Abstract
Two classes of spliceosome are present in eukaryotic cells. Most introns in nuclear pre-mRNAs are removed by a spliceosome that requires U1, U2, U4, U5, and U6 small nuclear ribonucleoprotein particles (snRNPs). A minor class of introns are removed by a spliceosome containing U11, U12, U5, U4atac, and U6 atac snRNPs. We describe experiments that demonstrate that splicing of exon 5 of the rat calcitonin/CGRP gene requires both U2 snRNA and U12 snRNA. In vitro, splicing to calcitonin/ CGRP exon 5 RNA was dependent on U2 snRNA, as preincubation of nuclear extract with an oligonucleotide complementary to U2 snRNA abolished exon 5 splicing. Addition of an oligonucleotide complementary to U12 snRNA increased splicing at a cryptic splice site in exon 5 from <5% to 50% of total spliced RNA. Point mutations in a candidate U12 branch sequence in calcitonin/CGRP intron 4, predicted to decrease U12-pre-mRNA base-pairing, also significantly increased cryptic splicing in vitro. Calcitonin/CGRP genes containing base changes disrupting the U12 branch sequence expressed significantly decreased CGRP mRNA levels when expressed in cultured cells. Coexpression of U12 snRNAs containing base changes predicted to restore U12-pre-mRNA base pairing increased CGRP mRNA synthesis to the level of the wild-type gene. These observations indicate that accurate, efficient splicing of calcitonin/CGRP exon 5 is dependent upon both U2 and U12 snRNAs.
Keywords: RNA, alternative splicing, U12 snRNA, calcitonin
INTRODUCTION
Alternative mRNA splicing plays an important role in the regulation of eukaryotic gene expression, allowing the production of functionally distinct protein products from a single gene. A large, and continually increasing number of genes have now been shown to be subject to differential mRNA splicing, and it is estimated that >35% of human genes undergo differential mRNA splicing (Sorek and Amatai 2001; Modrek and Lee 2002). In higher eukaryotes, alternative splicing has been shown to modulate gene expression in a developmental, sex-specific, and tissue-specific manner.
Two classes of introns are found in nuclear pre-mRNA. Splicing of the major, or U2-dependent type of intron requires U1, U2, U4, U5, and U6 snRNPs and other protein factors, whereas removal of the rare U12-dependent introns is carried out by spliceosomes containing U11, U12, U5, U4atac, and U6atac (Tarn and Steitz 1996a,b). All U12-dependent introns discovered thus far are found in pre-mRNAs that also contain several U2-dependent introns (Dietrich et al. 2001a). This suggests that recognition of U2-dependent and U12-dependent introns, in pre-mRNAs containing both classes of intron, requires cooperation between components of the major and minor type spliceo-some.
The removal of U12-dependent introns is similar in many ways to the removal of U2-dependent introns. Splicing of both types of intron is carried out by two sequential nucleophilic substitution reactions (Tarn and Steitz 1996a,b). U5 snRNP is common to both types of spliceo-some and U1, U2, U4, and U6 snRNPs all have functional homologs in U12-dependent splicing. In addition, U1 and U11, as well as U2 and U12 snRNPs contain several proteins in common (Will et al. 1999). One difference between U2-and U12-dependent splicing is that unlike U2-dependent introns, no U12-dependent intron has been shown to be spliced in an alternative fashion (Dietrich et al. 2001a). The potential for U12-dependent introns to undergo alternative mRNA splicing is suggested by the observation that purine-rich splice enhancers promote splicing of U12-dependent introns (Wu and Krainer 1998; Hastings and Krainer 2001) and artificial genes containing U12-dependent introns can be alternatively spliced in vivo (Dietrich et al. 2001a).
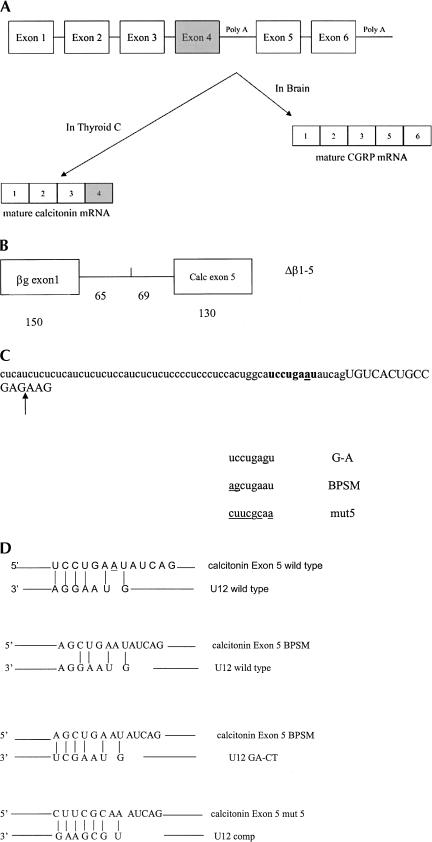
The mammalian calcitonin/CGRP gene has six exons and is alternatively processed in a tissue-specific fashion (Fig. 1). Splicing together the first four exons generates calcitonin mRNA, which encodes the serum calcium-regulating hormone calcitonin. In thyroid C cells, >98% of the mature mRNA derived from the calcitonin/CGRP gene encodes calcitonin (Sabate et al. 1985). In neurons, 99% of the transcripts from the calcitonin/CGRP gene are processed into an mRNA containing exons 1–3, 5, and 6. This mRNA is translated into calcitonin gene-related peptide (CGRP), a neuropeptide (Amara et al. 1982). Processing of the calcitonin/CGRP pre-mRNA has been shown to be regulated at the level of both polyadenylation and splicing (Leff et al. 1987; Roesser et al. 1993; van Oers et al. 1994; Lou et al. 1995, 1998, 1999).
FIGURE 1.
The rat calcitonin/CGRP gene. (A) Structure and alternative RNA processing of the rat calcitonin/CGRP gene, exon sequences are denoted by boxes. (B) The Δβ 1-5 minigene structure showing the sizes in nucleotides of the exon and intron segments. (C) Sequence of the rat calcitonin/CGRP gene at the exon 5 splice acceptor. Intronic sequence is in lowercase, and exon 5 sequence is in capitals. A candidate U12 branch sequence is in bold, and the position of cryptic splicing is indicated by the arrow. The sequence of the A-G, BPSM and mut5 base changes in the putative U12 branch sequence are also shown, with base changes underlined. (D) Predicted interactions between the candidate branch sequence upstream of exon 5 and U12 snRNA. The first alignment is between wild-type calcitonin/CGRP and wild-type U12, the second contains the calcitonin/CGRP BPSM base changes, the third calcitonin/CGRP BPSM with U12 containing compensatory base changes, and the fourth potential base-pairing between calcitonin/CGRP mut5 and U12comp.
Here, we demonstrate that the CGRP-specific exon 5 of the rat calcitonin/ CGRP gene requires both U2 snRNA and U12 snRNA for accurate inclusion into mRNA. In vitro, spliceosome recognition of exon 5 requires U2 snRNA, and accurate splicing of exon 5 is decreased greatly in the absence of free U12 snRNA. In vivo, base changes at a potential U12 interaction site in intron 4 (calcitonin/CGRP BPSM or calcitonin/ CGRPmut5) greatly decreased CGRP mRNA production without increasing calcitonin mRNA levels. Coexpression of U12 snRNAs with compensatory base changes predicted to restore base pairing with calcitonin/CGRP BPSM pre-mRNA or calcitonin/CGRP mut 5 pre-mRNA increased CGRP mRNA production to wild-type levels.
RESULTS
Splicing of calcitonin exon 5 in vitro
We are interested in examining the mechanisms that control tissue-specific alternative RNA processing of the mammalian calcitonin/CGRP gene (Fig. 1A). Several elements have been identified in and around exon 4 that control exon 4 inclusion into mRNA and use of the calcitonin-specific polyadenylation site in intron 4 (Leff et al. 1987; Adema et al. 1988, 1990; Roesser et al. 1993; Yeakley et al. 1993; van Oers et al. 1994; Lou et al. 1995, 1998, 1999; Zandberg et al. 1995; Coleman and Roesser 1998). To determine whether sequences in or around exon 5 also play a role in the regulation of calcitonin/CGRP mRNA processing, RNA from minigenes containing exon 5 were tested in in vitro splicing reactions. Pre-RNA synthesized from constructs containing calcitonin exon 3 fused to calcitonin intron 4 and exon 5; or β globin exon 1 fused to the last 170 nucleotides of calcitonin intron 4 and exon 5 were spliced very inefficiently in vitro (data not shown). Pre-mRNA from a hybrid minigene containing human β globin exon 1 and 65 nt of intron 1 fused to the last 62 nt of the rat calcitonin/CGRP intron 4 and the first 130 nt of exon 5 (Δβ1-5, Fig. 1B) was spliced efficiently in HeLa nuclear extract (Fig. 2). Transcripts synthesized from the Δβ1-5 minigene in vitro were thus used in further experiments as a splicing substrate for study of exon 5 inclusion. The putative Δβ1-5 splice product was amplified by reverse transcription and the PCR. DNA sequence analysis of the amplified fragment demonstrated that the RNA was accurately spliced (data not shown).
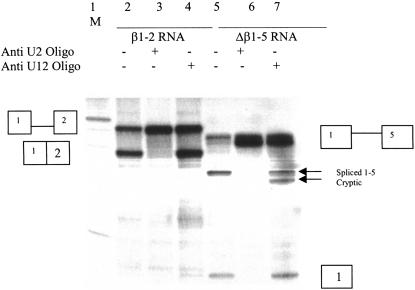
FIGURE 2.
U2 and U12 dependence of β1-2 and Δβ1-5 RNA splicing. In vitro splicing reactions containing radiolabeled β1-2 (lanes 2–4) or Δβ1-5 RNA (lanes 5–7). Splicing reactions were carried out for 1.5 h in 50% Hela nuclear extract, and splice products were separated on 8% polyacrylamide gels containing 8 M urea, and splice products were visualized by autoradiography. Reactions in lanes 3 and 6 contained 2 μM Anti-U2 2′ O-methyl oligonucleotide and 2 μM anti-U12 2′ O-methyl oligonucleotide was added to lanes 4 and 7. Oligonucleotides were added to HeLa nuclear extract and preincubated under splicng conditions for 20 min prior to addition of splicing substrates. A 100-nt DNA ladder (New England Biolabs) was 5′ radiolabeled and used as a size marker in lane 1. The prominent band is 500 nucleotides in length.
No strong match to the U2-dependent branchpoint consensus sequence is apparent upstream of calcitonin/CGRP exon 5 (Fig. 1C). There is a sequence 6–13 nt 5′ of exon 5 that is a good match to the U12-dependent branchpoint sequence, allowing six of seven base-pairing interactions between the putative branch sequence and U12 snRNA (Fig. 1D). To test whether Δβ1-5 splicing is U2 or U12 dependent, 2′ O-methyl oligonucleotides complementary to either U2 or U12 snRNAs (Tarn and Steitz 1996a, 1994), were added to in vitro splicing reactions containing either Δβ1-5 RNA, or β1-2 RNA. β1-2 RNA, which has the first exon, intron, and second exon of the human β globin gene, was used as a splice control. Pretreatment of nuclear extract with 2′ O-methyl oligonucleotide U2b (2 μM), which is complementary to nt 27–49 of U2 snRNA, abolished both β1-2 and Δβ1-5 splicing (Fig. 2, lanes 3,6). Preincubation of splicing extracts with a 2′ O-methyl oligonucleotide complementary to nt 11–28 of U12 snRNA had no effect on β1-2 splicing, but significantly increased cryptic splicing ofΔβ 1-5 (Fig. 2, lane 4 vs. 7). The cryptic Δβ1-5 splice product, which is barely detectable in the absence of the U1211–28 oligonucleotide, results from usage of an AG 13 nt downstream of the authentic start of exon 5 (Fig. 1C). The cryptically spliced RNA molecule was identified both by RT–PCR, followed by DNA sequence analysis and by RNAse protection assay (data not shown). These observations indicate that splicing of both β1-2 and Δβ1-5 RNAs requires U2 snRNA and suggest that U12 snRNA affects the accuracy of Δβ1-5, but not β1-2 splicing.
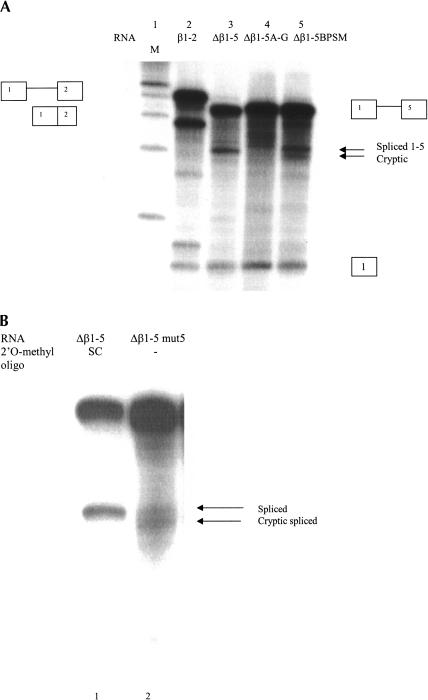
To further examine the possibility that U12 snRNP is involved in calcitonin/CGRP exon 5 splicing, two mutations in the potential U12 branchpoint sequence of Δβ1-5 were constructed. One, Δβ1-5 A-G, changed the potential U12-dependent branchpoint from an A to a G. The second, Δβ1-5 BPSM, changed the branchpoint sequence from UCCUGAAU to AGCUGAAU (Fig. 1C,D). Mutation of the U12 branchpoint consensus sequence at these positions has been demonstrated to disrupt U12-dependent splicing in intron F of the human P120 gene (Hall and Padgett 1996) and is predicted to have no effect on possible U2 snRNA interaction with the site. RNAs transcribed from the altered Δβ1-5 mini-genes were used as splice substrates in HeLa nuclear extract. Changing the potential branchpoint from an A to G abolished splicing of Δβ1-5 A-G RNA (Fig. 3, lane 4). Although no spliced RNA was detected, free exon 1 accumulated, suggesting that the first step of splicing was not affected, but that the second splicing step was inhibited. The UC-AG base changes led to significantly increased cryptic splicing of Δβ1-5 BPSM RNA (Fig. 3, lane 5). Splicing of Δβ1-5 BPSM RNA closely resembled Δβ1-5 RNA splicing in the presence of the 2′ O-methyl oligonucleotide complementary to U12 snRNA (cf. Fig. 3, lane 5 and Fig. 2, lane 7). These observations suggest that U12 snRNA base-pairing with this potential branchpoint sequence is neccessary for accurate splicing to exon 5.
FIGURE 3.
Base changes in a potential U12-dependent branchpoint affect calcitonin/CGRP exon 5 splicing. (A) Radiolabeled RNA splice substrates synthesized in vitro from β1-2, Δβ1-5, Δβ1-5 A-G, and Δβ1-5 BPSM minigenes were spliced in HeLa nuclear extract (see Fig. 1C for sequence of mutations). Splicing was carried out as in Figure 2. Lane 1 contains a radiolabeled 100-nt DNA ladder (New England Biolabs) as a size marker. (B) Radiolabeled Δβ1-5 RNA containing 2 μM SC oligonucleotide (lane 1) and Δβ1-5 mut5 (lane 2) RNA were spliced in Hela nuclear extract. Splicing was assayed as in Figure 2.
Both the A-G and BPSM base changes introduce an AG dinucleotide and disrupt a potential polypyrimidine tract in intron 4. It is possible that placing another AG into the vicinity of the splice junction or interrupting the polypyrimidine sequence is responsible for the changes in splicing that were observed. To examine this possibility, a Δβ1-5 minigene was constructed containing six base changes in the putative U12 branchpoint predicted to abolish U12 base pairing, but not disrupt the polypyrimidine tract or introduce an AG (Δβ1-5mut5, Fig. 1C). Splicing of Δβ1-5mut5 RNA in HeLa nuclear extract was very similar to splicing of Δβ1-5BPSM. Nearly equal amounts of spliced 1-5 and cryptic splice product were produced (Fig. 3B). Preincubation of extract with a 2′O-methyl oligonucleotide with a random sequence (oligonucleotide SC), had no effect on Δβ1-5 splicing (Fig. 3B, lane 1), suggesting that increased cryptic splicing is not due to a nonspecific effect of 2′O-methyl oligonucleotide addition.
Splicing of calcitonin exon 5 in vivo
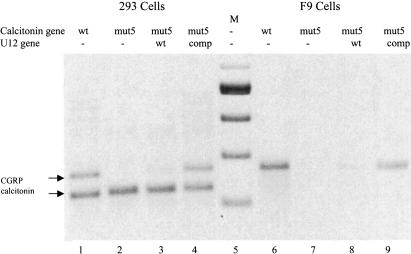
To test the possible involvement of U12 snRNA on exon 5 splicing in vivo, full-length calcitonin/CGRP genes containing the BPSM, A-G, and mut5 changes were constructed and transiently transfected into either 293 cells or mouse F9 cells. Expression of the calcitonin/CGRP gene in 293 cells leads to the production of 50% calcitonin mRNA and 50% CGRP mRNA, whereas F9 cells produce almost exclusively CGRP mRNA (Roesser et al. 1993; Coleman and Roesser 1998). Whole-cell RNA was isolated from transfected cells, and RNA was reverse transcribed into cDNA using random hexamers as primers. Calcitonin/CGRP splicing was assayed by competitive PCR, using a 5′ primer complementary to calcitonin/CGRP exon 3 and two 3′ primers, one complementary to exon 4 and one to exon 5 (Lou et al. 1995; Coleman and Roesser 1998; Tran and Roesser 2003). Significant amounts of both CGRP mRNA, calcitonin mRNA, and partially or aberrantly processed RNAs were produced from the wild-type rat calcitonin/CGRP gene in 293 cells (Fig. 4). The identity of the calcitonin and CGRP mRNAs was confirmed by RT–PCR of gel-excised RNAs and direct sequencing of the PCR products (data not shown). In F9 cells, CGRP mRNA was the predominant product, with small amounts of unprocessed RNA containing intron 3 (Fig. 4, band ≈930 nt in length).
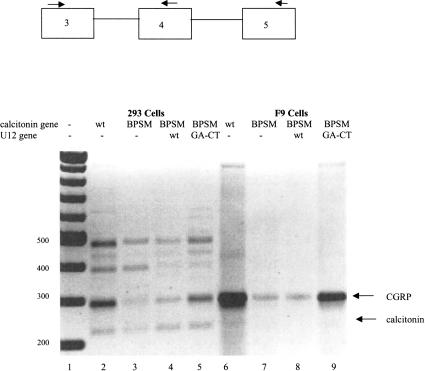
FIGURE 4.
Coexpression of U12GA-CT restores CGRP mRNA production in cells expressing the calcitonin/CGRP BPSM gene. A total of 2 μg of the mammalian expression vector SRα containing the indicated calcitonin/CGRP gene was transfected into 293 cells (lanes 2–5) or F9 cells (lanes 6–9). RNA was harvested 48 h post-transfection. Wild-type (WT) calcitonin/CGRP (lane 2 or 6) or calcitonin/CGRP BPSM (lanes 3–5,7–9) were cotransfected with 1μg of wild-type U12 snRNA gene (lanes 4,8) or U12 snRNA GA-CT, which contains compensatory base changes ( lanes 5,9). RT–PCR analysis of the splice products was performed using primers complementary to exons 3, 4, and 5. PCR fragments were separated on 1.5% agarose gels and visualized with ethidium bromide staining under UV light. Negative image is shown. A 100-nucleotide ladder (New England Biolabs) was used as a size marker in lane 1. Sizes are indicated in base pairs.
Expression of calcitonin/CGRP A-G led to greatly diminished levels of CGRP mRNA in both 293 cells or F9 cells, as compared with the wild-type gene, whereas the amount of calcitonin mRNA did not change (data not shown). Calcitonin/CGRP BPSM expression also led to a decreased CGRP:calcitonin mRNA ratio, as compared with the CGRP: calcitonin mRNA ratio produced from the wild-type calcitonin/CGRP gene (Fig. 4, lanes 2,6 vs. lanes 3,7). Cotransfection of the wild-type U12 snRNA gene, under control of the U1 promoter, with calcitonin/CGRP BPSM, increased the CGRP:calcitonin mRNA ratio by ≈10% (Fig. 4, lanes 4,8). Coexpression of a U12 snRNA gene with a GA-CT change at positions 24/25 of U12, which is predicted to restore base pairing between U12 snRNA and the altered branchpoint sequence of BPSM (Fig. 1D), restored the CGRP:calcitonin mRNA ratio to 75% of that of the wild-type gene (Fig. 4, lanes 5,9).
Expression of calcitonin/CGRP mut5 also produced very low levels of CGRP mRNA in both 293 and F9 cells (Fig. 5, lanes 2,7). Coexpression of wild-type U12 RNA increased the CGRP:calcitonin mRNA ratio by 10%, as compared with the wild-type calcitonin/CGRP gene (Fig. 5, lanes 3,8 vs. lanes 2,7). Coexpression of U12comp, which is predicted to be capable of base-pairing with calcitonin/CGRP mut5 pre-mRNA, fully rescued CGRP mRNA production.
FIGURE 5.
Coexpression of U12comp restores CGRP expression from the calcitonin/CGRP-mut5 gene. RNA from cells transfected with 2 μg of wild-type (lanes 2,6) or mut5 calcitonin/ CGRP genes (lanes 2–4,7–9) were analyzed by reverse transcriptase and PCR using primers complementary to calcitonin exons 3, 4, and 5 and visualized as in Figure 4. Cells were cotransfected with 1 μg of wild-type U12 (lanes 3,8) or U12comp (lanes 4,9). DNA size markers are in lane 5.
DISCUSSION
Tissue-specific processing of the calcitonin/CGRP pre-mRNA has been shown to be regulated at the level of both polyadenylation and splicing. Several sites in and around calcitonin/CGRP exon 4 are involved in control of calcitonin/CGRP pre-mRNA processing (Leff et al. 1987; Roesser et al. 1993; van Oers et al. 1994; Lou et al. 1995, 1998, 1999). Much less is known about sequences in and around exon 5 that could potentially regulate calcitonin/CGRP pre-mRNA processing. In vitro splicing systems using pre-mRNA substrates containing an upstream exon along with all or parts of calcitonin/CGRP exons 4 and 5, demonstrated that splicing to exon 5 was heavily favored over splicing to exon 4 in nuclear extracts made from either calcitonin or CGRP-producing cells (Bovenberg et al. 1988, 1989). Changing the noncanonical branchpoint upstream of exon 4 to the preferred A allowed usage of the exon 4 splice acceptor in nuclear extract from calcitonin-producing cells, but not in nuclear extract from CGRP-producing cells (Cote et al. 1991). Deletion of the exon 5 splice acceptor was shown to abolish CGRP splicing in CGRP-producing cells, but did not increase the level of calcitonin splicing (Yeakley et al. 1993).
To study the requirements for calcitonin/CGRP exon 5 usage, we constructed a hybrid minigene containing exon 1 of the human β globin gene and rat calcitonin/CGRP exon 5. RNA transcribed from Δβ1-5 was efficiently and accurately spliced in HeLa nuclear extract, although HeLa cells make predominantly calcitonin mRNA when expressing the calcitonin/CGRP gene (Roesser et al. 1993). We were unable to directly determine the site of branchpoint formation in the Δβ1-5 pre-mRNA. No RNA products were observed during splicing time courses that showed the proper kinetics or gel mobility to be potential lariat intermediates (Figs. 2, 3; data not shown). Branchpoint formation during splicing to human calcitonin/CGRP exon 5 was demonstrated to utilize an A residue 36 nt upstream of the exon 5 start at the sequence UACUCAC (Bovenberg et al. 1988). However, no candidate U2 branchpoint sequence occurs at this position in the rat intron 4 (Fig. 1C). The last 20 nt of rat intron 4 contain no obvious branchpoint sequence candidates and no pyrimidine tracts longer than 4 nt. From 21 to 164 nt upstream of rat exon 5, there are only five A residues and no Gs in the RNA. Upstream of this potential elongated poly-pyrimidine tract are several candidate U2 branchpoints.
Intron 4 of the rat calcitonin/CGRP gene does contain a potential U12-dependent branchpoint sequence UCC UGAAU, with the putative branchpoint separated from the beginning of exon 5 by 7 nt (Fig. 1C). It was previously observed that functional U12 branchpoints are found no closer than 10 nt from the 3′ splice acceptor (Dietrich et al. 2001b). Our observation that changing the A at position -7 (relative to exon 5) to a G abolishes splicing to exon 5 both in HeLa nuclear extract and 293 cells suggests that this may be the branchpoint utilized. A more likely explanation, however, is that this sequence binds U12 snRNA and acts as part of a complex to direct accurate splicing of exon 5. The U2-dependent branchpoint used may be located elsewhere.
The mechanism by which U12 sn-RNA might direct accurate exon 5 splicing is not clear. The β globin exon 1 splice donor is U1 dependent, and the sequence of the calcitonin exon 3 splice donor (AG/GUAAGG) suggests that it also is recognized by U1 (U1 donor consensus AG/GURAGU) and not U12 (consensus U12 donor/AUAUCCUU). This would seem to preclude recognition of the donor-acceptor pair by a U11-U12 di-snRNP. Glycerol gradient centrifugation suggests, however, that a fraction of U12 snRNPs are not associated with U11 snRNP (Montzka and Steitz 1988). It is possible that monomeric U12 snRNP or U12 in larger complexes not containing U11 is involved in calitonin/CGRP exon 5 splice recognition. In addition, it has been demonstrated that U12-dependent introns are removed more slowly than U2-dependent introns, and it has been proposed that U12-dependent introns may thus be targets for post-transcriptional regulation (Patel et al. 2002). Exon 5 dependence upon U12 may thus slow recognition of exon 5 by the splicing apparatus, allowing cell-specific regulatory mechanisms to compete. A similar situation is observed in calcitonin/CGRP exon 4, where the exon 4 branch-point is a noncanonical C (in the rat gene) or U (in humans). Changing the weakly recognized branchpoint to A uncouples calcitonin/CGRP splicing from cell-specific regulation, allowing calcitonin mRNA production in all cell types (Adema et al. 1988).
Two observations suggest that U12 snRNA is involved in exon 5 splicing during the second catalytic step of splicing. First, approximately equal amounts of free exon 1, a product of the first catalytic step of splicing, were released from Δβ1-5 RNA in HeLa nuclear extract with or without pre-treatment with the anti-U12 oligonucleotide (Fig. 2, lane 5 vs. 7). Second, Δβ1-5 A-G RNA, in which the U12 interaction site is disrupted, undergoes the first catalytic step of splicing in vitro, but not the second step to produce spliced RNA (Fig. 3, lane 4). These observations reinforce the idea that U12 is not acting in branchpoint selection during recognition of the splice acceptor at exon 5, but may function as a splice regulator at a later step.
Wherever the intron 4 branchpoint is located, there is strong evidence that U12 interacts with the UCCUGAAU sequence upstream of exon 5, and that this interaction is important for CGRP splicing. Pretreatment of nuclear extract with a 2′O-methyl oligonucleotide complementary to U12 snRNA, shown previously to disrupt U12-dependent splicing (Tarn and Steitz 1996a) or disrupting potential base-pairing interactions between U12 and the pre-mRNA by changing the potential branch point sequence, greatly increases the level of cryptic splicing in HeLa nuclear extract. In vivo, CGRP mRNA production significantly decreased when rat calcitonin/CGRP genes containing base changes predicted to disrupt the U12 interaction site were expressed in 293 cells. Expression of U12 genes containing compensatory base changes predicted to restore base pairing between U12 and the mutant pre-mRNA fully rescued CGRP mRNA production. The incomplete U12 snRNA allele specificity may be explained by the fact that U12 is being transcribed from the U1 promoter, and there may be much more U12 snRNA expressed from the transfected gene than is normally present in 293 cells. The observation that increasing the U12 gene calcitonin/CGRP gene ratio in cotransfections increases CGRP mRNA levels slightly, supports this possibility (data not shown).
The reason that base changes in the U12 interaction site have a somewhat different effect on splicing to exon 5 in vitro and in vivo is not clear. It may be that β globin exon 1 in the Δβ1-5 RNA used in vitro is a better splice donor than the calcitonin/CGRP exon 3 splice donor utilized in vivo. The stronger splice donor may allow some splicing, although much of it is to a cryptic splice acceptor site. An alternative explanation is that U12 snRNA binding is required in vivo but not essential in vitro because of splice site competition in vivo.
Taken together, our observations strongly suggest that accurate splicing to the rat calcitonin/CGRP exon 5 requires both U2 and U12 snRNAs. Future studies will focus on determining the makeup and mechanism of splicing complexes formed on Δβ1-5 pre-mRNA. Our data is also the first demonstration of an alternatively spliced intron that is U12 snRNA dependent. It will be important to determine the role, if any, that U12 plays in the regulation of alternative splicing of the calcitonin/CGRP gene.
MATERIALS AND METHODS
Plasmid construction
The Δβ1-5 hybrid minigene was constructed by PCR amplification of a 192-nt fragment of the rat calcitonin/CGRP gene containing the last 62 nt of intron 4 and the first 130 nt of exon 5, using the primers 5′-ACTGAATTCCCCTCATCTCTCTCATC-3′ and 5′-ACTGGATCCAAGGCTTCAGAGCCCAC-3′ (EcoRI and BamHI sites introduced by primers are underlined). The amplified fragment was cut with BamHI and EcoRI and ligated into BamHI–EcoRI cut pBS+ β1-2, which contains an EcoRI site engineered into human β globin intron 1 (Roesser et al. 1993). The resulting minigene contains β globin exon 1, the first 65 nt of β globin intron 1, the last 62 nt of calcitonin/CGRP intron 4, and the first 130 nt of exon 5. PCR mutagenesis was utilized to construct Δβ1-5 A-G, Δβ1-5 BPSM, and Δβ1-5 mut5 using primers 5′-CACTGGCATCCTGAG*TATCAGTGTCACTGC, 5′-CACTGGC AA*G*CTGAATATCCAGTGTCACTGC, and 5′-CACTGGCA C*T*T*C*GC*AA*TATCCAGTGTCACTGC, respectively (asterisks denote base changes from the wild-type sequence). The same primers were used for PCR mutagenesis of full-length rat calcitonin/CGRP genes to make calcitonin/CGRP A-G and calcitonin/ CGRP BPSM.
For expression in 293 cells, calcitonin/CGRP genes were sub-cloned into the HindIII–BamHI sites of HSRα as previously described (Roesser et al. 1993). Wild-type and mutant U12GA-CT genes were previously constructed by replacement of the U1 coding sequence in a functional U1 gene with U12 coding regions (Hall and Padgett 1996) and were a generous gift of Dr. Richard A. Padgett. The U12 comp gene was constructed by using the primers 5′-CTGAGCAGATCTATGCCTTAAACTTATGAGGCGAAGAAA TAACGATTCGGGGTG and 5′-GTTAGATCGTCGACCGGGCA GATCGCAACTCC to amplify the U12 gene by the PCR. The DNA fragment generated by PCR was digested with SalI and BglII and ligated into the U12 vector digested with the same enzymes to replace the wild-type U12 gene with U12 comp.
Splicing substrates
To synthesize transcripts for in vitro splicing, minigenes were digested with BamHI to completion and gel purified. In vitro transcription was carried out in the presence of 800 Ci/mmole [α-32P]UTP using T3 RNA polymerase, and all pre-mRNAs were purified on NucTrap columns (Stratagene).
In vitro splicing
Splicing in HeLa nuclear extract was carried out essentially as described (Roesser et al. 1993). Reactions were carried out at 30°C for 2 h in a total volume of 20 μL containing 50% HeLa nuclear extract. Splicing products were separated by electrophoresis on 8% polyacrylamide gels containing 8 M urea, and RNAs were visualized by autoradiography. Candidate Δβ1-5 splice products were excised from gels and eluted in 0.5 M ammonium acetate, 10 mM EDTA, 0.1% SDS at 37°C overnight. Eluted RNAs were reverse transcribed with Thermoscript reverse transcriptase (Invitrogen) using the primer 5′-AAGGCTTCAGAGCCC-3′, which is complementary to calcitonin/CGRP exon 5. The cDNAs were then amplified by PCR using the above primer and 5′-ATGGTGCACCT GACTCCTGA-3′, a primer complementary to β globin exon 1. The nucleotide sequence of the PCR products were determined directly. U2B 2′O-methyl oligonucleotide 5′-AUAAGAA CAGAUACUACACUUGA-3′, U12 2′O-methyl oligonucleotide 5′-AUUUCCUUACUCAUAAG-3′ and scrambled primer (SC) 2′O-methyl 5′-UGCAAUCGUAGCUAAUC-3′ (Dharmacon) were added to splicing reactions to a final concentration of 2 μM.
Transfection and cell culture
The 293 cells and F9 cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. Cells were transfected with Lipofectamine Plus (Invitrogen) and total cellular RNA was harvested 48 h post-transfection.
RNA analysis
Total RNA was isolated using Trizol (Invitrogen) and analyzed for splice products using reverse transcriptase–PCR. cDNAs were transcribed from RNA by Thermoscript reverse transcriptase (Invitrogen) using random hexamers as primers. PCR reactions were performed using 30 amplification cycles, the PCR products were extracted, separated by electrophoresis on 1.5% agarose gels, and visualized by ethidium bromide staining and under UV light. Figures 4 and 5 are negative images of the ethidium-stained gels. The primer complementary to calcitonin exon 4 used for PCR amplification was 5′-CTTGTTGAGGTCTTGTGTGTA-3′, the primer complementary to exon 5 was 5′-AAGGCTTCAGAGCCCAAC-3′ and the 5′-primer complementary to exon 3 was 5′-CCTACTG GCTGCACTGGTGC-3′. These competitive PCR reactions are run in the linear range, and the ratio of CGRP:calcitonin found in each sample was use to quantitate the effects of mutations on CGRP mRNA production (Lou et al. 1995; Tran and Roesser 2003).
Acknowledgments
I thank Dr. Richard A. Padgett for helpful discussions and for the generous gift of the U12 snRNA expression plasmids.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
snRNP, small nuclear ribonucleoprotein particle
snRNA, small nuclear RNA
CGRP, calcitonin gene-related peptide
pre-mRNA, pre-messenger RNA
PCR, polymerase chain reaction
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5210404.
REFERENCES
- Adema, G.J., Bovenberg, R.A., Janz, H.S., and Baas, P.D. 1988. Unusual branch point selection involved in splicing of the alternatively processed Calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 16: 9513–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema, G.J., Van Hulst, K.L., and Baas, P.D. 1990. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 18: 5365–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara, S.G., Jonas, V., Rosenfeld, M.G., Ong, E.S., and Evans, R.M. 1982. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298: 240–244. [DOI] [PubMed] [Google Scholar]
- Bovenberg, R.A.L., Adema, G.J., Jansz, H.S., and Baas, P.D. 1988. Model for tissue specific Calcitonin/CGRP-I RNA processing from in vitro experiments. Nucleic Acids Res. 16: 7867–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenberg, R.A.L., Moen, T.C., Jansz, H.S., and Baas, P.D. 1989. In vitro splicing analysis of mini-gene constructs of the alternatively processed human calcitonin/CGRP-I pre-mRNA. Biochim. Biophys. Acta 1008: 223–233. [DOI] [PubMed] [Google Scholar]
- Coleman, T.P. and Roesser, J.R. 1998. RNA secondary structure: An important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochemistry 37: 15941–15950. [DOI] [PubMed] [Google Scholar]
- Cote, G.J., Nguyen, I.N., Lips, C.J.M., Berget, S.M., and Gagel, R.F. 1991. Validation of an in vitro RNA processing system for CT/ CGRP precursor mRNA. Nucleic Acids Res. 19: 3601–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.C., Shukla, G.C., Fuller, J.D., and Padgett, R.A. 2001a. Alternative splicing of U12-dependent introns in vivo responds to purine-rich enhancers. RNA 7: 1378–1388. [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.C., Perris, M.J., Seyboldt, A.S., and Padgett, R.A. 2001b. Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 21: 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, S.L. and Padgett, R.A. 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271: 1716–1718. [DOI] [PubMed] [Google Scholar]
- Hastings, M.L. and Krainer, A.R. 2001. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 7: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff, S.E., Evans, R.M., and Rosenfeld, M.G. 1987. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/ CGRP gene expression. Cell 48: 517–524. [DOI] [PubMed] [Google Scholar]
- Lou, H., Yang, Y., Cote, G.J., Berget, S.M., and Gagel, R.F. 1995. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol. Cell. Biol. 15: 7135–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, H., Neugebauer, K.M., Gagel, R.F., and Berget, S.M. 1998. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 18: 4977–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, H., Helfman, D.M., Gagel, R.F., and Berget, S.M. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19: 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13–19.11753382 [Google Scholar]
- Montzka, K. and Steitz, J.A. 1988. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. 85: 8885–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A., McCarthy, M., and Steitz, J.A. 2002. The splicing of U12-type introns can be a rate limiting step in gene expression. EMBO J. 21: 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesser, J.R., Liittschwager, K., and Leff, S.E. 1993. Regulation of tissue-specific splicing of the calcitonin/calcitonin gene-related peptide gene by RNA binding proteins. J. Biol. Chem. 268: 8366–8375. [PubMed] [Google Scholar]
- Sabate, M.I., Stolarsky, L.S., Polak, J.M., Bloom, S.R., Varndell, I.M., Ghatei, M.A., Evans, R.M., and Rosenfeld, M.G. 1985. Regulation of neuroendocrine gene expression by alternative RNA processing. Colocalization of calcitonin and calcitonin gene-related peptide in thyroid C-cells. J. Biol. Chem. 260: 2589–2592. [PubMed] [Google Scholar]
- Sorek, R. and Amatai, M. 2001. Piecing together the significance of splicing. Nat. Biotechnol. 19: 196. [DOI] [PubMed] [Google Scholar]
- Tarn, W.Y. and Steitz, J.A. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes & Dev. 8: 2704–2717. [DOI] [PubMed] [Google Scholar]
- ———. 1996a. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84: 801–811. [DOI] [PubMed] [Google Scholar]
- ———. 1996b. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Tran, Q. and Roesser, J.R. 2003. SRp55 is a regulator of calcitonin/ CGRP alternative RNA splicing. Biochemistry 42: 951–957. [DOI] [PubMed] [Google Scholar]
- van Oers, C.C., Adema, G.J., Zandberg, H., Moen, T.C., and Baas, P.D. 1994. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol. Cell. Biol. 14: 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., Reed, R., and Luhrmann, R. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284: 2003–2005. [DOI] [PubMed] [Google Scholar]
- Wu, Q. and Krainer, A.R. 1998. Purine-rich enhancers function in the AT-AC pre-mRNA splicing pathway and do so independently of intact U1 snRNP. RNA 4: 1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley, J.M., Hedjran, F., Morfin, J.P., Merillat, N., Rosenfeld, M.G., and Emeson, R.B. 1993. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol. Cell. Biol. 13: 5999–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandberg, H., Moen, T.C., and Baas, P.D. 1995. Cooperation of 5′ and 3′ processing sites as well as intron and exon sequences in calcitonin exon recognition. Nucleic Acids Res. 23: 248–255. [PMC free article] [PubMed] [Google Scholar]