Abstract
Elucidation of the three-dimensional (3D) structures of the two sequential active sites in spliceosomes is essential for understanding the mechanism of premessenger RNA splicing. The mechanism is predicted to be catalyzed by the small nuclear RNA (snRNA) components of spliceosomes. To obtain new tertiary constraints between the RNA components, we produced and mapped crosslinks between U6 snRNA and the proximal RNAs of active yeast spliceosomes (“yeast“ in this report is Saccharomyces cerevisiae). Thus, specific sites in U6, when substituted with a photoreactive 4-thiouridine or 5-iodouridine, produced spliceosome-dependent crosslinks to U2 snRNA, or in one case, to the pre-mRNA substrate. One set of U2–U6 crosslinks formed before the Prp2p-dependent step of spliceosome assembly, whereas another set formed during or after this step but before the first chemical step of splicing. This latter set of crosslinks formed across U2–U6 helix I. Importantly, this set provides new tertiary constraints for developing 3D models of fully assembled yeast spliceosomes, which are poised for the first chemical step of splicing.
Keywords: 5-iodouridine, 4-thiouridine, photoaffinity, crosslinking, U6 snRNA, pre-mRNA splicing
INTRODUCTION
In eukaryotes, the linear sequence of a gene may be interrupted by one or more noncoding sequences, called introns. During gene expression, introns in premessenger RNA transcripts of genes must be recognized, cleaved, and removed, while the gene-coding sequences, called exons, are spliced together to yield a linear sequence of mRNA, free of introns. The accuracy of this process ensures that each mRNA gene has the correct sequence for expression. Pre-mRNA splicing occurs in the nucleus, and proceeds via two transesterification reactions per intron: (1) The first chemical step involves the 2′ OH group of a specific intronic adenine, called the branchpoint, undergoing nucleophilic substitution of the phosphoroyl group at the 5′ splice site, thus releasing the 5′ exon (Fig. 1). (2) The second chemical step involves the 3′ OH group of the cleaved 5′ exon undergoing nucleophilic substitution of the phosphoroyl group at the 3′ splice site, thus covalently linking the 5′ and 3′ exons and releasing the excised intron. The nucleotides (nts) of the excised intron are connected in the shape of a lariat. Splicing is carried out within large (45S) nuclear particles, called spliceosomes (for review, see Burge et al. 1999; Hastings and Krainer 2001; Brow 2002). Each spliceosome contains a pre-mRNA substrate, up to five associated small nuclear RNAs (U1, U2, U4, U5, and U6 snRNAs) and about 70 known proteins in Saccharomyces cerevisiae (Stevens et al. 2002) or more than 100 proteins in human spliceosomes (Jurica et al. 2002; Makarov et al. 2002; Rappsilber et al. 2002; Zhou et al. 2002).
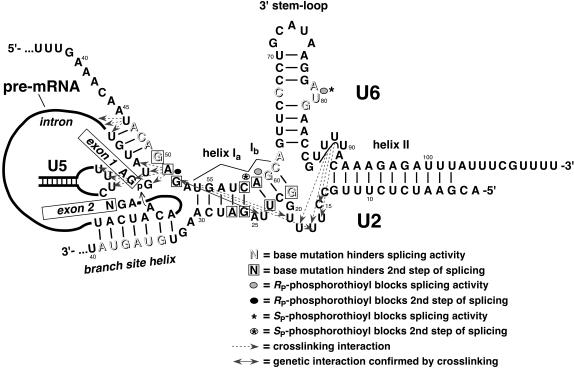
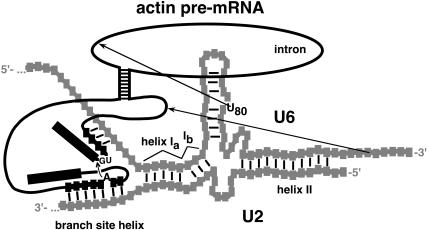
FIGURE 1.
Map of the important features of the RNAs in yeast spliceosomes. The map depicts our model of the secondary structures at a stage when yeast spliceosomes are fully assembled. Many of the mutational effects and/or tertiary interactions highlighted here occur at different stages of spliceosome assembly and splicing, when the secondary structures vary. New and previously reported crosslinks are illustrated with dashed arrows. Mutationally sensitive nucleotides and essential, phosphoryl oxygen atoms are highlighted. The solid, double-headed arrow represents a tertiary interaction between G52 of U6 snRNA and U24 and/or A25 of U2 snRNA (Madhani and Guthrie 1994; Valadkhan and Manley 2000).
To initiate spliceosome assembly, the presence of an intron in pre-mRNA triggers its association with a U1 small nuclear ribonucleoprotein particle (snRNP; Du and Rosbash 2002, 2001). In yeast, U1 snRNP is preassociated with U2, U5, and U4–U6 snRNPs in a pentasnRNP complex (Stevens et al. 2002; Malca et al. 2003). The conserved branchpoint sequence of the intron is recognized by and associates with BBP/SF1 protein initially, followed by U2 snRNA and p14/Snu17 protein (Berglund et al. 1997; Will et al. 2001). Spliceosome assembly is a dynamic process, involving several specific rearrangements and displacements of the snRNAs and spliceosomal proteins, according to a coordinated, allosteric cascade (described in Brow 2002). A map of RNA interactions and functional groups that contribute to spliceosome assembly and splicing in yeast is shown in Figure 1.
The catalytic mechanisms of the two chemical steps of splicing are not yet known; although some aspects have been elucidated (Moore and Sharp 1993; Sontheimer et al. 1997; Gordon et al. 2000; Newby and Greenbaum 2002). The prevailing hypothesis is that pre-mRNA splicing is catalyzed by the snRNA components of spliceosomes, based on mechanistic and structural similarities between spliceosomes and RNA-only, group II self-splicing introns (for comparisons, see Steitz and Steitz 1993; Padgett et al. 1994; Sontheimer et al. 1999; Gordon et al. 2000). The RNA catalysis model gained substantial corroboration recently when it was found that peptide bond formation in ribosomes is catalyzed primarily by the rRNA components—rather than the protein components—of ribosomes (Nissen et al. 2000; Hansen et al. 2002). To better understand how pre-mRNA splicing is catalyzed, one needs more detailed information about the dynamic molecular structure of a spliceosome as it proceeds through both chemical steps of splicing.
With this goal in mind, we recently assayed nearly all known splicing-defective mutations of U6 snRNA, to identify the stage of spliceosome assembly and splicing that is strongly inhibited by each mutation (Ryan and Abelson 2002; Ryan et al. 2002). In the present report, we performed UV crosslinking experiments to identify tertiary contacts between U6 snRNA and the other RNAs in active yeast spliceosomes, using established methods (Sontheimer and Steitz 1993; Kim and Abelson 1996). Thus, we found a set of U2–U6 crosslinks, which formed at the last defined stage of spliceosome assembly, just before the first chemical step of splicing. These crosslinks provide important tertiary constraints for 3D models of fully assembled yeast spliceosomes. They also provide reference parameters for studies of spliceosome structures.
RESULTS
Twenty-six synthetic U6 RNAs containing a single site-specific photo-crosslinking substituent catalyze wild-type levels of pre-mRNA splicing in yeast extract
We synthetically incorporated a photoreactive 4-thiouridine (4-thioU) substituent in U6 RNA at one of the following 16 sites and tested each separately: U6 nt 51, 54, 57, 61, 62, 64, 66, 67, 70, 72, 74, 80, 87, 88, 89, or 90 (see Fig. 1). Similarly, we incorporated at 5-iodouridine (5-iodoU) substituent at one of the following 12 sites: U6 nt 40, 41, 42, 43, 44, 45, 46, 53, 54, 55, 56, or 57. Selective activation of 5-iodoU for photo-crosslinking was achieved by irradiating spliceosomes with the 325 nm output of a HeCd laser, which pumps energy into the low-frequency absorption tail of 5-iodoU for crosslinking, without exciting A, G, U, and C electrons (Norris et al. 1996; see their Fig. 1). The modified U6 RNAs were tested for splicing activity in U6-reconstituted extract using our standard in vitro assay (Fabrizio et al. 1989). In addition to the known effect of U6.thioU54 in completely blocking the second chemical step of splicing (Kim and Abelson 1996), another modified U6 RNA, U6.thioU61, was found to inhibit splicing relative to synthetic wild-type U6 RNA control (data not shown). This modest inhibition was expected for a 4-thioU substituent at this position, as the mutation C61U was previously shown to inhibit splicing modestly in vitro (Fabrizio and Abelson 1990). Regardless, the U6.thioU61 RNA was screened for UV crosslinking in parallel with the other modified U6 RNAs.
A 5-iodoU substituent at U54 in yeast U6 snRNA crosslinks to U2 snRNA at nts U17, U18, and U19
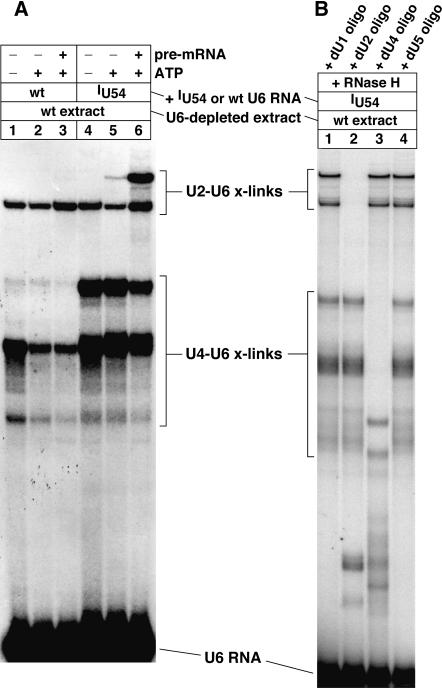
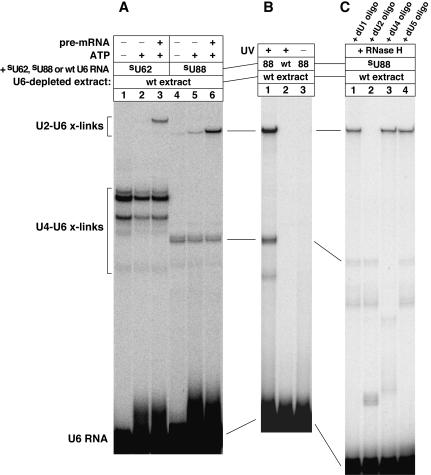
One of the 12 5-iodoU-substituted U6 RNAs, U6.iodoU54, generated a crosslinking product that was insensitive to proteinase K digestion (data not shown), suggesting that this crosslink was RNA–RNA type. Formation of the crosslink was completely dependent on UV irradiation, the presence of ATP, and the presence of the 5-iodoU substituent in U6 (Fig. 2A uppermost intense band; for UV dependence, see lane 3 of Fig. 8B). Furthermore, it was strongly dependent on the presence of actin pre-mRNA (Fig. 2A, cf. lanes 5 and 6). The ATP, pre-mRNA, and U6 dependencies imply that the crosslink formed within spliceosomes. Because yeast extracts may contain endogenous unspliced pre-mRNAs, formation of the crosslink may be more dependent on the presence of pre-mRNA than suggested by the control (lane 5), which lacked the addition of actin pre-mRNA but was not depleted of endogenous pre-mRNAs.
FIGURE 2.
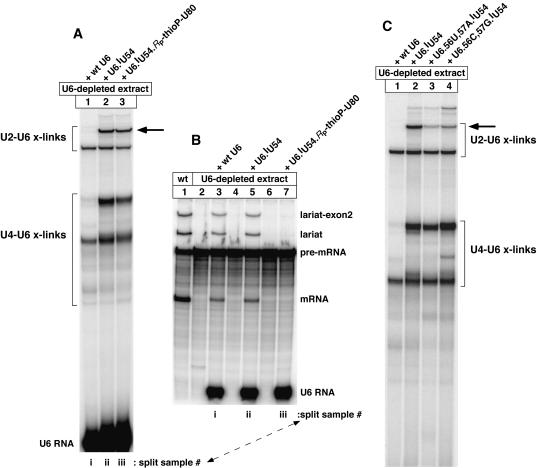
Formation of crosslinked U2–U6.iodoU54 RNA (lane 6, uppermost intense band) is dependent on ATP, pre-mRNA, and the 5-iodoU54 substituent of U6 (cf. lanes 1–6). U6-depleted yeast splicing extract was reconstituted with 32P-labeled U6.iodoU54 RNA and irradiated with UV light (325 nm) to generate photo-crosslinking products under splicing conditions. Pre-mRNA splicing activity was monitored in split samples (data not shown). (B) Small aliquots of the crosslinked product mixture for lane 6 of (A) were subjected to oligo-directed, RNase H digestions to identify which snRNAs were crosslinked to U6 snRNA (lanes 1–4).
FIGURE 8.
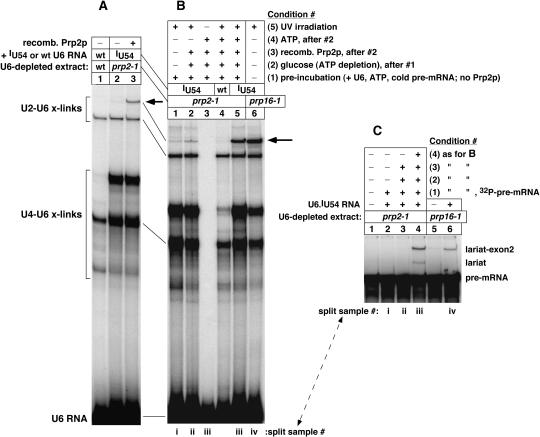
(A) Formation of crosslinked U2–U6.iodoU54 RNA (at arrow) depends on Prp2 protein (cf. lanes 2 and 3). We followed the protocol for Figure 2A but used heat-inactivated prp2–1 mutant extract. (B) Formation of crosslinked U2–U6.iodoU54 RNA depends on both Prp2p and ATP at the Prp2p-dependent step (cf. lanes 2 and 5). As usual, the presence or absence of free ATP was controlled by adding glucose (see Materials and Methods). (C) Samples in (B) were split, as described for Figure 7B, to monitor their pre-mRNA splicing activity (split as noted).
The identity of the nucleotides crosslinked to U6.iodoU54 was determined in three steps. First, the crosslinked RNA was identified by oligo-directed RNase H digestion using DNA oligonucleotides complementary to U1, U2, U4, and U5 snRNAs. Thus, we found that U6.iodoU54 was crosslinked to U2 snRNA (Fig. 2B, lane 2). In Figure 2A, another U2–U6 crosslink formed that was dependent on UV irradiation but was independent of the 5-iodoU substituent (lanes 1–3) as well as independent of actin pre-mRNA and ATP (lane 1). Therefore, this U2–U6 crosslink formed to a large extent outside of an actin pre-mRNA-containing spliceosome, perhaps forming in a penta-snRNP complex. This iodoU-independent crosslink involves unknown positions on both the U2 and U6 snRNAs. We also found that U6.iodoU54 produced two major crosslinks to U4 snRNA (Fig. 2B, lane 3): one with a dependence on the 5-iodoU substituent and the other without such dependence (Fig. 2A, cf. lanes 1 and 4). The yield of the 5-iodoU-independent U4–U6 crosslink was enhanced by withholding ATP (cf. lanes 1 and 2; also for lanes 4 and 5 on lighter exposure not shown), consistent with the knowledge that U4–U6 base-pairing dissociates during spliceosome assembly in an ATP-dependent manner (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998).
The second step for identifying the crosslinked nucleotides was to obtain an approximate mapping of the crosslinking sites in the U2 strand. This was carried out by oligo-directed RNase H digestion of the gel-purified crosslink, using oligonucleotides that were complementary to U2 nts 29–43 (the SRU2 oligonucleotide), to nts 104–121, or to nts 706–724. When the SRU2 oligonucleotide was used, the U6-radiolabeled crosslinked cleavage products migrated as a cluster on denaturing polyacrylamide gels, relatively close to a band of uncrosslinked U6 RNA in an adjacent lane (data not shown, although the same cleavage products are apparent near the bottom of lane 2 of Figure 2B for the SRU2 oligonucleotide). This implies that most of the high molecular weight, unlabeled U2 strand of the crosslink was cleaved off. Additional U2 oligonucleotides yielded cleavage products consistent with this mapping. Therefore, the iodoU-dependent crosslinks lie within the first ~ 30 nucleotides of the 5′ end of U2 snRNA. By the same analysis, the iodoU-independent U2–U6 crosslinks lie within the first ~ 30 nucleotides of U2 as well. The 5′ terminal sequence of U2 lies adjacent to the branchpoint interaction sequence in all eukaryotic U2 snRNAs, located at nts 33–39 in yeast (see Fig. 1). Unfortunately, we found no UV-dependent U2 or U6 primer extension stops for the 5-iodoU-independent crosslink (the uppermost band in lanes 1–3 of Fig. 2A). We can speculate about the UV laser-induced photochemistry that generated this crosslink: perhaps the 325-nm irradiation activated an endogenous 4-thioU, as yet unknown, or perhaps the UV absorption tail of a nucleotide was red-shifted by its local molecular environment. This puzzle remains to be solved.
The third step for identifying the crosslinked nucleo-tide(s) crosslinked was to map their positions on the U2 strand precisely via primer extension mapping (Sontheimer 1994). We confirmed the accuracy of this method by performing primer extension on the U6 strand of the U2–U6.iodoU54 crosslink. As expected, primer extension proceeded just through U6 nt 55, where the iodoU54 crosslinking site blocked further extension (data not shown). For each crosslinking site mapped, two different primers were used in separate mapping experiments: one complementary to U2 nts 104–121 and the other to U2 nts 119–136. In each experiment, sequencing products were generated using the U2 primer on a wild-type U2 gene (SNR20). The sequencing products provided sequence-defined mobility markers for all U2-dependent primer extension products on the gel.
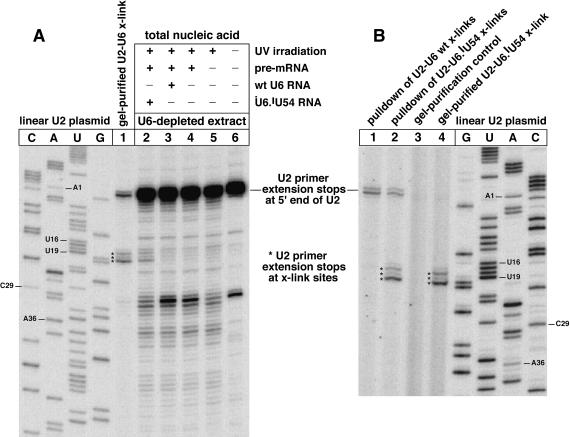
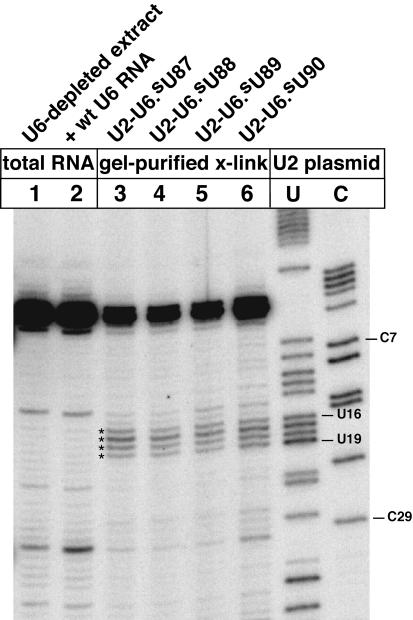
Both U2 primers gave the same mapping results for gel-purified crosslinked U2–U6.iodoU54 RNA. The crosslinked U2 nucleotides were identified by primer extension stops that were dependent on the 5-iodoU substituent of U6 (Fig. 3A, lane 1, marked with asterisks; compared with control lanes 2–6). The gel-purified crosslinked RNA (for lane 1) was isolated from fourfold more yeast extract than was used for each control (lanes 2–6). One can immediately see that the purified crosslinked RNA produced substantially more primer extension blocking at U2 nts 17–19 (lane 1, at asterisks) than that produced in a nonirradiated control (lane 6). The primer extension products of the control (lane 6) reveal that the primer extension stops at nts 17–19 (lane 1) are not typical stops on U2 snRNA, for the primer employed. The pair of bands closest to the top of lane 1 was produced by complete extension of the U2 primer to the 5′ end of U2 snRNA, plus occasional addition of a nontem-plated nucleotide. The primer extension stops due to U2 nts 17–19 (lane 1) were dependent on the 5-iodoU54 substituent of U6 (cf. lanes 2 and 3 for asterisked rows; entire samples were loaded quantitatively in lanes 2–6). Thus, primer extension mapping identified U2 nts 17–19 as the crosslinking sites in U2–U6.iodoU54 crosslinked RNA.
FIGURE 3.
Identification of the crosslinked nucleotides in U2–U6.iodoU54 crosslinked RNA. (A) Sites of crosslinking were identified by primer extension mapping using a radiolabeled primer complementary to U2 RNA nts 104–121 (data not shown) or to nts 119–136 (all lanes). To generate sequence-defined mobility markers, the primer was extended in sequencing reactions on a wild-type U2 gene (lanes C,A,U,G). The 5-iodoU54 substituent of U6 was crosslinked to U2 nts 17–19 in gel-purified material (lane 1; asterisks indicate crosslinking-dependent primer extension stops). In parallel controls, we used one-fourth as much yeast extract as for lane 1, and the crosslinked RNAs were not separated from total nucleic acid. The high yields of primer extension stops at U2 nts 17–19 (lanes 1–2) were dependent on the 5-iodoU substituent of U6 (cf. lanes 2 and 3). (B) The U2–U6.iodoU54 crosslink was purified in two additional ways: (1) Crude U2–U6.iodoU54 product mixture was separated on a gel with a higher ratio of acrylamide:bisacrylamide (29:1) and with a fourfold longer electrophoresis period than for (A). In a parallel control, wild-type U6-reconstituted extract was irradiated and electrophoresced on the same gel. Nucleic acid that migrated at the same Rf as the U2–U6.iodoU54 crosslink was eluted from the control lane and subjected to primer extension in parallel. This control produced no primer extension products (lane 3) near those for the U2–U6.iodoU54 crosslink (lane 4, at asterisks). Therefore, the primer extension stops in lane 4 depend on the 5-iodoU substituent of U6, lacking in lane 3. The stops in lane 4 match those in lane 1 of (A) for a less pure sample of the U2–U6.iodoU54 crosslink. (2) Affinity selection was used to purify the U2–U6.iodoU54 crosslinks from total nucleic acid using a 5′-biotinylated DNA oligonucleotide complementary to U6 nts 26–48. The affinity probe selects total U6 RNA on streptavidin-coated magnetic beads. Primer extension mapping was conducted in situ with the beads. The mapped crosslinking sites (lane 2, at asterisks) matched those in lane 4. For a parallel control lacking 5-iodoU, we used wild-type U6 RNA for crosslinking and affinity selection (lane 1). Affinity-purified U6 RNA was contaminated with free U2 snRNA (lanes 1–2), in contrast to gel-purified, crosslinked U2–U6 RNA (lanes 3–4).
The most prominent band in each lane of lanes 1–6 in Figure 3A is due to extension of the U2 primer to the 5′ end of endogenous U2 snRNA. We determined that the gel-purified U2–U6.iodoU54 crosslinked RNA mapped in lane 1 was contaminated with a low level of uncrosslinked endogenous U2 snRNA, which migrated near the crosslink during gel electrophoresis. We found that a fourfold longer electrophoresis time successfully separated noncrosslinked U2 snRNA from the crosslinked RNA, as assayed by U2 primer extension (Fig. 3B, lane 4, vs. lanes 1–2 which show noncrosslinked U2 contaminant).
The last necessary control was to check that the primer extension stops at U2 nts 17–19 (Fig. 3A, lane 1, asterisks) were not due to another nucleic acid that happened to comigrate with the gel-purified crosslink. Thus, the crosslinked U2–U6.iodoU54 RNA was prepared and gel-purified in parallel with a control that lacked the 5-iodoU substituent. A gel slice of equal size and migratory distance was cut from each lane. The total nucleic acid of each slice was eluted, precipitated, and subjected to the U2 primer extension assay in parallel. Primer extension stops were produced at U2 nts 17–19 for the crosslinked RNA, as expected (Fig. 3B, lane 4, at asterisks). For the control gel slice, no equivalent primer extension stops were produced (lane 3). One can see in lane 4 that U6.iodoU54 may also have crosslinked to nucleotide U16 of U2 snRNA (see band just above asterisks), but we cannot be certain of such crosslinking, because controls that lacked 5-iodoU (Fig. 3A, lanes 3–6) also generated a primer extension stop at U16 in yields at least as high as that of the 5-iodoU-containing control (lane 2; in row just above asterisks). Therefore, we conclude that U6.iodoU54 was crosslinked to U2 nts 19, 17, and 18 (highest to lowest yield; illustrated in Fig. 1).
All primer extension products generated on gel-purified crosslinked RNA migrated slightly faster on denaturing polyacrylamide gels than equivalent products generated on crosslinked RNA in total nucleic acid (Fig. 3A, cf. lanes 1 and 2). This is because the gel-purified RNA was accompanied by visibly higher levels of coprecipitated salts. The salt effect on electrophoresis mobility was confirmed by purifying the same crosslinked RNAs via affinity purification, instead of gel purification. For affinity purification, a buffered solution of total nucleic acid, containing crosslinked U6 RNA, was hybridized to a biotinylated DNA oligo-nucleotide complementary to U6 nts 26–48. Unbound nucleic acids and excess salts were washed away, thereby eliminating the salt effect on primer extension mapping gels (Fig. 3B, cf. lanes 2 and 4). A drawback of the affinity purification method was coisolation of considerable amounts of noncrosslinked U2 snRNA, which contaminates U2 primer extension mappings (lanes 1 and 2, apparent near top of lanes). Nonetheless, as a result of using appropriate controls and alternative methods to confirm our mapping results, we are confident that the U2–U6.iodoU54 crosslinking sites are accurately mapped to U2 nts 17–19.
A 4-thioU substituent at U87, U88, U89, or U90 in yeast U6 RNA crosslinks to U2 snRNA at nucleotides C15, U16, U17, and U18
We prepared 16 synthetic U6 snRNAs that have a single 4-thioU substituent as listed in the first paragraph of the Results, and we followed the same procedures for generating and mapping U2–U6 crosslinks. Four of these 4-thioU substituents, at U87, U88, U89, or U90, each generated a U2–U6 crosslinking product that comigrated with the other such products in parallel lanes on a denaturing polyacryl-amide gel (Fig. 4A, lane 6; data not shown). The U2–U6 crosslinks were strongly dependent on both ATP and actin pre-mRNA (cf. lanes 4–6). Furthermore, the crosslinks were completely dependent on UV irradiation and the presence of the 4-thioU substituent in U6 (Fig. 4B; data not shown). Because yeast extracts may contain ATP that is tightly bound by spliceosomal proteins, formation of the crosslinks may be more strictly dependent on ATP than suggested by the ATP-depleted (ΔATP) controls (Fig. 4A, lane 4; data not shown). A trace of the crosslink is faintly visible in lane 4 and replicates, even though a common stock of ΔATP extract was sufficiently depleted of ATP to prohibit U2–U6.thioU62 crosslinking (lane 1) and pre-mRNA splicing (not shown) in parallel experiments.
FIGURE 4.
(A) Formation of crosslinked U2–U6.thioU62 RNA (lane 3) is dependent on ATP and pre-mRNA (cf. lanes 1–3). The same is true for crosslinked U2–U6.thioU88 RNA (lane 6; cf. lanes 4–6). We followed the protocol for Figure 2A but used U6.thioU62 or U6.thioU88 RNA and ~ 365 nm UV light. (B) Crosslinking of U2–U6.thioU88 RNA is dependent on UV irradiation and the 4-thioU substituent of U6 (cf. lanes 1–3). (C) Small aliquots of the crosslinked product mixture for lane 1 of (B) were subjected to oligo-directed, RNase H digestions to identify which snRNAs were crosslinked to U6 snRNA (lanes 1–4).
The identities of the nucleotides crosslinked to U6.thioU87/88/89/90 were determined in three steps, as for our U2–U6.iodoU54 crosslinks (above). RNase H mapping revealed that each of these 4-thioU substituents was crosslinked to U2 snRNA within the first ~ 30 nucleotides of the U2 strand (data not shown). Primer extension mapping showed that each of the four 4-thioU substituents was crosslinked to the same set of U2 nucleotides: C15, U16, U17, and U18 (Fig. 5, lanes 3–6, in asterisked rows). The primer extension stops at U2 nts 15–18 (lanes 3–6) were dependent on the 4-thioU substituents of U6 (lane 2). We conclude that each U6.thioU87/88/89/90 RNA was cross-linked to U2 nts 16, 15, 17, and 18 (highest to lower yield, 15 ≈ 17; illustrated in Fig. 1).
FIGURE 5.
Identification of the crosslinked nucleotides in gel-purified, U2–U6.thioU87/88/89/90 crosslinked RNA. Primer extension mapping was performed as for Figure 3A using a primer complementary to U2 nts 89–111 (data not shown) or to U2 nts 104–121 (all lanes). Primer extension stops due to 4-thioU-dependent U2–U6 crosslinking are marked with asterisks; equivalent stops were found across lanes 3–6. These stops depend on the presence of U6 RNA (lane 1) and on the 4-thioU substituent of U6 (lane 2). The controls for lanes 1–2 were prepared as described for lanes 4–3 of Figure 3A, respectively. Sequence-defined mobility markers were also prepared as before (lanes U,C).
In Figure 5, lanes 3–6, the ratio of yields of the four thioU-dependent primer extension stops in each lane is nearly constant across these four lanes, despite the fact that the 4-thioU substituent was located at either U87, U88, U89, or U90. This implies that U6 and/ or U2 snRNA had sufficient degrees of conformational freedom during cross-linking for a 4-thioU substituent at any of the four U6 sites to interact with a constant set of four U2 nucleotides in a similar manner and geometry.
A 4-thioU substituent at U80 in yeast U6 RNA crosslinks to the actin pre-mRNA substrate at nucleotide A(+209) of the intron
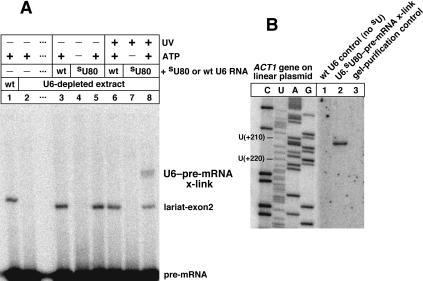
Of the 16 synthetic U6 snRNAs carrying a single 4-thioU substituent (see first paragraph of Results), the one with a 4-thioU at U80 generated a crosslink to actin pre-mRNA (Fig. 6A, lane 8), by a mechanism dependent on ATP, UV irradiation, and the presence of the 4-thioU substituent in U6 (cf. lanes 5–8). In these experiments, actin pre-mRNA was radiolabeled, whereas the U6.thioU80 RNA was not. This allowed us to assay pre-mRNA splicing as well as crosslinking in split samples: nonirradiated samples (lanes 3–5) were split from irradiated samples (lanes 6–8). On the 5% denaturing polyacrylamide gel for Figure 6A, unreacted pre-mRNA had the same mobility as excised lariat intron. Nonetheless, the lariat intron was produced in all splicing-competent samples (lanes 1, 3, 5, 6, and 8), as observed on a higher percentage gel (not shown). When endogenous splicing activity (lane 1) was depleted of ATP before U6 reconstitution, splicing was prohibited as expected (lanes 4 and 7), and U6.thioU80 did not crosslink to pre-mRNA (lane 7). Conversely, when splicing was active and the 4-thioU80 substituent was present and UV irradiated, a crosslink formed between U6.thioU80 RNA and 32P-labeled actin pre-mRNA (lane 8).
FIGURE 6.
(A) Formation of U6.thioU80-pre-mRNA crosslinks (lane 8) is dependent on ATP, UV irradiation and the 4-thioU substituent of U6 RNA (cf. lanes 5–8). A stock of U6-depleted yeast extract (lane 2) was used for U6 reconstitutions (lanes 3–8). Splicing was monitored in nonirradiated samples (lanes 3–5) split from the irradiated samples (lanes 6–8). The protocol was the same as for Figure 4A, except that actin pre-mRNA was 32P-labeled, whereas U6 RNA was not, and the denaturing gel was 5% polyacrylamide (29:1). (B) Identification of the crosslinked nucleotides in gel-purified, U6.thioU80-pre-mRNA crosslinked RNA. Primer extension mapping was performed as for Figure 3A, except the primer, 3′F, was complementary to nucleotides +57–74 of the 3′ exon of actin pre-mRNA. A primer extension stop due to crosslinking of intron A(+209) (lane 2) was dependent on the 4-thioU substituent of U6 (lane 1; this control was not gel-purified before primer extension). Another wild-type U6-reconstituted control (lane 3) was gel-purified in parallel with the crosslink, as described for lane 3 of Figure 3B(1). Sequence-defined mobility markers were prepared as before (lanes C,U,A,G).
The identity of the crosslinked nucleotide(s) of actin pre-mRNA was determined using steps 2 and 3 of our protocol (above). The radiolabeled primer, 3′F, was complementary to the +57–74th nucleotides of the 3′ exon in actin pre-mRNA. Primer extension mapping revealed that the +209th nucleotide of the intron was crosslinked (Fig. 6B, lane 2; band mobility is equivalent to primer extension through U(+210) in lane U). The crosslinked nucleotide, A(+209) of the intron, is 57 nucleotides upstream of the branchpoint adenine in yeast actin pre-mRNA. The primer extension stop at intron A(+209) was dependent on the 4-thioU80 substituent of U6, as revealed for the total nucleic acid of a control reconstituted with wild-type U6 RNA (lane 1). Also, the nucleic acid of a gel slice control (in step 3 of our protocol) did not yield the A(+209) stop (lane 3). The results indicate that the 4-thioU80 substituent of U6 RNA crosslinked to A(+209) of the actin intron.
Primer extension mapping also indicated that U6.thioU80 was crosslinked to unspliced pre-mRNA substrate in spliceosomes. The crosslinked nucleotide, intron A(+209), lies upstream of the branchpoint adenine of actin pre-mRNA. We know from previous experience that primer extension on lariat-intermediate substrates is always blocked at the branchpoint of the lariat structure, including previous use of the 3′F primer (Kim and Abelson 1996). Therefore, the progression of primer extension to A(+209) demonstrates that U6.thioU80 was crosslinked to unspliced pre-mRNA.
A 4-thioU substituent at A62 and U64 of yeast U6 RNA crosslinks to the 5′ end of U2 snRNA, 5′ of the branchpoint interaction sequence
The two synthetic U6 snRNAs carrying a 4-thioU at A62 or at U64 each generated crosslinks to U2 snRNA (Fig. 4A, lane 3; data not shown), by a mechanism dependent on ATP, actin pre-mRNA, UV irradiation, and the 4-thioU substituent of U6 (cf. lanes 1–3; data not shown). The crosslinks produced by U6.thioU64 (not shown) looked the same as those in lane 3, but in ~ 33% lower yield. RNase H mapping of the thioU-dependent crosslinks in U2–U6.thioU62/64 revealed that these were located at the 5′ end of U2, within its first 40 nts (data not shown). Unfortunately, we could not identify which U2 nucleotides were crosslinked, because the yield of crosslinked RNA was insufficient for primer extension mapping in our hands, despite our efforts to scale up the yield and combine gel-purified product.
A 4-thioU substituent at U54, U57, or U80 of yeast U6 RNA crosslinks to U4 snRNA in an ATP-dependent manner
Of the 16 synthetic U6 snRNAs carrying a single 4-thioU substituent (described in the first paragraph of Results), all but those with a 4-thioU at C66 or U70 generated crosslinks to U4 snRNA. Most of these U4–U6 crosslinks were independent of ATP and actin pre-mRNA. However, five U6 snRNAs, with a 4-thioU at A51, U54, U57, C61, and U80, generated U4–U6 crosslinks in a manner dependent on ATP but not on actin pre-mRNA (Fig. 2A, cf. lanes 1–3 and lanes 4–6; data not shown). We did not determine which U4 nucleotides were crosslinked, because the 4-thioU substituents at nts 54, 57, 61, and 80 of U6 are located within the duplex region of U4–U6 di-snRNP, a complex formed during spliceosome assembly. The secondary structure of the U4–U6 duplex shows the proximity of U6 nts 51, 54, 57, 61, and 80 to the U4 strand (Brow and Guthrie 1988). It is known that crosslinking of 4-thioU is enhanced at the ends of RNA duplexes and at G-U wobble pairs (Dubreuil et al. 1991), precisely where nts 51, 54, 57, 61, and 80 of U6 are located in the U4–U6 duplex. We expect that these crosslinks reflect the well-known secondary structure of the duplex. The U4–U6.thioU51 and U4–U6.thioU80 crosslinks were especially strong and can serve as ATP-dependent markers for the U4–U6 duplex.
Formation of the U2–U6.iodoU54 crosslinks is strongly promoted by the ATP-dependent activity of Prp2p, whereas the U2–U6.thioU88 crosslinks form independently of Prp2p
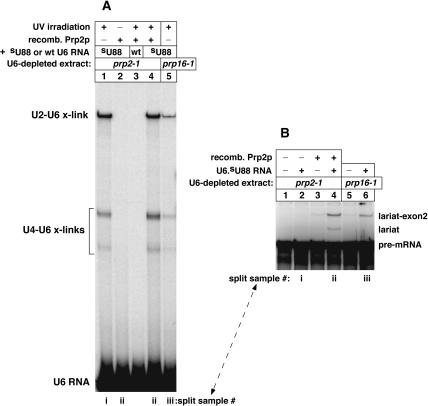
Prp2 protein (Prp2p) is an RNA-dependent ATPase and putative RNA helicase that is essential for yeast growth and for the last stage of spliceosome assembly, just prior to the first chemical step of splicing (Cheng and Abelson 1987; Kim and Lin 1996). To determine whether the U2–U6.iodoU54 crosslinks formed before or after the Prp2p-dependent step of spliceosome assembly, we used a haploid, temperature-sensitive prp2–1 extract that allows one to heat-inactivate the mutant Prp2–1 protein in splicing samples at 34°C for 30 min. After heat inactivation, and simultaneous depletion of endogenous U6 snRNA, addition of synthetic U6 RNA and pre-mRNA was not sufficient to restore splicing activity (Fig. 7B, lane 2). Addition of recombinant Prp2p without synthetic U6 RNA restored a very low level of pre-mRNA splicing (lane 3), indicating that some residual endogenous U6 snRNA was still present after its depletion. In these experiments, we needed to deplete just enough endogenous U6 RNA for subsequent U6 reconstitution to produce appreciable levels of U2–U6 crosslinks. Addition of both U6 RNA and Prp2p prompted splicing activity, as expected (lane 4). Using split aliquots of the samples, we found that the U2–U6.thioU88 crosslinks formed before the Prp2p step, as heat inactivation of Prp2–1p had no effect on this crosslinking (Fig. 7A, cf. lanes 1 and 4). In contrast to the U2–U6.thioU88 crosslinks, we found that U2–U6.iodoU54 crosslinking occurred efficiently after the Prp2p step, as heat inactivation of the Prp2–1 protein strongly inhibited this crosslinking, whereas addition of recombinant Prp2p promoted crosslinking (Fig. 8A, cf. lanes 2 and 3, for band at arrow).
FIGURE 7.
(A) Formation of crosslinked U2–U6.thioU88 RNA occurs before the Prp2p-dependent step of spliceosome assembly (cf. lanes 1 and 4). We followed the protocol for Figure 4A but used heat-inactivated prp2–1 mutant extract (lanes 1–4), as well as prp16–1 mutant extract that is strongly inhibited at the second chemical step of splicing (lane 5; see lane 6 in B). (B) Samples in (A) were split after U6 reconstitution, just before addition of actin pre-mRNA, which was 32P-labeled for (B) but not for (A). Splicing assays were conducted in parallel for the split samples. The splicing yields of (B) can be compared with the crosslinking yields of (A) (split samples as noted).
To determine whether formation of the U2–U6.iodoU54 crosslinks depends on both Prp2p and ATP at the Prp2p-dependent step, or whether active Prp2p alone is sufficient for crosslinking, an aliquot of prp2–1 mutant extract was depleted of endogenous U6 snRNA and Prp2–1p, as above. The U6-depleted Prp2–1p-inactivated (ΔU6,ΔPrp2–1p) extract was not competent for splicing (Fig. 8C, lane 1), even after addition of U6.iodoU54 RNA (lane 2). The sample for lane 2 was split prior to loading, and half was UV irradiated to assay U6.iodoU54 crosslinking. Only a low level of the U2–U6.iodoU54 crosslinks was produced in this splicing-incompetent sample (Fig. 8B, lane 1, band in row at arrow; split sample as noted). A threefold larger sample of ΔU6,ΔPrp2–1p extract was divided: A ⅙ portion was treated with wild-type U6 RNA and pre-mRNA, and a ⅚ portion was treated with iodoU54-substituted U6 RNA and pre-mRNA, to allow spliceosome assembly to proceed up to the Prp2p-dependent step. After a brief incubation ATP was depleted, and recombinant Prp2p was added alone or in combination with ATP. The larger samples were split into individual samples. Addition of Prp2p without ATP was not sufficient to activate splicing in the stalled spliceosomes (Fig. 8C, lane 3), whereas addition of both Prp2p and ATP promoted splicing, as expected (lane 4). These splicing effects were compared with the ability of these samples to form U2–U6.iodoU54 crosslinks, in the split samples. Thus, the addition of Prp2p without ATP allowed only a low level of the U2–U6.iodoU54 crosslinks to form (Fig. 8B, lane 2, band in row at arrow). In contrast, the addition of both Prp2p and ATP substantially enhanced formation of these crosslinks, as expected (lane 5, band in row at arrow). The controls confirmed that this crosslinking was dependent on UV irradiation (lane 3) and on the 5-iodoU54 substituent of U6 (lane 4). We conclude that formation of the U2–U6.iodoU54 crosslinks in yeast spliceosomes is strongly enhanced by, or dependent on, the ATP-dependent activity of Prp2p in yeast.
The U2–U6.iodoU54 crosslinks form just before the first chemical step of splicing
To further define when the U2–U6.iodoU54 crosslinks formed during the splicing pathway, we incorporated a chemical modification in synthetic U6.iodoU54 RNA that blocks the first chemical step, but not the Prp2p-dependent step, of splicing. This chemical modification is a nonbridging, RP-phosphorothioyl at the A79–U80 linkage in U6.iodoU54 RNA. This particular phosphorothioyl blocks splicing following the Prp2p step, just before and/or during the first chemical step (Fabrizio and Abelson 1992; Yean et al. 2000). We observed this splicing block, as expected (Fig. 9B, lane 7); yet despite this block, the phosphorothioyl substituent allowed formation of the U2–U6.iodoU54 set of crosslinks (Fig. 9A, lane 3, band in row at arrow; split sample as noted). Indeed, a relatively high level of the crosslinks formed, comparable to the level formed in a control that lacked the RP-phosphorothioyl substituent (Fig. 9A, cf. lanes 2 and 3, bands in row at arrow). The high yield of crosslinking for the RP-phosphorothioyl substituent is in stark contrast to the block to such crosslinking imposed by heat-inactivated Prp2p (see Fig. 8A, lane 2). Therefore, formation of the iodoU54-dependent crosslinks occurs during or after the Prp2p step, and just before the first chemical step. Our combined results demonstrate that nucleotide U54 of U6 snRNA directly contacts U2 nts 17–19 at the last stage of spliceosome assembly.
FIGURE 9.
Crosslinking of U6.iodoU54 RNA to U2 nts 17–19 occurs just before the first chemical step of pre-mRNA splicing. (A) Synthetic U6 RNA was prepared with a nonbridging RP-phosphorothioyl substituent (thioP) at the A79–U80 linkage, to strongly block the first chemical step of splicing (lane 7 in B) but not the Prp2p step (Yean et al. 2000). 5-IodoU was incorporated at position U54 to generate crosslinks, as described for Figure 2A. The doubly substituted U6 RNA generated the typical U2–U6.iodoU54 band of crosslinks (at arrow) to the same extent as a control lacking the thioP substituent (cf. lanes 2 and 3); therefore, the crosslinks formed before the first chemical step of splicing. (B) Samples in (A) were split, as described for Figure 7B, to monitor their pre-mRNA splicing activity (split as noted). (C) Individual pairs of splicing-inhibitory mutations were incorporated in U6.iodoU54 RNA, prior to crosslinking. The yield of iodoU54-dependent U2–U6 crosslinks (at arrow) decreased in direct proportion to the splicing yield previously assayed for each pair of mutations (Ryan and Abelson 2002). This suggests that crosslinking occurred in splicing-competent spliceosomes.
U6 mutations within U2–U6 helix Ia inhibit U2–U6.iodoU54 crosslinking to the same extent as they inhibit pre-mRNA splicing
The crosslinking sites in U2–U6.iodoU54 span base-complementary sequences of U6 and U2 snRNAs, namely those that form U2–U6 helix Ia during spliceosome assembly (see Fig. 1). We previously showed that mutational disruption of base-pairing within U2–U6 helix Ia inhibits spliceosome assembly at the Prp2p-dependent step (Ryan et al. 2002). Hence, we were curious whether these same mutational disruptions of helix Ia would enhance, inhibit, or completely block formation of our U2–U6.iodoU54 crosslinks. This information could tell us whether these crosslinks form in the presence or absence of base-paired U2–U6 helix Ia.
For these crosslinking experiments we constructed a set of synthetic U6.iodoU54 RNAs that contained U6 mutations known to disrupt a portion of U2–U6 helix Ia base-pairing (Madhani and Guthrie 1992). Rather than observe an enhancement or complete block of crosslinking due to these helix-disrupting mutations, we observed that the mutations inhibited iodoU54-dependent U2–U6 crosslinking to the same extent as they inhibited pre-mRNA splicing. In one case, U6.56C,57G.iodoU54 RNA produced a 31% yield of the iodoU54-dependent crosslinks (Fig. 9C, lane 4, band at arrow) relative to that produced for U6.iodoU54 without mutations (lane 2). The relative splicing yield for these mutations was previously determined to be 28% (for U6.56C,57G RNA; Ryan and Abelson 2002). The 5-io-doU54 substituent has no detectable effect on splicing activity (Fig. 9B, cf. lanes 3 and 5; data not shown). In another case, U6.56U,57A.iodoU54 RNA produced a 16% yield of the iodoU54-dependent U2–U6 crosslinks (Fig. 9C, lane 3, band in row at arrow) relative to that produced for U6.iodoU54 without mutations (lane 2). The relative splicing yield for these mutations was previously determined to be 10% under the same conditions (for U6.56U,57A RNA; Ryan and Abelson 2002). Thus, an approximately direct correlation was observed: as the yield of pre-mRNA splicing increased, the yield of iodoU54-dependent U2–U6 cross-links increased nearly proportionately. This correlation suggests that the iodoU54-dependent crosslinking of U2 and U6 snRNAs captured tertiary interactions in spliceosomes that were competent for pre-mRNA splicing.
DISCUSSION
The long-range objective of our research is to define a sufficient number of tertiary constraints in spliceosomes to develop an accurate 3D model of the RNA interactions in and around the two active sites of spliceosomes. In progress toward this goal we screened a large number of individual 4-thioU substituents in U6 snRNA for their ability to generate UV-dependent site-specific crosslinks between U6 and the other RNAs in active yeast spliceosomes. This screen yielded three sets of spliceosomal crosslinks between U2 and U6 snRNAs as well as one crosslink between U6 snRNA and actin pre-mRNA. All of these crosslinks were dependent on the presence of ATP, pre-mRNA, and the 5-iodoU or 4-thioU substituent of U6; therefore, these crosslinks putatively formed within spliceosomes.
The U2–U6.iodoU54 set of crosslinks trapped tertiary contacts in fully assembled yeast spliceosomes, during and/ or after the Prp2p step and before the first chemical step of splicing. A nearly direct correlation between the crosslinking yield and splicing yield suggests that these crosslinks formed in splicing-competent spliceosomes. The crosslinks trapped physical interactions between U54 of U6 and the U2 sequence that connects U2–U6 helix I and U2–U6 helix II (see Fig. 1). This connector sequence in U2 is thought to be single-stranded, and its structure is not sufficiently defined to allow prediction of the orientation of helix II relative to helix I. Therefore, our current 3D model of fully assembled yeast spliceosomes does not include helix II, nor does it include the single-stranded sequences between helices I and II. We know that neither U2–U6 helix II nor the sequence of U6 nts 86–108 is required for splicing activity in vivo or in vitro (Ryan et al. 2002). However, helix II is thought to provide redundancy for the function of U2–U6 helix Ib in spliceosomes, thus serving to protect splicing activity (Field and Friesen 1996). The crosslinked U2–U6.iodoU54 interactions are in the vicinity of a putative tertiary interaction, first proposed by Madhani and Guthrie (1994) and confirmed by Valadkhan and Manley (2000). This tertiary interaction occurs between G52 of U6 snRNA and U24, A25 of U2 snRNA. Our current 3D model predicts that this tertiary interaction can occur in fully assembled yeast spliceosomes (manuscript in preparation). Hence, the Guthrie–Manley interaction and the crosslinked U2–U6.iodoU54 interactions are expected to be present in fully assembled yeast spliceosomes.
For the U2–U6.thioU87–89 set of crosslinks, a 4-thioU substituent at each position from U87 through U90 in U6 RNA was, in all four cases, crosslinked to U2 nts 16, 15, 17, and 18 (highest to lower yield, 15 ≈ 17). Further analysis of this set of crosslinks showed that crosslinking occurred during spliceosome assembly, before the Prp2p-dependent step. These crosslinks contribute to our understanding of the RNA structures in assembling yeast spliceosomes prior to the Prp2p step. The striking feature of these crosslinks is that each individual 4-thioU from U87 to U90 of U6 crosslinked to the same four pyrimidines in U2 RNA, C15 to U18, and produced the same ratio of four crosslinked products in each case. The crosslinks span structurally undefined sequences of U6 and U2 snRNAs—the sequences that connect U2–U6 helices Ib and II. Perhaps the four pyrimidines of U2 at nts 15–18 provide a binding site for the 4-thioU substituent in each case. This binding site might also bind the 5-iodoU substituent of U6.iodoU54, thus directing its crosslinking to two of the same four pyrimidines of U2, namely U17 and U18, as observed.
For the U6.thioU80–pre-mRNA crosslink, a 4-thioU substituent at U80 in U6 RNA crosslinked to the +209th nucleotide of the yeast actin intron. Crosslinking was dependent on spliceosome assembly and occurred just before the first chemical step of splicing. The crosslinked nucleotide, A(+209), lies 15 nucleotides upstream of a 9-nt sequence in the actin intron which is complementary to a 9-nt sequence at +37–45 of the intron (Parker and Patterson 1987). These complementary sequences can form an intramolecular duplex within the intron. A similarly positioned duplex can form in the introns of yeast RP51A, RP51B, and CYH2 pre-mRNAs (Newman 1987; Goguel and Rosbash 1993). Such complementary sequences are a typical feature of long introns in S. cerevisiae (Parker and Patterson 1987). Duplex formation effectively shortens the intron and somehow promotes its assembly into a spliceosome. Formation of the RP51B intron duplex, or formation of a randomized duplex in its place, enhances incorporation of these introns into spliceosomes, thus enhancing their splicing efficiency (Libri et al. 1995). Likewise, the two complementary sequences in the RP51A intron must be present to enhance its splicing efficiency (Goguel and Rosbash 1993). In CYH2 introns, the ability to form the duplex is necessary for splicing in vitro and in vivo, if the upstream complementary sequence is present (Newman 1987). Further experiments suggested that the complementary sequences of the actin intron (of ACT1) function similarly in yeast.
The phosphoroyl group of the A79-U80 linkage in U6 snRNA is the only functional group in spliceosomal snRNAs known to bind to a divalent metal ion that is required for pre-mRNA splicing (Yean et al. 2000). Therefore, it may be of interest that the U80 base makes physical contact with the 209th nucleotide of the actin intron, near the nine base-pair intramolecular duplex of this intron. Other U6–pre-mRNA crosslinks were generated in yeast spliceosomes by Chan et al. (2003). Their oligo-directed RNase H mapping showed that U6 RNA was crosslinked to actin pre-mRNA near the intramolecular duplex of the intron, somewhat near the location of our U6.thioU80–intron crosslink (Fig. 10). Specifically, their crosslinks were mapped to U6 nts 100–112, at the 3′ end of U6, and to nts +1–40 of the actin intron. (We note that the crosslinked nucleotides were not individually identified in their Figure 2C, as the primer extension stops proposed to be crosslink-specific in their lane 5 also appeared in lanes 6 and 7 for different gel-purified crosslinks to an appreciable extent; however, their Figure 2B shows convincingly that the actin intron was crosslinked within the first 40 nucleotides of its 5′ end.) One of the oligonucleotides used by Chan et al. (2003) for RNase H mapping was specifically inhibited by the presence of the crosslinks. This inhibited oligonucleotide was complementary to nts +20–40 of the actin intron, suggesting that the crosslinks were located within this intronic sequence. They also found that crosslinking depended on the presence of the Prp19p complex, which is required for spliceosome assembly in yeast (Tarn et al. 1993).
FIGURE 10.
A hypothetical scenario suggests that our U6.thioU80-pre-mRNA crosslink (see arrow) may have formed when the actin pre-mRNA intron contained a putative intramolecular duplex, as depicted. The U80-intron contact and the intron duplex are both present in actin pre-mRNA in yeast spliceosomes (Newman 1987; see Results), although perhaps not concurrently. This scenario includes another U6-intron contact, involving the 3′ end of U6 snRNA (see arrow), as recently described by Chan et al. (2003). This tertiary contact is also present in actin pre-mRNA in yeast spliceosomes, perhaps when the intron duplex is present. It will be interesting to learn how many of the depicted interactions are present concurrently and at which stages of spliceosome assembly.
If the nine base-pair, actin intron duplex were present at the moment of U6–intron crosslinking detected by Chan et al. (2003), and if the tertiary interaction of our U6.thioU80-intron crosslink were also present concurrently, then the intron duplex could hold all of these U6–intron interactions relatively close together in space, as illustrated in Figure 10. This hypothesis also suggests that the intron duplex might pack against U6 snRNA in assembling spliceosomes. Chan et al. (2003) determined that their U6–intron crosslinks formed during spliceosome assembly, like our U6–intron crosslink. As more structural features of spliceosome assembly are characterized, it will be interesting to learn whether the actin intron duplex and the U6–intron tertiary interactions, depicted in Figure 10, are present concurrently during assembly or not.
In conclusion, the tertiary contacts trapped by our U2–U6.thioU87/88/89/90 crosslinks and by our U6.thioU80-pre-mRNA crosslink occur during spliceosome assembly. Therefore, these tertiary constraints are useful for assessing structural data of yeast spliceosomes during assembly and for developing structural models of the dynamic allosteric cascade of assembly. Our U2–U6.iodoU54 crosslinks provide the first data regarding tertiary snRNA-snRNA contacts in fully assembled yeast spliceosomes.
MATERIALS AND METHODS
Site-specifically modified U6 RNAs were prepared using template-directed ligation of synthetic oligonucleotide pieces of U6 RNA, as previously described (Ryan and Abelson 2002). 4-ThioU-containing oligonucleotide pieces of U6 were synthesized as described (Adams et al. 1994; Murray et al. 1995). 5-IodoU-containing oligonucleotide pieces were synthesized using commercially available phosphoramidites (Glen Research). 32P-labeled actin pre-mRNA was prepared according to Lin et al. (1985), except the final concentrations of the following were: 100 μM for UTP; 500 μM each for ATP, GTP, and CTP; and 3.75 μCi/μL for [α-32P] UTP. (This specific activity of 32P-labeled actin pre-mRNA was used in all previous experiments reported by D.E.R. and J.A.) Our procedures for reconstituting yeast extract with synthetic U6 RNA, and for assaying pre-mRNA splicing in U6-reconstituted extract, were previously described (Ryan and Abelson 2002). ATP depletions were achieved by adding superstoichiometric amounts of glucose to activate endogenous hexokinase, as described by Horowitz and Abelson (1993). Autoradiography was collected on a PhosphorImager, except for Figure 2A, which was developed on X-ray film.
Crosslinking of 5-iodoU-substituted U6 RNA to spliceosomal RNAs
Our typical 5-μL sample for depletion of endogenous U6 snRNA in splicing extract was scaled up fivefold to 25 μL. For the U6 reconstitution step we added 50 fmole of 5-iodoU-substituted U6 RNA containing a site-specific 32P radiolabel (7500 cpm/fmole) in 5 μL of 1× splicing buffer. To initiate splicing we added 5 μL of 10 fmole/μL cold actin pre-mRNA in 1× splicing buffer. After 15 min of splicing at 23°C, the sample was placed on ice and transferred to a watertight-fitting hole in a vertical side of an open Lucite box. The Lucite box was filled with ice and water to keep the sample at ~ 0°C during UV irradiation with the 325 nm line of a HeCd laser (Melles Griot). The laser beam (15–30 mW power) was aimed at the center of the aqueous sample (at the bottom of the Eppendorf tube with lid open); thus, the sample was irradiated on ice for 1.5 h. After irradiation, the sample was treated with proteinase K and was phenol extracted and ethanol precipitated. The crosslinked RNA of interest was gel-purified on a 5% denaturing polyacrylamide gel, in parallel with a wild-type U6-reconstituted control, for further analysis by RNase H treatment or primer extension mapping to map the site(s) of crosslinking (see below). Additional control samples were prepared in parallel but were not gel-purified prior to loading alongside the purified control and crosslinked RNA (perhaps further treated) on a 5% analytical denaturing polyacrylamide (19:1) gel.
Crosslinking of 4-thioU-substituted U6 RNA to spliceosomal RNAs
Samples were prepared as above using 4-thioU-substituted U6 RNAs, except that after the 15 min splicing reaction, the samples were placed on ice and transferred to a 96-well plate, resting on an ice and water slurry, and covered with a Pyrex Petri dish (to cutoff UV light below 285 nm). The samples were irradiated for 15 min at a distance of ~10 cm with a ~365-nm UV lamp (Model B-100AP, UV Products).
RNase H assay to identify the RNA crosslinked to U6 snRNA
Crosslinked RNA was prepared as above without gel purification for the initial RNase H assay of step 1 in our crosslink mapping scheme (see Results). UV-irradiated precipitated nucleic acid was dissolved in 100 μL of RNase H buffer (20 mM HEPES pH 7.8, 0.2 mM EDTA, 50 mM KCl, 1 mM DTT) and divided into 5 × 20-μL samples for RNase H digestion. Each sample was treated with 0.2 μg of DNA oligonucleotide complementary to U1, U2, U4, U5, or actin pre-mRNA. The DNA oligonucleotides were complementary to nts 110–124 of U1 snRNA, nts 29–43 of U2 snRNA (the SRU2 oligonucleotide), nts 76–91 of U4 snRNA, and nts 124–139 of U5 snRNA. These oligonucleotides had been tested for specificity and efficacy of RNase H digestion by Hitoshi Sawa. For the annealing step, each sample was heated at 70°C for 3 min and allowed to slowly cool in ambient air at 23°C for 1 h. Each sample was treated with 2 μL of 25 mM MgCl2, 1 μL of 6 μg/μL tRNA carrier (Sigma), and 2.5 units of RNase H (TaKaRa Bio Inc). Samples were incubated at 34°C for 10 min, followed by addition of 0.1 volume of 3 M NaOAc pH 5.3 and 3 volumes of ethanol to precipitate total nucleic acid at −20°C overnight.
Primer extension assay and accompanying sequencing
Crosslinked RNA was prepared as above. The gel-purified precipitated RNA was dissolved in 35 μL of TE pH 7.5. A 7 μL aliquot was subjected to primer extension by adding 50 fmole of gel-purified 32P-labeled DNA primer (7500 cpm/fmole) in 1 μL of TE pH 8 and 2 μL of 5× hybridization buffer (150 mM KCl, 50 mM Tris pH 7.5, 1 mM EDTA). The three U2 primers were complementary to nts 89–104, nts 104–121, and nts 119–136. Samples were incubated at 90°C for 1.5 min and allowed to cool in ambient air at 23°C for 10 min. During this step, 1 μL of AMV reverse transcriptase (25 units/μL; Roche) was diluted into 200 μL of 1.5× reaction buffer (50 mM TrisCl pH 8.5; 1.5 mM each of dATP, dGTP, dTTP, dCTP; 15 mM MgCl2, 15 mM DTT, 0.75 mM spermidine). After the 10 min cooling period, 20μL of the diluted AMV RT in 1.5× buffer was added to the cool 10-μL sample. The treated sample was incubated at 42°C for 30 min. At this point, total RNA in the sample was digested by adding 0.5 μL of 0.5 M EDTA and 2 μL of 2 M NaOH, with incubation at 65°C for 1 h. Total DNA was precipitated by adding 6 μg of mussel glycogen as carrier (Roche) and 0.1 volume of 3 M NaOAc pH 5.3 and 3 volumes of ethanol followed by cooling to −20°C overnight.
Affinity purification of U2–U6.iodoU54 crosslinked RNA
A gel-purified DNA oligonucleotide complementary to U6 nts 26–48 was synthesized with a biotin moiety attached via a linker arm to its 5′ end (Glen Research). A fresh batch of U2–U6.iodoU54 crosslinked RNA was prepared as above. After phenol extraction and ethanol precipitation, total nucleic acid was dissolved in 15 μL of 0.5× SSC, treated with 1.5 pmole of the gel-purified biotinylated DNA oligonucleotide in 0.5× SSC, and annealed to the oligonucleotide at 90°C for 1.5 min, followed by cooling in ambient air at 23°C for at least 10 min. Ten microliters of 1 mg/mL Streptavidin paramagnetic beads (Promega) were washed three times with 0.5× SSC. The annealed mixture was transferred to the washed beads and incubated at 23°C for 30 min. Subsequently, the beads were washed four times with 0.5× SSC. To this bead isolate was added 15 μL of ddH2O. The slurry was heated at 37°C for 2 min to elute the affinity-selected crosslinked RNA into aqueous solution. The eluate was treated with 3 μg of tRNA carrier, for ethanol precipitation of the purified crosslinks.
Preparation of U6.iodoU54 RNA containing a 5′ RP-phosphorothioyl (thioP) substituent at U80
This site-specifically modified U6 RNA was prepared using template-directed ligation of synthetic oligonucleotide pieces of U6 RNA, as described (Ryan and Abelson 2002). Full-length U6 was ligated from the following four RNA pieces: nts 1–38, 39–59.iodoU54, 60–76, and 77–112.RP-thioP-U80. The 5-iodoU-substituted piece was synthesized at Caltech using commercially available phosphoramidites (Glen Research). The U6.77–112.RP-thioP-U80 piece was made by in vitro transcription using a synthetic DNA template and T7 RNA polymerase (Amersham) as previously described (Milligan et al. 1987; see their Optimization of Yields). The transcription included 4 mM GMP, 1 mM GTP, 1 mM ATP, 1 mM CTP, 1 mM α-thioP UTP (Amersham; no longer commercially available), and 0.62 μCi/μL of [α-32P] CTP. The full-length transcript was gel-purified prior to U6 ligation.
Acknowledgments
We thank present and past members of the Abelson and Guthrie laboratories for helpful discussions, and Tamara Brenner and Stephen Rader for comments on the manuscript. This work was supported by National Institutes of Health Grant GM32637 to J.A., and by a Caltech Division of Biology Fellowship to D.E.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7060404.
REFERENCES
- Adams, C.J., Murray, J.B., Arnold, J.R.P., and Stockley, P.G. 1994. A convenient synthesis of S-cyanoethyl-protected 4-thiouridine and its incorporation into oligoribonucleotides. Tetrahedron Lett. 35: 765–768. [Google Scholar]
- Berglund, J.A., Chua, K., Abovich, N., Reed, R., and Rosbash, M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUACC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. and Guthrie, C. 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334: 213–218. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world (eds. R.F. Gestland, T.R. Cech, and J.F. Atkins), pp. 525–560, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chan, S.-P., Kao, D.-I., Tsai, W.-Y., and Cheng, S.-C. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Cheng, S.-C. and Abelson, J. 1987. Spliceosome assembly in yeast. Genes & Dev. 1: 1014–1027. [DOI] [PubMed] [Google Scholar]
- Du, H. and Rosbash, M. 2001. Yeast U1 snRNP-pre-mRNA complex formation without U1 snRNA-pre-mRNA base pairing. RNA 7: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–92. [DOI] [PubMed] [Google Scholar]
- Dubreuil, Y.L., Expert-Bezancon, A., and Favre, A. 1991. Conformation and structural fluctuations of a 218 nucleotides long rRNA fragment: 4-thiouridine as an intrinsic photolabeling probe. Nucl. Acids Res. 19: 3653–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., and Abelson, J. 1990. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science 250: 404–409. [DOI] [PubMed] [Google Scholar]
- ———. 1992. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 20: 3659–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., McPheeters, D.S., and Abelson, J. 1989. In vitro assembly of yeast U6 snRNP: A functional assay. Genes & Dev. 3: 2137–2150. [DOI] [PubMed] [Google Scholar]
- Field, D.J. and Friesen, J.D. 1996. Functionally redundant interactions between U2 and U6 spliceosomal snRNAs. Genes & Dev. 10: 489–501. [DOI] [PubMed] [Google Scholar]
- Goguel, V. and Rosbash, M. 1993. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell 72: 893–901. [DOI] [PubMed] [Google Scholar]
- Gordon, P.M., Sontheimer, E.J., and Piccirilli, J.A. 2000. Metal ion catalysis during the exonligation step of nuclear pre-mRNA splicing: Extending the parallels between the spliceosome and group II introns. RNA 6: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.L., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2002. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 99: 11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, M.L. and Krainer, A.R. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13: 302–309. [DOI] [PubMed] [Google Scholar]
- Horowitz, D.S. and Abelson, J. 1993. Stages in the second reaction of pre-mRNA splicing: The final step is ATP independent. Genes & Dev 7: 320–329. [DOI] [PubMed] [Google Scholar]
- Jurica, M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., and Moore M.J. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H. and Abelson, J. 1996. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA 2: 995–1010. [PMC free article] [PubMed] [Google Scholar]
- Kim, S.-H. and Lin, R.-J. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16: 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer, B., Achsel, T., and Lührmann, R. 1998. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. 95: 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Stutz, F., McCarthy, T., and Rosbash, M. 1995. RNA structural patterns and splicing: Molecular basis for an RNA-based enhancer. RNA 1: 425–436. [PMC free article] [PubMed] [Google Scholar]
- Lin, R.-J., Newman, A.J., Cheng, S.-C., and Abelson, J. 1985. Yeast pre-mRNA splicing in vitro. J. Biol. Chem. 260: 14780–14792. [PubMed] [Google Scholar]
- Madhani, H.D. and Guthrie, C. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817. [DOI] [PubMed] [Google Scholar]
- ———. 1994. Randomization-selection analysis of snRNAs in vivo: Evidence for a tertiary interaction in the spliceosome. Genes & Dev. 8: 1071–1086. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Lührmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Malca, H., Shomron, N., and Ast, G. 2003. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 23: 3442–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J. and Sharp, P.A. 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Adams, C.J., Arnold, J.R.P., and Stockley, P.G. 1995. The roles of the conserved pyrimidine bases in hammerhead ribozyme catalysis. Evidence for a magnesium ion-binding site. Biochem. J. 311: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby, M.I. and Greenbaum, N.L. 2002. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 9: 958–965. [DOI] [PubMed] [Google Scholar]
- Newman, A. 1987. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 6: 3833–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Norris, C.L., Meisenheimer, P.L., and Koch, T.H. 1996. Mechanistic studies of the 5-iodouracil chromophore relevant to its use in nucleoprotein photo-crosslinking. J. Am. Chem. Soc. 118: 5796–5803. [Google Scholar]
- Padgett, R.A., Podar, M., Boulanger, S.C., and Perlman, P.S. 1994. The stereochemical course of group II intron self-splicing. Science 266: 1685–1688. [DOI] [PubMed] [Google Scholar]
- Parker, R. and Patterson, B. 1987. Architecture of fungal introns: Implications for spliceosome assembly. In Molecular biology of RNA: New perspectives (eds. B. Dudock and M. Inouye), pp. 133–149. Academic Press, New York.
- Raghunathan, P.L. and Guthrie, C. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8: 847–855. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, D.E. and Abelson, J. 2002. The conserved central domain of yeast U6 snRNA: Importance of U2–U6 helix Ia in spliceosome assembly. RNA 8: 997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, D.E., Stevens, S.W., and Abelson, J. 2002. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA 8: 1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer, E.J. 1994. Site-specific RNA crosslinking with 4-thiouridine. Mol. Biol. Rep. 20: 35–44. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J. and Steitz, J.A. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. [DOI] [PubMed] [Google Scholar]
- Sonthiemer, E.J., Sun, S., Piccirilli, J.A. 1997. Metal ion catalysis during splicing of pre-messenger RNA. Nature 388: 801–805. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Gordon, P.M., and Piccirilli, J.A. 1999. Metal ion catalysis during group II intron self-splicing: Parallels with the spliceosomes. Genes & Dev. 13: 1729–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T.A. and Steitz, J.A. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 90: 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Ryan, D.E., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2002. Composition and functional characterization of the yeast spliceosomal Penta-snRNP. Mol. Cell 9: 31–44. [DOI] [PubMed] [Google Scholar]
- Tarn, W.-Y., Lee, K.-R., Cheng, S.-C. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. 22: 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan, S. and Manley, J.L. 2000. A tertiary interaction detected in a human U2–U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA 6: 206–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., MacMillan, A.M., Katopodis, N.F., Neubauer, G., Wilm, M., Lührmann, R., and Query, C.C. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20: 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean, S.-L., Wuenschell, G., Termini, J., and Lin, R.-J. 2000. Metalion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]