the gp64 deletion in AcMNPV (Table 1). Expression of Thogoto virus GP75 in transfected Sf9 cells and the presence of GP75 in pseudotyped virions were confirmed by Western blot analysis (Fig. 3A and B). Moreover, when Sf9Op1D cells were transfected with either the vAcgp64−/Thogp75 or the vAcgp64−/pPolh-Thogp75 bacmid, the virus could be propagated (Table 1), confirming that the bacmid was otherwise replication competent. Because Thogoto virus GP75 was expressed in infected Sf9 cells and can be incorporated into AcMNPV budded virions, these results suggest that Thogoto virus GP75 is not able to functionally substitute for GP64 in the context of an AcMNPV infection. To determine whether GP75 was functional in Sf9 cells, we performed a membrane fusion (syncytium formation) assay in Sf9 cells, using the donor plasmid construct that expresses GP75 under the control of the AcMNPV gp64 promoter. Transiently expressed GP75 did not mediate detectable low-pH-triggered membrane fusion (data not shown). Since Thogoto virus is a tick-borne virus that replicates in a variety of mammals (6), we also tested GP75 for syncytium formation in mammalian cells. When GP75 was expressed in CHO cells and similarly assayed, pH-triggered membrane fusion was readily detected (Fig. 3C and D). Thus, GP75 mediated membrane fusion in mammalian cells but did not appear to be functional in insect Sf9 cells. This may explain the inability of GP75 to rescue the gp64-null bacmid.
FIG. 3.
Thogoto virus GP75 protein expression, detection in AcMNPV budded virions, and membrane fusion activity. (A) Western blot analysis of GP75 expression in Sf9 cells that were either mock transfected (lane 1) or transfected with the vAcgp64−/Thogp75 (lane 2) or vAcgp64− (lane 3) bacmid and examined at 72 h posttransfection. GP75 was detected by using an affinity-purified antibody generated against a GP75 peptide (N-KERAHEKSKDLPFGNK-C). (B) Western blot detection of GP75 protein in purified budded virions (vAcgp64− or vAcgp64−/Thogp75) generated in Sf9Op1D cells. An anti-capsid protein (VP39) antibody was used as an internal control (lower panel). (C and D) Syncytium formation assay. CHO cells were transfected with a plasmid (pRC-THO) expressing GP75 under the control of a cytomegalovirus early promoter. At 24 h posttransfection, cells were incubated in phosphate-buffered saline at pH 7.0 (C) or pH 5.0 (D) for 15 min; then they were examined for syncytium formation after 90 min. Large syncytial masses containing many nuclei are indicated by arrowheads. Cells transfected with the empty vector (pRC-CMV) did not show syncytium formation (data not shown).
Infectious virion production by pseudotyped viruses.
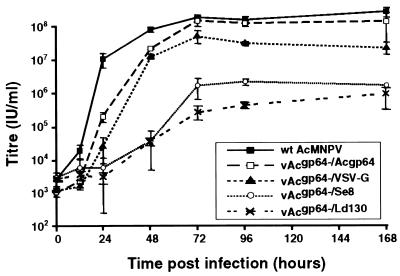
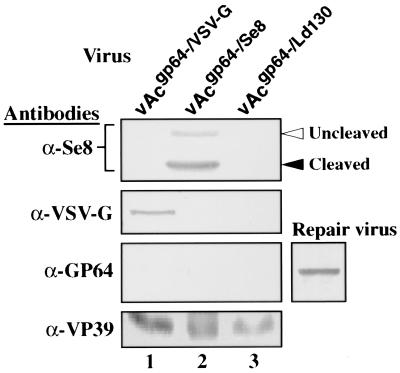
To more thoroughly characterize the ability of certain envelope proteins to substitute for GP64 in pseudotyped gp64-null viruses, we performed one-step growth curves. Infectious virus production from the repair virus (containing wild-type gp64 reinserted into the polyhedrin locus) was similar to that from wild-type AcMNPV (Fig. 4), indicating that the use of the bacmid itself and the introduction of envelope genes into the polyhedrin locus did not have a significant negative effect on infectious virion production. Virus production by the VSV-G-pseudotyped virus was approximately 1 log unit lower than that by the wild-type AcMNPV (Fig. 4). Virus production by viruses pseudotyped with the F protein from either SeMNPV or LdMNPV was approximately 2 log units lower than that by control viruses (Fig. 4). The presence of VSV-G and SeMNPV F proteins in purified budded virions was also confirmed (Fig. 5, lanes 1 and 2). Although Ld130 antibodies were not available, the GP64-null phenotype was confirmed (Fig. 5, lane 3), and the genotypes of all viruses were confirmed by PCR analysis (data not shown).
FIG. 4.
One-step growth curves are shown for wild type AcMNPV, the repair virus vAcgp64−/Acgp64, and AcMNPV gp64-null viruses pseudotyped with VSV-G (vAcgp64−/VSV-G), Se8 (vAcgp64−/Se8), or Ld130 (vAcgp64−/Ld130). Sf9 cells were infected at a multiplicity of infection of 5, and supernatants were harvested at the indicated times postinfection and titered on Sf9Op1D cells. Each data point represents the average from three separate infections (error bars, standard deviations), except for the data point for the Se8-pseudotyped virus at 168 h postinfection, which represents a single infection.
FIG. 5.
Western blot analysis of pseudotyped virions. A series of Western blots of purified virions from pseudotyped viruses vAcgp64−/VSV-G (lane 1), vAcgp64−/Se8 (lane 2), and vAcgp64−/Ld130 (lane 3) were probed with antibodies α-Se8 (against SeMNPV F1 protein, the 60-kDa Se8 cleavage product) (33), α-VSV-G (20), and α-GP64 (monoclonal antibody AcV5) (14). An anti-nucleocapsid antibody (α-VP39) was used as an internal control for each preparation of purified budded virions (20). GP64 was not detected from pseudotyped viruses but was detected in a positive control shown on the right (Sf9 cell lysates infected with the repair virus, vAcgp64−/Acgp64). Positions of uncleaved and cleaved SeMNPV F are indicated.
Proteolytic cleavage of the SeMNPV F protein is essential for rescue of the gp64-null bacmid.
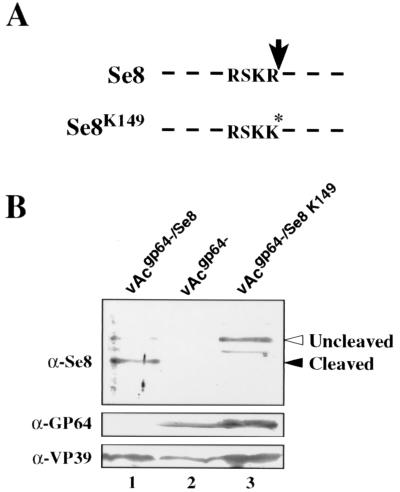
Recently, a proprotein convertase (PPC or furin) cleavage site was identified in a number of baculovirus F proteins (15). The consensus furin cleavage site [R-X-(R/K)-R] is conserved at a similar position in F proteins from group II NPVs and GVs. Recent biochemical and mutagenic studies of the SeMNPV F protein strongly suggested that the SeMNPV F protein is cleaved by a PPC or furin protease (33). To determine whether cleavage at the furin cleavage site is essential for rescue of the gp64-null virus by the SeMNPV F protein, we generated a gp64-null bacmid expressing a SeMNPV F protein with a point mutation in the furin cleavage site (Se8K149) (Fig. 6A). This point mutation, an arginine-to-lysine substitution in a critical residue in the consensus furin cleavage site, was previously shown to prevent F protein cleavage (33). Using two protocols for testing gp64 complementation, we found that the F protein containing the Se8K149 mutation did not complement the gp64-null bacmid (see Table 1). However, the Se8K149-pseudotyped virus can be propagated in Sf9Op1D cells, and titers of approximately 2 × 107 IU/ml can be achieved in these cells (Table 1, vAcgp64−/Se8K149). Western blot analysis of Se8K149-pseudotyped virions shows that Se8K149 can be detected in these preparations (Fig. 6B, lane 3), indicating that Se8K149 can be incorporated into the virion. In these preparations, Se8K149 was detected as a protein of approximately 75 kDa. In contrast, a 60-kDa processed form of F (Se8) was the major protein form detected in virions pseudotyped with the wild-type SeMNPV F protein (Fig. 6B, lane 1) and in wild-type SeMNPV virions (15). These results demonstrate that the putative furin cleavage site is necessary both for processing of this F protein and for complementation of the gp64-null virus in Sf9 cells.
FIG. 6.
(A) Diagram of the furin cleavage site in the SeMNPV F protein (Se8) and the Se8K149 mutant. RSKR represents the furin cleavage site in the wild-type SeMNPV F protein (Se8), and RSKK represents the furin cleavage site containing a single amino acid substitution (Se8K149). (B) Western blot analysis of gp64-null budded virions pseudotyped with Se8 or Se8K149. Virion preparations (lanes 1 to 3) were probed with antibodies directed against the SeMNPV F protein (α-Se8), GP64 (α-GP64), and VP39 (α-VP39). Virions of vAcgp64−/Se8 were generated in infected Sf9 cells (lane 1), whereas virions of vAcgp64− (lane 2) and vAcgp64−/Se8K149 (lane 3) were generated in infected Sf9Op1D cells. The latter virions therefore contain Op MNPV GP64 (lanes 2 and 3). The VP39 capsid protein was included as an internal control. Positions of cleaved and uncleaved forms of Se8 are indicated.
DISCUSSION
The GP64 envelope protein is necessary for the critical first steps (attachment and entry) and the essential final steps (assembly and budding) in the infection cycle of AcMNPV. In this study, we examined functional similarities between GP64 and the recently discovered baculovirus F proteins. We show that an AcMNPV gp64-null virus can be rescued by F proteins from LdMNPV and SeMNPV (group II NPVs). The gp64-null viruses pseudotyped with F proteins from LdMNPV and SeMNPV generated substantially lower virus titers than wild-type AcMNPV. This suggests that the degree of compatibility between these F proteins and other AcMNPV or host proteins may not be optimal. Such incompatibilities might result in (i) inefficient virion budding, (ii) inefficient virus binding to host cell receptors, or (iii) inefficient membrane fusion and virus release from endosomes during virus entry. In spite of the differences in efficiency, these F proteins were able to substitute at least partially for all of the essential functions of GP64 and resulted in the production of infectious viruses. Thus, F proteins are functionally analogous to GP64. In contrast to the F protein genes from SeMNPV and LdMNPV, the PxGV gene did not rescue the gp64-null AcMNPV. Although it is possible that the PxGV F protein was inefficiently expressed, we believe this is unlikely because both the LdMNPV and SeMNPV F proteins were expressed in quantities sufficient to rescue the gp64-null bacmid, and expression of an epitope-tagged PxGV F protein was detected (data not shown). Therefore, we speculate that the inability of the PxGV F protein gene to rescue the gp64-null AcMNPV may be related to the greater evolutionary distance between AcMNPV and PxGV, resulting in less-compatible interactions between the F protein and other AcMNPV or host cell proteins. Consistent with this hypothesis, LdMNPV and SeMNPV F proteins are similar in length (676 and 665 amino acids, respectively) and more highly conserved (38% identity), whereas the PxGV F protein is only 544 amino acids long, and the PxGV and LdMNPV F proteins are only 24% identical. Thus, our results from pseudotyping experiments with heterologous F proteins suggest that functional complementation reflects evolutionary distance.
It is of special interest that group I NPVs such as AcMNPV and OpMNPV (which carry gp64 genes) also have genes encoding F protein homologs (Ac23 and Op21, respectively). One hypothesis to explain the existence of two different but functionally similar envelope proteins (GP64 and F proteins) within the family Baculoviridae, plus the presence of both GP64 and F protein homologs in certain group I NPVs, is the possibility that gp64 is a recent acquisition by an ancestor of the group I NPVs (29). In this scenario, many or most of the functions of the ancestral AcMNPV F protein homolog may have been replaced by the GP64 protein, thus releasing all or portions of the AcMNPV F protein homolog from selection pressure. Consistent with this hypothesis, AcMNPV and OpMNPV F protein homologs appear to be less well conserved than F proteins from group II NPVs and GVs. Even though group I and II NPVs are more closely related phylogenetically (13), the AcMNPV F protein (Ac23) and the LdMNPV F protein (Ld130) are only 13% identical. In contrast, the LdMNPV and PxGV F proteins are approximately 24% identical (29). In addition, the essential furin cleavage site that is present in LdMNPV and SeMNPV F proteins is absent in the predicted Ac23 and Op21 proteins. Furthermore, while F proteins from group II NPVs can functionally replace AcMNPV GP64, the F protein homolog of AcMNPV (Ac23) cannot compensate for a gp64 deletion in AcMNPV (22, 24). While F proteins from group II NPVs and GVs likely serve an essential role in infectivity, the function of the more divergent F protein homologs in group I NPVs remains unknown. However, the observation that AcMNPV and OpMNPV F gene homologs are intact ORFs suggests that they may provide a beneficial function. Consistent with this hypothesis, Pearson et al. (27a) recently reported that the OpMNPV F homolog (Op21) is expressed and colocalized with GP64 in the budded virion membrane. Baculovirus F protein homologs were also recently identified in a subgroup of apparent retroviruses. This subgroup, referred to as insect errantiviruses (19), appears to have evolved from invertebrate long terminal repeat-containing retrotransposons that acquired an envelope protein gene, to become infectious retroviruses (30, 31). The envelope protein genes of the Ty3/Gypsy group of retrotransposons from Drosophila melanogaster and the TED retrotransposon from T. ni (26) are homologs of the baculovirus F protein genes, and it has recently been suggested (19) that these invertebrate retrotransposons acquired their envelope protein gene from a baculovirus. In addition, a possible cellular homolog of the baculovirus F protein gene has been identified in Drosophila (29). Exchange of genetic material is known to occur between baculoviruses and their hosts, and also between coinfecting baculoviruses (8, 18, 21). A recent study by Herniou et al. (13) suggested that baculovirus genomes were highly fluid, with gene rearrangements and multiple gene content changes occurring during their evolution. Of 17 genes that are thought to be unique to group I NPVs, 4 encode structural proteins (13). Thus, gp64 and possibly other structural protein genes in group I NPVs may represent gene acquisitions from the host or from other viruses. The acquisition of new envelope proteins represents a mechanism by which some viruses are known to enhance their fitness. For example, orthomyxoviruses use reassortment to increase their diversity, and the acquisition of new envelope protein genes permits them to avoid immune detection. Although it is not clear if or why GP64 would provide a selective advantage, we might speculate that acquisition of GP64 could have increased the efficiency of virus-receptor interactions, virus entry, or virus budding. Alternatively, GP64 may have facilitated the recognition of a different host cell receptor(s) and increased the range of cells or tissues infected. Thus, acquisition of this gene may have resulted in either more productive infections, or an increase or change in cell types infected.
In addition to F proteins, we also asked whether GP75 from Thogoto virus could functionally complement GP64. The fact that Thogoto virus GP75 shows 28% amino acid sequence identity to AcMNPV GP64 (23) is striking and suggests that GP64 and GP75 proteins have a common evolutionary origin, even though orthomyxoviruses and baculoviruses do not appear to share a common evolutionary origin. Since Thogoto virus is vectored to mammals by arthropods (ticks), an intriguing possible scenario is that an ancestor of the Thogoto virus acquired a GP64 homolog either from the arthropod host genome or from a host coinfected with a baculovirus (23). However, other possible scenarios cannot be eliminated. Since it is likely that the GP64 and GP75 proteins have a common evolutionary origin, we asked in the present study whether GP75 could substitute for GP64 in AcMNPV. We found that GP75 was expressed and appropriately transported in infected Sf9 cells, and GP75 was also found in pseudotyped virions. However, GP75 did not rescue the gp64-null AcMNPV. In subsequent experiments we were unable to detect membrane fusion activity in Sf9 cells expressing GP75, while, in contrast, GP75 mediated membrane fusion in mammalian CHO cells. These observations suggest that while GP75 and GP64 share functional similarity (i.e., they are both membrane fusion proteins), GP75 may be somehow incompatible with the Sf9 host cells or with the AcMNPV entry or budding apparatus. This incompatibility may reflect GP64 and GP75 adaptations to the different host and viral systems.
In summary, we have developed and used a versatile genetic system to examine the function and compatibility of heterologous envelope protein genes in the context of an AcMNPV baculovirus infection. Using this system, we demonstrated that F proteins from group II NPVs can rescue infectivity and budded virus production in the absence of gp64. We also found that Thogoto virus GP75 did not substitute for GP64 in AcMNPV, although the two proteins share 28% amino acid identity. In contrast, the unrelated but highly promiscuous VSV G protein effectively rescued a gp64-null AcMNPV. The bacmid-based pseudotyping system developed for these studies will be useful for future studies aimed at understanding the nature of envelope protein interactions necessary for efficient entry and budding, and for pseudotyping AcMNPV for potential medical and biotechnological applications. Examples of such applications might include pseudotyping AcMNPV with modified envelope proteins in order to modulate the host range of AcMNPV as a biocontrol agent, or to target specific cells or tissues for efficient or restricted delivery of genes for insect transgenesis or for gene therapy.
Acknowledgments
We thank T. Oomens, P. Nuttall, Y. Hashimoto, and J. Slavicek for supplying plasmids containing envelope protein genes and D. Bideshi, B. Federici, and T. Shen for valuable advice on methods.
This work was supported by NIH grant AI33657 and Boyce Thompson Institute project 1255-17.
REFERENCES
- 1.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 2.Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T.ni. J. Gen. Virol. 81:1593-1599. [DOI] [PubMed] [Google Scholar]
- 3.Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. E. Volkman. 2000. Baculoviridae: taxonomic structure and properties of the family, p. 195-202. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 4.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulach, D. M., C. A. Kumar, A. Zaia, B. Liang, and D. E. Tribe. 1999. Group II nucleopolyhedrovirus subgroups revealed by phylogenetic analysis of polyhedrin and DNA polymerase gene sequences. J. Invertebr. Pathol. 73:59-73. [DOI] [PubMed] [Google Scholar]
- 6.Davies, C. R., L. D. Jones, and P. A. Nuttall. 1986. Experimental studies on the transmission cycle of Thogoto virus, a candidate orthomyxovirus, in Rhipicephalus appendiculatus. Am. J. Trop. Med. Hyg. 35:1256-1262. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, M. J., G. E. Smith, and M. D. Summers. 1983. Acquisition of host cell DNA sequences by baculoviruses; relationships between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis virus. J. Virol. 47:287-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen, P. D., and L. K. Miller. 2001. Insect viruses, p. 599-628. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 10.Hashimoto, Y., T. Hayakawa, Y. Ueno, T. Fujita, Y. Sano, and T. Matsumoto. 2000. Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275:358-372. [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa, T., G. F. Rohrmann, and Y. Hashimoto. 2000. Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278:1-12. [DOI] [PubMed] [Google Scholar]
- 12.Hefferon, K., A. Oomens, S. Monsma, C. Finnerty, and G. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455-468. [DOI] [PubMed] [Google Scholar]
- 13.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole-genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 15.IJkel, W. F. J., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275:30-41. [DOI] [PubMed] [Google Scholar]
- 16.Leikina, E., H. O. Onaran, and J. Zimmerberg. 1992. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 304:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda, S., S. G. Kamita, and A. Kondo. 1993. Host range expansion of Autographa californica nuclear polyhedrosis virus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating from Bombyx mori NPV. J. Virol. 67:6234-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik, H. S., S. Henikoff, and T. H. Eickbush. 2000. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 10:1307-1318. [DOI] [PubMed] [Google Scholar]
- 20.Mangor, J. T., S. A. Monsma, M. C. Johnson, and G. W. Blissard. 2001. A GP64-null baculovirus pseudotyped with vesicular stomatitis virus G protein. J. Virol. 75:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, D. W., and L. K. Miller. 1982. A virus mutant with an insertion of a copia-like transposable element. Nature (London) 299:562-564. [DOI] [PubMed] [Google Scholar]
- 22.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morse, M. A., A. C. Marriott, and P. A. Nuttall. 1992. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein gp64. Virology 186:640-646. [DOI] [PubMed] [Google Scholar]
- 24.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64. to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 25.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman and Co., New York, N.Y.
- 26.Ozers, M. S., and P. D. Friesen. 1996. The env-like open reading frame of the baculovirus-integrated retrotransposon TED encodes a retrovirus-like envelope protein. Virology 226:252-259. [DOI] [PubMed] [Google Scholar]
- 27.Pearson, M. N., C. Groten, and G. F. Rohrmann. 2000. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J. Virol. 74:6126-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Pearson, M. N., R. L. Russell, and G. F. Rohrmann. 2001. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology 291:22-31. [DOI] [PubMed] [Google Scholar]
- 28.Plonsky, I., M. S. Cho, A. G. P. Oomens, G. W. Blissard, and J. Zimmerberg. 1999. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology 253:65-76. [DOI] [PubMed] [Google Scholar]
- 29.Rohrmann, G. F., and P. A. Karplus. 2001. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC E vol. Biol. 1:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, S. U., T. Gerasimova, M. Kurkulos, J. D. Boeke, and V. G. Corces. 1994. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 8:2046-2057. [DOI] [PubMed] [Google Scholar]
- 31.Tanda, S., J. L. Mullor, and V. G. Corces. 1994. The Drosophila Tom retrotransposon encodes an envelope protein. Mol. Cell. Biol. 14:5392-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkman, L. E., P. A. Goldsmith, R. T. Hess, and P. Faulkner. 1984. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology 133:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westenberg, M., H. Wang, W. F. IJkel, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]