Abstract
Conformational changes in the troponin/tropomyosin complex significantly alter the mechanical properties of cardiac muscle. Phosphorylation of cardiac troponin I, part of the troponin/tropomyosin complex, reduces calcium affinity, which leads to increased relaxation of cardiac muscle. Because cardiac troponin I plays a central role in tuning the heart to different work demands, detailed knowledge of troponin I protein regulation is required. Our group previously detected naturally occurring antisense RNA for troponin I in human and rat hearts, and here, attempt to unravel the structure of rat cardiac troponin I antisense RNA. We performed rapid amplification of cDNA ends (RACE) experiments and discovered antisense sequences identical to a copy of the sense mRNA, which led us to conclude that the antisense RNA must be transcribed from troponin I mRNA in the cytoplasm. Moreover, we isolated RNA structures comprising sense and antisense sequences in one continuous molecule. As we found no homolog structures described in the literature, we called this “hybrid RNA.” Because a duplex formation was demonstrated previously we concluded that hybrid RNA is a consequence of a tight interaction between sense and antisense troponin I RNA in vivo, which we discuss in the article.
Keywords: endogenous antisense RNA, hybrid RNA, cardiac troponin I, sense-antisense RNA interaction
INTRODUCTION
A growing number of natural antisense RNA transcripts of different eukaryotic genes have been detected over the last few years and the widespread existence of natural antisense RNA in eukaryotes is now well accepted (Yelin et al. 2003; Røsok and Sioud 2004). Although the function(s) of such natural antisense RNAs are as yet mostly unclear, growing evidence indicates an involvement in gene regulation (for review, see Kumar and Carmichael 1998). For instance, two recent publications from Tufarelli et al. (2003) and Haddad et al. (2003) demonstrated a direct involvement of natural antisense RNA in down-regulation of gene expression.
Our group is focusing on natural antisense RNAs from cardiac tissues and we were previously able to isolate different antisense RNAs, including α and β myosin heavy chain (MyHC; Luther et al. 1998) and cardiac troponin I (cTnI) antisense RNAs (Podlowski et al. 2002). Our investigations with cTnI antisense RNA have so far revealed cTnI antisense–sense duplex structures in vivo and reduced protein expression with oligonucleotides directed against the sense RNA in vitro (Podlowski et al. 2002). These results point to a possible role of cTnI antisense in the regulation of cTnI protein expression at the translational level, as postulated for other antisense products.
Cardiac TnI, as part of the heterotrimeric troponin complex comprising cTnI, troponin T, and troponin C, plays a major role in the actin/myosin interaction by regulating conformational changes in the troponin/tropomyosin complex (Tobacman 1996; Solaro and Rarick 1998). Phosphorylation of cTnI upon β-adrenergic stimulation causes increased relaxation of cardiac muscle by reducing the calcium affinity of the troponin complex (Holroyde et al. 1980). Thus, proper regulation of cTnI expression is clearly essential for normal heart function, with cTnI antisense RNA possibly playing an important role.
Interestingly, natural antisense RNAs described so far in the literature are either cis-encoded, meaning they are transcribed from the opposite strand in the genome and correspond to the complete or partial hnRNA sequence of the gene, or they are trans-encoded and therefore originate from a different locus in the genome (e.g., pseudogenes). The latter share only partial identity with the sense RNA (for reviews, see Kumar and Carmichael 1998; Vanhée-Brossollet and Vaquero 1998; Good 2003). No concrete examples of antisense RNA transcribed from the mRNA in the cytoplasm have been reported so far. Because our previous data from analyzing cTnI antisense RNA pointed to possible antisense transcription in the cytoplasm, it was important to determine the exact structure of the antisense RNA, to resolve whether it is derived from the mRNA or the genome. We therefore decided to perform 5′ and 3′ RACE experiments on the cTnI antisense RNA. Surprisingly, the further characterization of cTnI antisense RNA revealed a new sense/antisense RNA molecule structure, which may provide more insights into the regulatory pathway involving cTnI antisense RNA.
RESULTS
Characterization of antisense cTnI transcripts by RACE
Because precise characterization of the cTnI antisense RNA should provide more information about the origin of the antisense RNA, rapid amplification of cDNA ends (RACE) experiments were performed with total and poly(A)+ RNA. To amplify the antisense-specific cTnI sequence, in 5′ RACE a cTnI forward primer (631F; for primer details, see Fig. 1 and Table 1), and in 3′ RACE a cTnI reverse primer (363R) were used together with the universal primer mix supplied in the RACE kit.
FIGURE 1.
Overview of the positions of the cTnI primers used. The cTnI cDNA sequence is shown schematically as published in the GenBank database (accession no. X58499). The open box represents the open reading frame, whereas the black bars represent the 5′ and 3′ nontranslated parts of the sequence. The positions of the introns are indicated. The primers are shown in their respective positions and orientations (arrows). Sequence information on the primers is given in Table 1.
TABLE 1.
Sequences of primers used for reverse transcription and PCR
| Forward primers | Primer name | Sequence (5′ to 3′) | Used for |
| 255F | ATTGCGAAGCAGGAGATGGAGCGTGAG | PCR, 5′RACE | |
| 631F | ATTGAGAAGGAAAACCGGGAGGTGGGAG | 5′RACE |
| Reverse primers | Primer name | Sequence (5′ to 3′) | Used for |
| 110R | ATCGCTGCTCTCATCCGCCAT | 3′RACE | |
| 363R | TTCAAAGCCCAGCCCATCCAACACCA | PCR, 3′RACE | |
| 407R | TGAAGCTGCCGGCATAGGTC | PCR | |
| 419R | TCTTCATCCACTTTGTCCACACGAGCGT | PCR | |
| 599R | CCTGCATCATGGCATCTGCTGA | PCR | |
| 731R | GCCATGGCTCAGCCCTCAAAC | PCR |
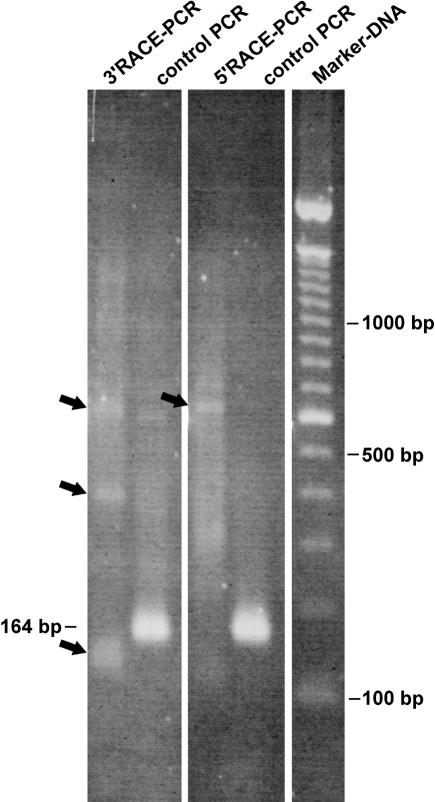
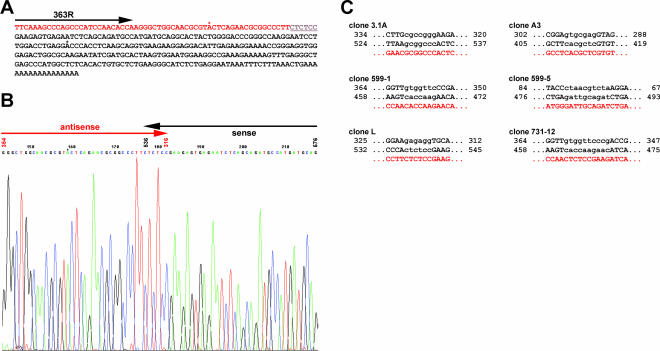
In all cases the RACE-PCR produced faint bands and a smear in the background, whereas the control reaction gave the expected fragment of about 160 bp. One example of an ethidium-bromide-stained agarose gel is shown in Figure 2. In spite of the poor performance, we cloned and sequenced those RACE-PCR reactions. The results showed that the RACE experiments did indeed amplify antisense cTnI. The sequences obtained with 5′ RACE showed the expected antisense orientation and revealed a sequence identical to the sense RNA including a reverse poly(A), that is, 5′ poly(U) tail. One clone out of eight was shorter and ended just before the expected reverse poly(A) tail. However, 3′ RACE did not show any linear cTnI antisense sequence, but instead produced antisense/sense cTnI sequences in one continuous RNA molecule. Figure 3, A and B, shows an example of the sequence derived from clone cTnI (3′RACE)-L (Fig. 3A) and a section from the original sequence data showing the junction between antisense and sense sequences (Fig. 3B). No anomalies were identified in the sequence, apart from two point mutations (GC to AT transversions, marked by asterisks) that could not be found in other sequenced clones. We therefore concluded that these mutations are PCR artefacts produced during RACE-PCR. Interestingly, the transition point between sense and anti-sense sequences was next to an inverted repeat in the cTnI sequence (gray underlined sequence in Fig. 3A). In Figure 3C other examples of crossing points detected in sequenced hybrid cTnI clones are shown. In all cases, these transition points are located next to inverted repeats in the cTnI sequence.
FIGURE 2.
Example of a RACE-PCR result. An ethidium-bromide-stained agarose gel giving an example of a cTnI RACE-PCR result. Antisense-specific RACE was done with primer 631F for 5′RACE and with primer 363R for 3′RACE. As a control, an intern fragment was amplified using primer 255F and 419R. The control PCR resulted in the expected fragment at 164 bp with the 5′ and 3′ RACE-ready cDNA as template, whereas the RACE-PCR produced only some faint bands (arrows) and a smear in the background. Because optimization was not effective, we cloned the complete RACE-PCR.
FIGURE 3.
Original sequence of a cTnI clone obtained after antisense 3′ RACE and examples of different crossing points of hybrid-RNA. (A) The complete sequence of clone pCRII-cTnI(3′RACE)-L. Black letters represent sense, and red letters antisense sequences. An inverted repeat sequence region is underlined and shown in gray. Note that there are only two mismatches within the obtained cTnI sequence (G/C to A/T transversions, marked by asterisks) not found in other clones sequenced and therefore assumed to be a PCR artefact. (B) A section of the original ABI sequence data including the switch from antisense to sense is shown in detail. The vertical numbers over the sequence correspond to the numeration in the published cTnI mRNA sequence (GenBank accession no. X58944). One of the mutations described above is again marked by an asterisk. (C) Examples of crossing points detected in different hybrid-RNA clones. The hybrid-RNA sequences are shown in red letters beneath the corresponding cDNA sequence (black letters). Inverted repeats are displayed in lowercase letters. The nucleotide numbers refer to GenBank accession number X58499. Importantly, all crossing points detected were located in or next to inverted repeats of the cTnI sequence.
To exclude positional effects of the primers used in the first round of RACE-PCR, we repeated both 5′ and 3′ RACE with different primers. We used forward primer 255F in 5′ RACE and the reverse primer 110R for 3′ RACE to produce longer products in the 5′ RACE and shorter products in the 3′ RACE. But in both cases the overall picture did not change: We detected linear antisense cTnI products in the 5′ RACE and antisense/sense sequences in the 3′ RACE (data not shown). As we could find no corresponding description of this or an equivalent structure in the literature, we decided to call these new antisense/sense RNA sequence structures “hybrid RNA” (see discussion).
Direct detection of hybrid RNA with PCR
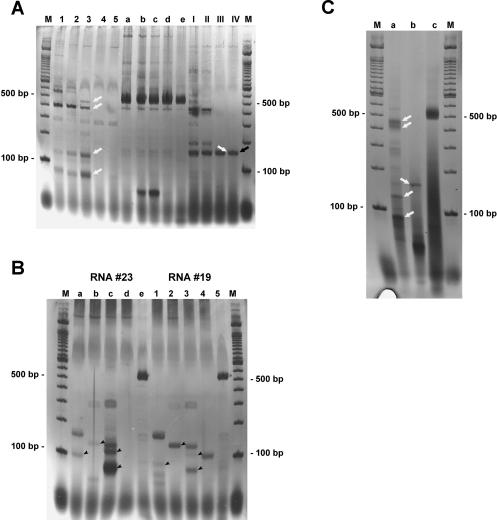
Because it is known that reverse transcription (RT) can produce artefacts by strand switching during the RT step (Mader et al. 2001; Zaphiropoulos 2002; Zeng and Wang 2002), and despite the fact that it is unclear whether RT with MMLV RNaseH(−) enzyme used for our RACE experiments could also produce these artefacts (Brincat et al. 2002; Zhu et al. 2002), we decided to perform direct PCR experiments with two cTnI reverse primers on cDNA reverse transcribed using simply a poly(dT) primer and Thermoscript RNaseH(−) reverse transcriptase at higher temperatures. As described by Mader et al. (2001), artefacts resulting from strand switching by reverse polymerase were not observed when RT was performed at 60°C. We therefore reverse transcribed cDNA at 45°C, 50°C, 55°C, and 60°C. As a positive control we performed normal cTnI PCR with 255F as forward and 731R as reverse primers. Negative controls were performed as described in Materials and Methods and remained without any visible fragments (not shown). The results with two different reverse primer pairs and the positive control are shown in Figure 4A as a silver-stained polyacrylamide gel. After 30 cycles of PCR the positive control (Fig. 4A, a–e) shows a clear, intense band with the expected size of about 500 bp at all temperatures, perhaps slightly reduced with RT at 60°C. Note, if antisense RNA is not polyadenylated, RT with a poly(dT) oligo-nucleotide can only prime the sense mRNA template. Subsequent PCR with two reverse primers, as with 3′ RACE, will again select for cDNA species where strand switching to the antisense RNA template has occurred. Interestingly, PCR with two reverse primers clearly produced an RT temperature-dependent reduction in the complex band pattern seen at 45°C to a single visible band at 60°C for PCR performed with the two reverse primers 419R and 599R (lanes I–IV) and two faint bands with the two reverse primers 363R and 731R (lanes 1–5).
FIGURE 4.
cTnI PCR bands on a silver-stained polyacrylamide gel. (A) A hybrid PCR performed with the reverse primer pairs 363R/731R (lanes 1–5) and 419R/599R (lanes I–IV) and cDNA synthesized at different temperatures (45°C–60°C). As a control for cDNA quality, PCR with forward primer 255F and reverse primer 731R was performed in parallel (lanes a–e). Clearly visible, the complex band pattern obtained with cDNA synthesized at 45°C (lanes 1,a,I) is gradually reduced when cDNA is produced at higher temperatures (50°C, lanes 2,b,II; 55°C, lanes 3,c,III; 60°C, lanes 4,5,d,e,IV; lanes 5,e show an independent experiment at 60°C) leaving only a few bands, whereas the control PCR shows the expected band at around 500 bp with more or less the same intensity in all lanes. The product from lane IV (black arrow) was shotgun cloned and sequenced. The clone obtained showed the expected antisense/sense hybrid structure (see text). (B) A reverse PCR band pattern achieved with cTnI reverse primer pairs 419R/599R (lanes a,1), 407R/599R (lanes b,2), 363R/731R (lanes c,3), and 363R/599R (lanes d,4). cDNA was derived from different RNA preparations (called #23, lanes a–e, and #19, lanes 1–5) and synthesized at 60°C with Thermoscript (Invitrogen). Clearly, the different cDNAs give comparable band patterns with identical primer pairs, although some differences in intensity exist (arrowheads). Only PCR with 363R/599R (lanes d,4) gave no visible band in RNA #23 but one band in RNA #19. The control amplification of cTnI with the forward primer 255F and reverse primer 731R (lanes e,5) gave the expected 500 bp fragment in both cases. (C) The results of a one-tube RT-PCR with Tth-Polymerase on total RNA are shown. The RT step was done for 20 min at 60°C. (Lane a) The two reverse primer 363R and 731R had been used. (Lane b) 419R and 599R. The control PCR with the forward primer 255F and the reverse primer 731R is shown in lane c. When compared to part A of this figure, a nearly identical fragment pattern is visible (white arrows). Cloning and sequencing of this RT-PCR identified hybrid RNA as expected.
After cloning and sequencing the relatively abundant fragment obtained from the PCR with primer pair 419R/ 599R (Fig. 4A, lane IV, arrow) we were able to detect the expected cTnI hybrid sequences. The cloned fragment (clone 3.1A), with a total length of 160 bp, was identical in size to the band observed in Figure 4A, and comprised 115 bp antisense and 45 bp sense sequence with perfect homology to the published cTnI sequence. The transition point was different compared to other sequenced hybrid clones (see Fig. 3C), but the relative sequence orientation was the same in all cases.
As another control, we performed reverse PCR with various primer pairs on different cDNAs from two individual RNA preparations (called #19 and #23) reverse transcribed at 60°C with Thermoscript and a poly(dT) primer. With the exception of PCR using the two reverse primers 363R/599R (lanes d,4), which gave no visible band with RNA #23 but a weak band with RNA #19, the two sets of RT-PCRs resulted in comparable band patterns, only differing in band intensities (Fig. 4B, arrowheads).
When using Tth-polymerase instead of MMLV, artefacts in reverse transcription should be avoided (Mader et al. 2001). Because Tth-polymerase is able to act as reverse transcriptase and DNA polymerase at the same time, we tried a one-tube RT-PCR using Tth-polymerase as the only enzyme. The results after reverse transcription at 60°C and 35 cycles of PCR are shown in Figure 4C. In lane a the RT-PCR was done with 363R/731R and in b with 419R/599R. The positive control PCR with 255F/731R is shown in lane c. The negative controls showed no visible fragments (not shown). Compared with Figure 4A, the band pattern is nearly identical (white arrows in Fig. 4A,C) for both reverse PCR reactions. Additionally, after cloning and sequencing of the RT-PCR reaction using Tth-polymerase, we were again able to identify cTnI hybrid clones.
Northern blot
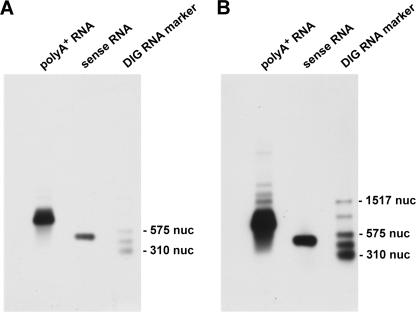
To test whether the hybrid structure could be detected by another independent experiment, we tried Northern blots. We used 1.5 μg rat poly(A)+ RNA and hybridized with in vitro transcribed DIG-marked cTnI hybrid RNA using clone 3.1A as template. As a positive control, 10 ng in vitro transcribed cTnI sense RNA was also loaded on the gel. In Figure 5 the results are shown (Fig. 5A after 1 min exposition time, Fig. 5B after 10 min exposition time). A clear signal in the RNA lane is visible and also the control RNA gave an intensive signal. In Figure 5B some further signals are visible that could point to hybrid RNA.
FIGURE 5.
Example of a Northern blot. Shown is the Northern blot of 1.5 μg rat poly(A)+ RNA after the incubation with cTnI hybrid RNA (clone 3.1A, DIG-marked) as probe. A and B are the same blots after different exposition times (1 min in A and 10 min in B). A prominent band at about 800 nucleotides (nuc) is visible in both cases. Also, the positive control (in vitro transcribed cTnI sense RNA) gives the expected signal at 500 nuc. After 10 min of exposition (B), several additional bands are visible that could represent hybrid RNA. As molecular weight marker the DIG-RNA marker III (Roche) was used.
DISCUSSION
For a detailed characterization of sequence, structure, and subcellular localization of cTnI antisense RNA we performed RACE experiments with rat heart RNA. The cloned products revealed complete homology between the anti-sense RNA sequences and published cTnI mRNA sequence with no signs of intron-specific sequences or other sequence deviations. Because transcription of antisense RNA from the noncoding strand in the genome would lead to an unspliced antisense RNA molecule, and because intron/exon boundaries in this antisense sequence would not match the sense sequence, splicing identical to the sense RNA should be impossible. Thus, we conclude that the cTnI antisense RNA is not transcribed from the opposite strand of the coding region in the genome, as described by Tufarelli et al. (2003), Haddad et al. (2003), or Rossignol et al. (2002), for example. Additionally, we found only a few point mutations at different positions in the sequence of different clones, which we attributed to classical PCR artefacts produced during RACE-PCR with nonproofreading DNA polymerase. This means that transcription of the cTnI antisense RNA from a putative spliced retropseudogene sequence can be excluded, because antisense RNA arising from a pseudogene should show a number of mutations. For example, the pseudogene-encoded nNOS antisense RNA has only about 80% homology to nNOS in the core region (Korneev et al. 1999). Additional screening for cTnI homolog sequences in the rat genome database available so far (about 92% of the genome is available online) also showed no evidence of a cTnI pseudogene. Moreover, 3′ RACE revealed no evidence of polyadenylation (see below and Fig. 6). Taken together, these facts strengthen our assumption that the most likely mechanism producing antisense RNA of cTnI is transcription from the processed cTnI mRNA in the cytoplasm. Therefore, an enzyme homolog of the known RNA-dependent RNA polymerase must be predicted. This is in accordance with other authors, who have investigated a systematic identification of antisense transcripts (Røsok and Sioud 2004). The authors found potential antisense RNA derived from sense mRNA and conclude the presence of an RNA-dependent RNA polymerase. However, direct evidence for such an enzyme is up to now missing in mammalian cells (Dillin 2003; Stein et al. 2003).
FIGURE 6.
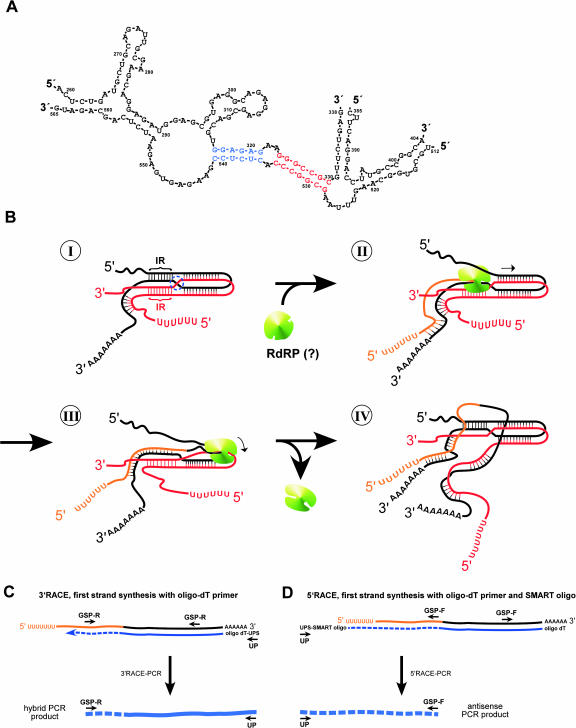
Schematic model of how cTnI hybrid-RNA molecules could arise from 3′ RACE-PCR. (A) Part of a predicted RNA secondary structure of cTnI generated by computer analysis (DNASIS, Hitachi Software Engineering). This structure includes two inverted repeats that had been found in different hybrid-RNA clones next to the crossing points. The blue sequence corresponds to the crossing point found in clone L, the red sequence to the crossing point of clone 3.1A (see Fig. 3C). These inverted repeats could be involved in the hybrid-RNA formation (see B). (B) Hypothetical model of hybrid-RNA formation. Starting with a partial antisense–sense duplex RNA (antisense RNA sequences shown in red, sense RNA sequences in black), an intramolecular loop could form due to inverted repeats (IR) in the cTnI sequence (I). This could cause a four-strand-like structure with a crossover between the sense and antisense RNA sequences (blue dotted circle in I). If such a structure is used as a template for another round of antisense RNA formation (II), starting at the poly(A) sequence of the sense RNA, the enzyme (e.g., an RNA-dependent RNA polymerase, RdRP, shown in green) would reach this crossing point. Here, the enzyme could switch between sense and antisense RNA (III). This would lead to a hybrid structure that is composed of a 5′ poly(U) antisense–sense 3′ poly(A) sequence (antisense sequence in orange, sense sequence in gray) in one continuous molecule (IV). This hybrid structure is, besides the sense RNA, the only product that is polyadenylated and therefore suitable for a RACE experiment using oligo-dT primer. (C) 3′ RACE on hybrid RNA. First strand synthesis (shown in small blue line) with oligo-dT primer including the universal primer sequence (UPS) leading to a hybrid cDNA, that results in a PCR product consisting of antisense sequences (broken blue line) and sense sequences (continuous blue line) in the following 3′ RACE PCR step with reverse gene-specific primer (GSP-R) and forward universal primer (UP). (D) 5′ RACE on hybrid RNA. First-strand synthesis with oligo-dT primer and SMART-oligo. The following 5′ RACE PCR with forward GSP (GSP-F) and reverse UP results in the expected antisense PCR product (dashed blue line) with an oligo(dT) sequence at the 5′ end. Note that a polyadenylation of the antisense RNA would additionally lead to hybrid 5′ RACE and antisense 3′ RACE products.
The RACE-PCRs revealed the antisense sequence including an inverse poly(A) tail with 5′ RACE, but failed to give linear antisense sequences in 3′ RACE, confirming that the antisense RNA is not polyadenylated at the 3′ end. Instead, we found a sequence that “switches” from antisense to sense RNA, which we called “hybrid RNA.” It is known that template switching can be caused by reverse transcriptase, as is common in retroviruses (Goodrich and Duesberg 1990; Negroni and Buc 2001), but this template switching seems to require RNaseH activity (Brincat et al. 2002) and lower temperatures (Mader et al. 2001). So it is still controversial whether reverse transcriptase without RNaseH activity is actually able to produce artefacts by template switching (Zeng and Wang 2002; Zhu et al. 2002).
However, by applying 3′ RACE conditions to antisense RNA, we were essentially selecting for precisely such events if antisense RNA is not polyadenylated, because the 3′ RACE cDNA-specific primer needs a 3′poly(A) sequence and cannot work with a 5′ poly(T) end (or poly (U)), nor can the reverse primer prime first strand synthesis on an-tisense RNA—only on sense RNA. Thus 3′ RACE will “select” for strand switching events, reverse primed synthesis using the sense strand as template before switching to use the antisense strand as template and copying poly(U) to produce 3′ poly(A) (Fig. 6). This need only happen infrequently, because any product will subsequently be efficiently amplified by the reverse primer and poly(dT) primer to produce a hybrid sequence with precisely the kind of structure shown in Figure 1. 5′ RACE-PCR, however, should proceed quite normally with the forward primer (Fig. 6D).
To investigate the hybrid RNA further, it was important to verify our results with different control experiments. By performing experiments with cDNA synthesized at different temperatures using a poly(dT) primer, we were indeed able to reduce the complexity of the hybrid PCR performed with two reverse primers from multiple bands at lower temperatures (45°C) to only some bands at 60°C (Fig. 4). This suggests that some template strand switching could occur during reverse transcription, although the intensity and integrity of some of the bands imply that it must be relatively efficient at particular sites. In contrast, the cTnI control PCR in all cases produced a major band of more or less comparable intensity.
Cloning and sequencing of one product derived from RT at 60°C confirmed our described hybrid-RNA structure. Also, in another control, we obtained comparable band patterns with two independently produced cDNA templates, synthesized at 60°C with a poly(dT) primer, in PCR experiments with different reverse primer pairs. This is hard to explain by a random process, again suggesting that either the hybrid RNAs represent no artefact, but really exist in vivo, or that highly efficient, site-specific template strand swapping occurs during reverse transcription.
As another control, we tried to amplify the hybrid RNA by one-tube RT-PCR using not MMLV reverse polymerase, but Tth-polymerase instead. This polymerase can act as a reverse transcriptase in the presents of Mn2+ ions. Reverse transcription was done for 20 min at 60°C using the gene-specific reverse primer for first strand synthesis, followed by a PCR with two reverse primers. Also in this case, as in all other experiments before, fragments of comparable sizes are detectable (Fig. 4). Additionally, by sequencing of the cloned RT-PCR fragments we were able to identify cTnI hybrid clones as expected. Because template switching is not observed with RT performed with Tth at 60°C (Mader et al. 2001), hybrid RNAs should exist in vivo.
The hybrid RNA must be produced from a structure that consists of sense and antisense RNA sequences. Because we knew that cTnI sense and antisense RNAs form duplexes in vivo (Podlowski et al. 2002), this duplex structure could serve as template for the hybrid-RNA formation. If the above postulated RNA-dependent RNA polymerase sometimes switches between strands during antisense transcription from those sense–antisense RNA duplexes, this would generate a 5′ poly(U)/antisense/sense/3′ poly(A) structure exactly as we have observed. Because this hybrid-RNA structure is 3′ polyadenylated, it is a perfect template for antisense 3′ RACE, but would not generate any evidence of strand switching in antisense 5′ RACE (see Fig. 6 and discussion below). Strand swapping would be simpler to imagine, and accommodate gaps in the sequence, as shown in Figure 3B (nt 316 joined to 542) if one postulates a double-stranded RNA loop structure. The crossover region could have hot spots due to short inverted repeat sequences that line up and form temporary four-strand structures, as in site-specific recombination (Fig. 6B). Importantly the sequences from different clones that have been generated by RACE or direct PCR did show that the switch between sense and antisense RNA is always following such a inverted repeat in the cTnI sequence (Figs. 3C, 6A). Because these inverted repeats are only short, it is not very likely that they are stable enough to promote strand switching by MMLV reverse transcriptase in vitro at temperatures of 50°C or even 60°C (as performed in our investigations). On the other hand, these repeats could very well be responsible for strand swapping under in vivo conditions.
As we searched the Medline database for publications dealing with sense/antisense RNA interaction, we found an interesting structure that is observed during the interaction of antisense CopA and sense CopT RNA in Escherichia coli (Kolb et al. 2000, 2001). Here, in the formation of the antisense–sense RNA duplex, a four-strand pattern is formed with intra- and intermolecular interactions between sense and antisense RNA leading to a crossover. This structure is stuck at the four-way junction because of inverted repeats as it could be for the cTnI sense–antisense duplex. Because this model is similar to the one we postulated for the formation of hybrid RNA, we used and modified it for Figure 6B. The process starts with a partial antisense–sense RNA duplex that has formed an intramolecular loop because of inverted repeats (Fig. 6B, I). This leads to a crossover between the sense and antisense RNAs (blue dotted circle in Fig. 6B, I). If the postulated RNA-dependent RNA polymerase (shown in green in Fig. 6B) uses this template for a new round of antisense formation, the enzyme proceeds just to the predicted crossing point (Fig. 6B, II). Here, it could sometimes switch from the sense RNA sequence to the antisense RNA sequence as template (Fig. 6B, III). At the end, this process would lead exactly to the kind of RNA that we have observed for cTnI (5′ poly(U)/antisense/sense/3′ poly(A), Fig. 6B, IV). When this hybrid RNA is used for RACE-PCR, the resulting structure is a hybrid sequence in 3′ RACE (Fig. 6C) and an antisense sequence in 5′ RACE (Fig. 6D) as we found in our RACE-PCR.
Although this schema fits well to our observed hybrid structure, some questions remain unsolved. So, the sense and antisense RNA outside the four-way structure could form a rigid sense–antisense duplex that would not be able to serve as template for antisense transcription. On the other hand, RNA-binding proteins could be associated with the RNA, keeping the sense and antisense RNAs separated from each other. Interestingly, in recent times the role of RNA-directed evolution is coming more and more into the scientific focus. One discussed mechanism is the generation of de novo sequences by template switching events during reverse transcription (Herbert 2004) like we have observed in our case.
Most of the hybrid-RNA sequences we obtained differed in terms of sequence length and crossover point between the sense and antisense sequences. Such heterogeneity should lead to several additional bands or even a background smear in Northern blots. In our Northern blot we used a hybrid RNA as probe together with 1.5 μg of poly(A)+ RNA. Here, we were able to show indeed some additional bands above the cTnI RNA band, but it is not clear whether these signals could arise, for example, from hnRNA as well (Fig. 5). So it is not unambiguous whether these bands are results of cTnI hybrid RNA. However, it is very likely that such hybrid-RNA structures are relatively rare but selected by RACE and PCR strategies.
In conclusion, the structure of the antisense cTnI in Rattus norvegicus provided good evidence that this antisense RNA is produced from processed cTnI mRNA templates in the cytoplasm. In addition, we found a new, up to now unknown structure comprising antisense and sense sequences in one molecule that we called “hybrid RNA.” This structure is most probably the result of a close interaction between sense and antisense cTnI RNAs in vivo and is always associated with a inverted repeat in the cTnI sequence. It is tempting to suggest that such a recombination process leading to hybrid RNA might be part of a RNA-directed evolution as described by Herbert (2004). Further investigations concerning the generation of cTnI hybrid RNA and its possible significance are in progress.
MATERIALS AND METHODS
Animals and RNA isolation
Female Wistar rats weighing approximately 200 g were used. After excision, the hearts were immediately frozen in liquid nitrogen and pulverized in a cooled mortar. The isolation of total RNA was performed using Trizol (Invitrogen), whereas poly(A)+ RNA was isolated using the μMacs mRNA isolation system (Mylteni Bio-tech). After isolation, RNA was treated with DNase I (Ambion) for 30 min at 37°C. DNase I efficacy was checked by using genome-specific primers in PCR reactions. The quality and quantity of the RNA preparations were tested by agarose gel electrophoresis and measurement of absorption at 260 and 280 nm. The ratio of optical density at 260 nm and 280 nm was ≥1.7 in all cases.
RACE-PCR
RACE-PCR was performed with the SMART RACE cDNA amplification kit (Clontech/BD Bioscience) as described in the users’ manual. In brief, total or poly(A)+ RNA was reverse transcribed with PowerScript reverse transcriptase for 1.5 h at 42°C. The supplied oligo-dT primers specific for 3′ or 5′ RACE were used. The SMART oligonucleotide provided was added for 5′ RACE-Ready cDNA. The resulting 5′ and 3′ RACE-Ready cDNAs were used for RACE-PCR, using the supplied universal primer (UP) mix in combination with cTnI forward primer for antisense-specific 5′ RACE (631F, 255F) and cTnI reverse primer for antisense-specific 3′ RACE (363R, 110R). RACE-PCR was done for 45 cycles using the following protocol. It started with 5 cycles at 72°C annealing temperature, followed by a touch-down from 72°C to 62°C for 20 cycles (−0.5°C/cycle) and ended with 20 cycles at 62°C. The RACE-PCR products were analyzed on an ethidium-bromide-stained agarose gel.
Generation of cDNA
For the generation of cDNA we used either Thermoscript reverse transcriptase (Invitrogen) or Tth-polymerase (Epicentre). When using Thermoscript, RT was performed with a poly(dT) primer at temperatures ranging from 42°C to 60°C and 1μg total RNA in a volume of 20 μL.
Reverse transcription with Tth-polymerase was performed as a one-tube RT-PCR. The RT step was done in the presence of 0.5 mM MnSO4 for 20 min at 60°C with two cTnI-specific reverse primers and 100 ng total RNA. The PCR step was performed as described in the following section.
PCR
Standard PCR was carried out with 1 μL cDNA, 200 μM dNTPS, and 0.5 μM primer in 1× PCR buffer with Advantage2 polymerase (BD-Bioscience) in a hot lid thermocycler for 30 cycles. Primers used are listed in Table 1. For the direct amplification of hybrid cDNA two reverse primers were used. Irrespective of the fact that Advantage2 is a hot start enzyme, the thermocycler was heated to 95°C before the samples were added to minimize any unspecific background. For specific annealing, a touch-down PCR protocol was used, starting at 70°C annealing temperature and ending at 62°C (−0.5°C/cycle for 16 cycles). In the following 14 cycles the annealing temperature was constant at 62°C. This cycling protocol was also used for the PCR step when using Tth-polymerase in a one-tube RT-PCR. Only PCR with primer 407R (in combination with 599R) was down with a touch-down from 63°C to 58°C (−0.5°C/cycle for 10 cycles). The remaining 20 cycles were performed at 58°C. As a negative control, PCR was done with no primer but with cDNA and also with primer, but without cDNA.
Gel electrophoresis and blotting of RNA
To detect cTnI-specific RNAs, we performed a Northern blot as described previously (Luther et al. 2001). Up to 1.5 μg of isolated rat poly(A)+ RNA was heat denatured for 5 min at 65°C and subsequently loaded on a 1% agarose gel (1× MOPS, 2% formaldehyde). The gel was run at 60 V for approximately 4 h at room temperature. The RNA was blotted onto a positively charged nylon membrane in 20× SSC for 18 h. Subsequently, the membrane was UV cross-linked with 120 mJ/cm2 immediately followed by pre-hybridization in DIG-EasyHyb (Roche) for 1 h at 65°C.
Cloning of cTnI for in vitro transcription
Rat cTnI (The rat cTnI cDNA sequence used here as reference can be found in the GenBank database; accession no. X58499) was amplified by RT-PCR with mRNA isolated from adult rat hearts and primer 87F-BamHI (ttgacagaatTCAGCATGGCGGATGAGA GCAGCGAT) and 731R-EcoRI (cctccaggatccGCCATGGGCTCAG CCCTCAAAC; cTnI sequence shown in capital letters) carrying restriction enzyme recognition sequences for subcloning. The resulting fragment was eluted from the gel and subcloned into the two vectors pSPT18 and pSPT19 (Roche), resulting in a sense orientation with regard to the T7 promoter for pSPT18 and in an antisense cTnI direction for pSPT19.
Nonradioactive detection of RNA
Detection of RNA was performed with the DIG nonradioactive detection system (Roche). For sense and antisense RNA detection DIG-labeled cTnI hybrid RNA was transcribed in vitro with T7-polymerase and DIG-UTP from the plasmid pCRII-cTnI-3.1A (clone 3.1A). As a positive control for the Northern blot, sense RNA had been in vitro transcribed with T7 polymerase using plasmid pSPT18-cTnI. Instead of DIG-UTP, normal UTP was used. The resulting RNA was 500 nt. Hybridization was carried out overnight with 100 ng/mL of DIG-labeled RNA at 65°C in DIG-EasyHyb (Roche). After several washing steps including a stringent washing with 0.1× SSC, 0.1% SDS at 60°C, the hybrid RNA were detected by incubation with an anti-DIG/alkaline phosphatase conjugate and a subsequent chemiluminescence reaction with CSPD or CPDstar. A blot stripping procedure, used so that we might use the same blot for another detection, involved heating the blot two times at 80°C for 1 h in 50% formamide, 5% SDS, and 50 mM Tris/HCl (pH 7.5). The blot was afterward washed in 2× SSC for 10 min at room temperature and used immediately for hybridization or stored at 4°C in the dark.
Cloning and sequencing
The products of the different PCRs were cloned in the T/A-cloning vector pCRII-Topo (Invitrogen) according to the manufacturer’s notes. The next day well-isolated clones were picked and cultivated in LB-ampicillin medium overnight. Plasmid DNA was isolated and sequenced in both directions using the BigDye Terminator mix (ABI) by a commercial provider.
Acknowledgments
We thank Anke Stach for excellent technical assistants. This work was supported by the Deutsche Forschungsgemeinschaft Lu 694/1-1.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5261204.
REFERENCES
- Brincat, J.L., Pfeiffer, J.K., and Telesnitsky, A. 2002. RNase H activity is required for high-frequency repeat deletion during Moloney Murine Leukemia Virus replication. J. Virol. 76: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin, A. 2003. The specifics of small interfering RNA specifity. Proc. Natl. Acad. Sci. 100: 6289–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, L. 2003. Translation repression by antisense sequences. Cell. Mol. Life Sci. 60: 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, D.W. and Duesberg, P.H. 1990. Retriviral recombination during reverse transcription. Proc. Natl. Acad. Sci. 87: 2052–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad, F., Bodell, P.W., Qin, A.X., Giger, J.M., and Baldwin, K.M. 2003. Role of antisense RNA in coordinating cardiac myosin heavy chain gene switching. J. Biol. Chem. 278: 37132–37138. [DOI] [PubMed] [Google Scholar]
- Herbert, A. 2004. The four Rs of RNA-directed evolution. Nat. Genet. 36: 19–25. [DOI] [PubMed] [Google Scholar]
- Holroyde, M.J., Robertson, S.P., Johnson, J.D., Solaro, R.J., and Potter, J.D. 1980. The calcium and magnesium binding sites on cardiac troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J. Biol. Chem. 255: 11669–11593. [PubMed] [Google Scholar]
- Kolb, F.A., Engdahl, H.M., Slagter-Jager, J.G., Ehresmann, B., Ehresmann, C., Westhof, E., Wagner, E.G.H., and Romby, P. 2000. Progression of a loop–loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 19: 5905–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb, F.A., Westhof, E., Ehresmann, C., Ehresmann, B., Wagner, E.G.H., and Romby, P. 2001. Bulged residues promote the progression of a loop–loop interaction to a stable and inhibitory antisense–target RNA complex. Nucleic Acids Res. 29: 3145–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev, S.A., Park, J.H., and O’Shea, M. 1999. Neural expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 19: 7711–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M. and Carmichael, G.G. 1998. Antisense RNA: Function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62: 1415–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther, H.P., Haase, H., Hohaus, A., Beckmann, G., Reich, J., and Morano, I. 1998. Characterization of naturally occurring myosin heavy chain antisense mRNA in rat heart. J. Cell. Biochem. 70: 110–120. [PubMed] [Google Scholar]
- Luther, H.P., Podlowski, S., Hetzer, R., and Baumann, G. 2001. Analysis of sense and naturally occurring antisense transcripts of myosin heavy chain in the human myocardium. J. Cell. Biochem. 80: 596–605. [DOI] [PubMed] [Google Scholar]
- Mader, R.M., Schmidt, W.M., Sedivy, R., Rizovski, B., Braun, J., Kalipciyan, M., Exner, M., Steger, G.G., and Mueller, M.W. 2001. Reverse transcriptase template switching during reverse transcriptase-polymerase chain reaction: Artificial generation of deletions in ribonucleotide reductase mRNA. J. Lab. Clin. Med. 137: 422–428. [DOI] [PubMed] [Google Scholar]
- Negroni, M. and Buc, H. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35: 275–302. [DOI] [PubMed] [Google Scholar]
- Podlowski, S., Bramlage, P., Baumann, G., Morano, I., and Luther, H.P. 2002. Cardiac troponin I sense–antisense RNA duplexes in the myocardium. J. Cell. Biochem. 85: 198–207. [PubMed] [Google Scholar]
- Røsok, Ø. and Sioud, M. 2004. Systematic identification of sense–antisense transcripts in mammalian cells. Nat. Biotechnol. 22: 104–108. [DOI] [PubMed] [Google Scholar]
- Rossignol, F., Vaché, C., and Clottes, E. 2002. Natural antisense transcripts of hypoxia-induceable factor 1α are detected in different normal and tumour tissues. Gene 299: 135–140. [DOI] [PubMed] [Google Scholar]
- Solaro, R.J. and Rarick, H.M. 1998. Troponin and tropomyosin. Circ. Res. 83: 471–480. [DOI] [PubMed] [Google Scholar]
- Stein, P., Svobode, P., Anger, M., and Schultz, R.M. 2003. RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacman, L.S. 1996. Thin filament-mediated regulation of cardiac contraction. Annul. Rev. Physiol. 58: 447–481. [DOI] [PubMed] [Google Scholar]
- Tufarelli, C., Stanley, J.A.S., Garrick, D., Sharpe, J.A., Ayyub, H., Wood, W.G., and Higgs, D.R. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genetics 34: 157–165. [DOI] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379–386. [DOI] [PubMed] [Google Scholar]
- Vanhée-Brossollet, C. and Vaquero, C. 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211: 1–9. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos, P.G. 2002. Template switching generated during reverse transcription? FEBS Lett. 527: 326. [DOI] [PubMed] [Google Scholar]
- Zeng, X.C. and Wang, S.X. 2002. Evidence that BmTXKβ-BmKCT cDNA from Chinese scorpion Buthus martensii Karsch is an artifact generated in reverse transcription process. FEBS Lett. 520: 183–184. [DOI] [PubMed] [Google Scholar]
- Zhu, S., Li, W., and Cao, Z. 2002. Does MMLV-RT lacking RNase H activity have the capability of switching templates during reverse transcription? FEBS Lett. 520: 185. [Google Scholar]