Abstract
The cellular protein, poly(rC) binding protein 2 (PCBP2), is known to function in picornavirus cap-independent translation. We have further examined the RNA binding properties and protein–protein interactions of PCBP2 necessary for translation. We have studied its putative multimerization properties utilizing the yeast two-hybrid assay and in vitro biochemical methods, including glutathione S-transferase (GST) pull-down assays and gel filtration. Through genetic analysis, the multimerization domain has been localized to the second K-homologous (KH) RNA binding domain of the protein between amino acids 125 and 158. To examine the function of multimerization in poliovirus translation, we utilized the truncated protein, ΔKH1-PCBP2, which is capable of multimer formation, but does not bind poliovirus stem–loop IV RNA (an interaction required for translation). Utilizing RNA binding and in vitro translation assays, this protein was shown to act as a dominant negative, suggesting that PCBP2 multimerization functions in poliovirus translation and RNA binding. Additionally, PCBP2 containing a deletion in the multimerization domain (ΔKH2-PCBP2) was not able to bind poliovirus stem–loop IV RNA and could not rescue translation in extracts that were depleted of endogenous PCBP2. Results from these experiments suggest that the multimerization of PCBP2 is required for efficient RNA binding and cap-independent translation of poliovirus RNA. By examining the functional interactions of the cellular protein PCBP2, we have discovered a novel determinant in the mechanism of picornavirus cap-independent translation.
Keywords: poliovirus, poly(rC) binding protein, translation, IRES, multimerization
INTRODUCTION
Picornaviruses are positive-strand RNA viruses that cause human diseases such as poliomyelitis, myocarditis, hepatitis, and the common cold. The genomic RNAs of picorna-viruses are ~7–8 kb in length and contain long (~600–1300 nt), highly structured 5′ noncoding regions (NCRs) that are essential for viral translation and RNA replication. Picor-naviruses utilize an alternative mechanism of translation initiation that is independent of the 5′ m7 GTP cap required for translation of most cellular mRNAs (Jang et al. 1988; Pelletier et al. 1988). Cap-independent translation occurs on other viral and some cellular RNAs (Hellen and Sarnow 2001), and the required sequences for internal ribosome binding have been defined and termed Internal Ribosome Entry Sites (or IRES elements). On the basis of sequence and structure homology, the IRES elements in picornaviruses have been divided into two groups; the Type I IRES elements are found in the genomic RNAs of enteroviruses (poliovirus and coxsackievirus) and rhinoviruses, and the Type II elements are found in those of aphthoviruses and cardioviruses. In addition to cis-acting RNA elements required for internal ribosome entry, essential trans-acting factors have been identified in cap-independent translation of picornaviruses. Several cellular proteins have been shown to bind to picornavirus IRES elements and have been implicated in translation. Examples of these proteins include polypyrimidine tract binding protein (PTB), La autoantigen, and upstream of n-ras protein (unr; for review, see Jackson 2002). One cellular protein, poly(rC) binding protein 2 (PCBP2), has been shown to bind to the 5′ noncoding region of poliovirus RNA and to be necessary for the translation of picornaviruses containing type I IRES elements (Blyn et al. 1996, 1997; Gamarnik and Andino 1997; Walter et al. 1999).
PCBP2 was first identified as a 38-kDa cellular protein that interacts with the stem–loop IV domain within the 5′ noncoding region of poliovirus RNA and is required for poliovirus translation (Blyn et al. 1996, 1997). Depletion of PCBP2 from HeLa cytoplasmic extracts severely decreased poliovirus translation, and the addition of recombinant PCBP2 to depleted extracts restored translation to mock depleted levels, confirming that PCBP2 is necessary for efficient poliovirus translation. In a subsequent study, Walter et al. (1999) showed that PCBP2 interacts with the 5′ non-coding region of picornavirus RNA containing both Type I and Type II IRES elements, but is only necessary for the translation of Type I IRES-containing picornaviruses. PCBP2 has also been implicated in poliovirus RNA replication. Andino and colleagues first reported a cellular protein that bound to the poliovirus RNA cloverleaf structure (stem–loop I) in conjunction with the viral protein 3CD and was necessary for RNA replication (Andino et al. 1990, 1993; Blyn et al. 1996). This cellular protein was later identified as PCBP2 and was confirmed to have a role in polio-virus RNA replication (Gamarnik and Andino 1997; Parsley et al. 1997; Walter et al. 2002).
PCBP2 belongs to the family of RNA binding proteins that contain a conserved RNA binding motif, the hnRNP K-homologous (KH) domain, which was originally identified in hnRNP K and consists of the βααββαstructural motif (Siomi et al. 1993). The four members of the PCBP family are located on separate chromosomes within the mouse and human genomes, but have high amino acid similarity and contain three KH domains that mediate their RNA binding activities (Tommerup and Leffers 1996; Makeyev and Liebhaber 2000 Makeyev and Liebhaber 2002). KH domain-containing proteins have been shown to multimerize and interact with other proteins. The Xenopus Vg1RBP protein contains four KH domains that contribute cooperatively to RNA binding and also mediate self-association (Git and Standart 2002). hnRNP K interacts with proteins containing SH domains, including src and vav, through a proline-rich domain found between KH2 and KH3. hnRNP K has also been shown to interact with SRp20 and 9G8 splicing proteins (Shnyreva et al. 2000) similar to the murine form of PCBP, which interacts with 9G8 (Funke et al. 1996). PCBP2 interacts with other hnRNP proteins, including other poly(C) binding proteins, in the yeast two-hybrid assay; however, the domains necessary for these interactions and the role of multimerization in protein function were not determined in these studies (Kim et al. 2000).
PCBPs mediate mRNA stability and down-regulation of translation by binding to the 3′ noncoding regions of α-globin, collagen, tyrosine hydroxylase, and 15-lipoxygenase mRNAs, imparting stability on these cellular messages (Weiss and Liebhaber 1994, 1995; Holcik and Liebhaber 1997; Stefanovic et al. 1997) or alternatively inhibiting translation of cellular RNAs, including α-globin and 15-lipoxygenase (Ostareck-Lederer et al. 1994; Kiledjian et al. 1995; Lindquist et al. 2000). The function of PCBPs in cap-independent translation was first identified in picorna-viruses, and these proteins do not have a role in canonical translation initiation of several capped mRNAs (Blyn et al. 1997; Gamarnik and Andino 1997; Walter et al. 1999). Although PCBP2 has been determined to be necessary for translation of picornaviruses containing Type I IRES elements, the mechanism by which this cellular protein mediates internal ribosome entry is not understood.
Recent studies of PCBP2 have shown that distinct KH domains are involved in viral translation and replication. Full-length PCBP2 is required for optimal RNA binding and translation; however, the KH1 domain is necessary and sufficient for binding to stem–loop I and stem–loop IV of poliovirus RNA (at ~10 μM recombinant protein) and can act as a dominant negative for poliovirus translation in Xenopus oocytes (Silvera et al. 1999). The KH3 domain increases RNA binding efficiency and is important for polio-virus translation, but not RNA replication (Walter et al. 2002). In the work presented here, we demonstrate that PCBP2 multimerization is mediated by a portion of the KH2 domain (amino acids 125–158). Utilizing PCBP truncations, we have mapped the multimerization domain of PCBP2 and have shown that multimerization is required for efficient binding to poliovirus stem–loop IV RNA and translation initiation.
RESULTS
PCBP2 multimerization
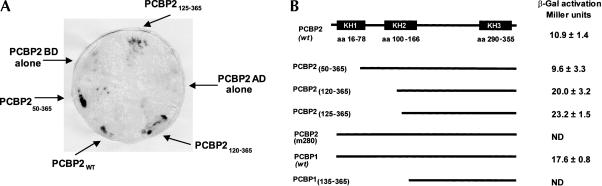
During a yeast two-hybrid screen originally aimed at identifying cellular proteins that interact with PCBP2, we discovered several versions of PCBP that interact with PCBP2 (shown in Fig. 1B). Seven of the 10 clones isolated from the HeLa cDNA library screen contained PCBP sequences, providing the first genetic evidence for PCBP2 multimerization. In addition to wild-type PCBP2, several N-terminally truncated forms of PCBP2 interacted with PCBP2 in a colorimetric β-gal assay (Fig. 1A). PCBP1, which shares >90% amino acid identity with PCBP2, was also isolated from the library, indicating that heteromultimers of PCBP2 and PCBP1 can form. Liquid β-gal assays were performed to evaluate the strength of PCBP multimerization in the yeast two-hybrid assay; β-gal activation of PCBP2 multimerization was ~10- to 23-fold above background, demonstrating that this is a significant interaction (Fig. 1B).
FIGURE 1.
Multimerization of PCBP clones isolated from a HeLa cDNA library. (A) Colorimetric β-gal assay on clones selected from the yeast-two hybrid screen. β-Gal activation was determined for yeast colonies selected on medium lacking His, Trp, and Leu. β-Gal activation causes a blue color phenotype in the presence of X-gal substrate. AD and BD refer to the Gal 4 activation domain and the binding domain plasmids, respectively. (B) Schematic of PCBP clones isolated following yeast two-hybrid screening. The K-homologous RNA binding domains (KH1, KH2, and KH3) in the full-length PCBP2 protein are shown, and the amino acid positions of these domains are cited from Makeyev and Liebhaber (2002). The schematic represents the domains of PCBP constructs isolated from the yeast-two hybrid screen that interact with PCBP2. The amino acids contained in each clone are indicated by the numbers in subscript. PCBP2m280 contains a single alanine insertion at amino acid position 280 that may have occurred during the synthesis of the HeLa cDNA library. β-Gal activation of each PCBP2 clone was determined using the liquid assay and values are represented as Miller Units (1 U equals the amount that hydrolyzes 1 μmole of ONPG substrate per minute). Quantitative β-gal assays were not performed for two of the clones (denoted as ND in the figure); however, these clones did exhibit an interaction with wild-type PCBP2 in the colorimetric assay.
On the basis of the clones isolated from the library screen, we predicted the domains of PCBP2 that are necessary for multimerization (Fig. 1B). Several PCBP clones contain deletions of the KH1 domain, indicating that this domain is not necessary for the interaction. The KH1 domain, which is known to be crucial for RNA binding, does not appear to be necessary for multimerization, providing the first evidence that distinct domains of PCBP2 are involved in RNA binding and multimerization. Wild-type PCBP2 multimerization activates β-gal synthesis ~10-fold above background; however, the clones that contain a deletion in the N-terminal 125 amino acids activate β-gal synthesis up to 23-fold, suggesting that the multimerization domain may be more accessible in these proteins. The results from the yeast two-hybrid assay suggest that PCBP2 is capable of multimerization and give preliminary evidence that the N-terminal 125 amino acids (KH1 and a portion of the KH2 domain) are not required for this interaction.
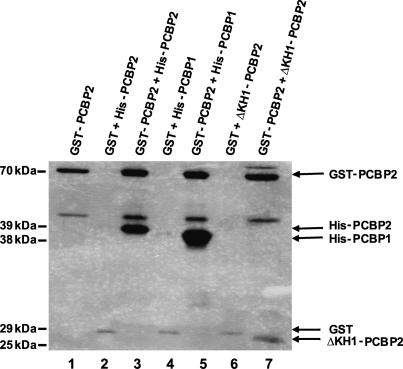
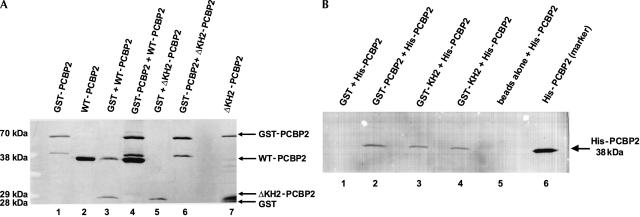
To further examine PCBP2 multimerization, we utilized a GST pull-down assay. A GST-PCBP2 fusion protein was used to test multimerization with recombinant wild-type PCBP2, PCBP1, and the N-terminal truncated PCBP2 (ΔKH1-PCBP2), containing amino acids 125–365, which was isolated from the HeLa cDNA library in the yeast two-hybrid screen. All three versions of recombinant PCBP interacted with GST-PCBP2 (Fig. 2, lanes 3,5,7) to levels above the nonspecific interaction with GST alone (Fig. 2, lanes 2,4,6), confirming PCBP multimerization. Following treatment with RNase A and T1, PCBP multimerization was still observed in GST pull-down assays, demonstrating that this interaction is not RNA dependent (data not shown).
FIGURE 2.
Biochemical evidence for PCBP multimerization. GST pull-down assays were carried out with recombinant wild-type PCBP2, wild-type PCBP1, ΔKH1-PCBP2 (containing an N-terminal deletion through amino acid 119), and GST-PCBP2 to test PCBP multimerization (lanes 3,5,7). Equal amounts of recombinant proteins, measured by Bradford assay, were used in the experiment. Proteins that interacted with GST-PCBP2 were collected on glutathione Sepharose, subjected to electrophoretic separation on a SDS-polyacrylamide gel, and visualized by silver staining. To determine nonspecific protein–protein interactions in this assay, each reaction was repeated with GST alone as a negative control (lanes 2,4,6). The molecular weights shown on the left side of the gel correspond to the size of each recombinant protein used in the assay.
ΔKH1-PCBP2 acts as a dominant negative in poliovirus translation and RNA binding
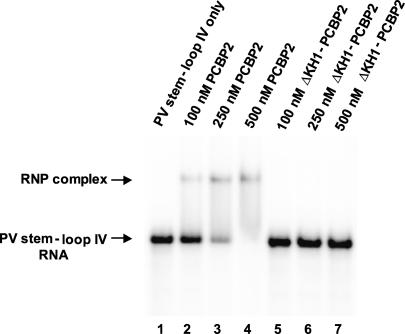
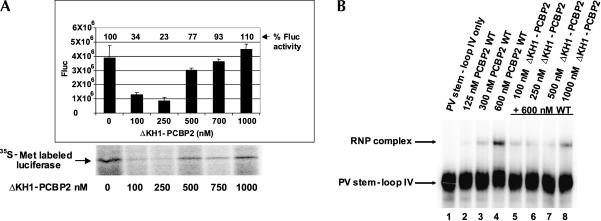
We tested the function of PCBP2 multimerization in picornavirus translation by utilizing the N-terminally truncated protein, ΔKH1-PCBP2, which is able to multimerize with wild-type PCBP2. In electrophoretic mobility shift assays, ΔKH1-PCBP2 was unable to form a complex with poliovirus stem–loop IV RNA when 500 nM of recombinant protein was added; however, wild-type PCBP2 formed a distinct complex with stem–loop IV RNA when 100–250 nM protein was added (Fig. 3). This confirms previous studies showing that the KH1 domain is required for poliovirus stem–loop IV RNA binding (Silvera et al. 1999; Walter et al. 2002). If ΔKH1-PCBP2 interacts with wild-type PCBP2, but does not bind to stem–loop IV RNA, we hypothesized that ΔKH1-PCBP2 would act as a dominant negative in polio-virus translation. To determine the effect of ΔKH1-PCBP2 on poliovirus protein synthesis, we carried out in vitro translation assays in HeLa cytoplasmic extracts (containing endogenous PCBP2) using transcribed RNA encompassing the 5′ NCR of poliovirus and the luciferase coding region. The addition of ΔKH1-PCBP2 to HeLa cytoplasmic extracts inhibited poliovirus translation 4.5-fold, suggesting that this mutated protein may be acting as a poison subunit in the PCBP2 multimer complex (Fig. 4A). In contrast, the addition of wild-type PCBP2 to HeLa cytoplasmic extracts has no effect on poliovirus translation (Walter et al. 1999). At higher concentrations (500–1000 nM) of ΔKH1-PCBP2, there is a reduced activity of this protein to inhibit translation, possibly due to insolubility, aggregation of ΔKH1-PCBP2, or the formation of ΔKH1-PCBP2 homomultimers that cannot interact with wild-type PCBP2. We have also analyzed the in vitro production of the luciferase protein in translation reactions that were incubated with [35S]methionine and analyzed by SDS-PAGE (Fig. 4A). These results confirm that ΔKH1-PCBP2 inhibits poliovirus translation, as there is a decrease in the synthesis of labeled luciferase protein (Fig. 4A).
FIGURE 3.
ΔKH1-PCBP2 does not bind poliovirus stem–loop IV RNA. Electrophoretic mobility shift assays were carried out with labeled PV stem–loop IV RNA and recombinant PCBP2 proteins. Recombinant wild-type PCBP2 (a positive control for RNA binding) or ΔKH1-PCBP2 (containing an N-terminal deletion through amino acid 119) was incubated with labeled stem–loop IV RNA and subjected to electrophoresis on a nondenaturing polyacrylamide gel. Each reaction contained 0.1 pmole of labeled stem–loop IV RNA and between 100 and 500 nM protein (indicated above each lane). Wild-type PCBP2 forms a complex with stem–loop IV RNA as observed by the shift in the labeled RNA probe; however, ΔKH1-PCBP2 does not form a complex with the same RNA probe.
FIGURE 4.
ΔKH1-PCBP2 acts as a dominant negative in poliovirus translation and binding to stem–loop IV RNA. (A) In vitro translation assays of poliovirus luciferase RNA. In vitro translation assays in HeLa cytoplasmic S10 extracts were carried out using luciferase reporter RNAs preceded by the poliovirus 5′ NCR. Luciferase production was determined using a luminometer. Translations without ΔKH1-PCBP added were set as 100%. (Bottom) The [35S]methionine translations with ΔKH1-PCBP. Translation reactions were carried out using luciferase reporter RNAs driven by the poliovirus 5′ NCR in HeLa S10 in the presence of [35S]methionine. These translation reactions also contained recombinant ΔKH1-PCBP indicated below each lane. The labeled luciferase translations were subjected to electrophoresis on a SDS-polyacrylamide gel and analyzed by autoradiography for protein production. (B) Electrophoretic mobility shift assays of PV stem–loop IV RNA with wild-type PCBP2 and ΔKH1-PCBP2. Mobility shift assays were carried out using 600 nM wild-type protein and increasing amounts of ΔKH1-PCBP (indicated above each lane). Wild-type PCBP2 complex formation with PV stem–loop IV RNA is shown in lanes 2,3,4.
To determine whether ΔKH1-PCBP2 inhibits wild-type PCBP2 binding to stem–loop IV RNA, we utilized electrophoretic mobility shift assays (Fig. 4B). The electrophoretic mobility shift assays contained labeled poliovirus stem–loop IV RNA, 600 nM wild-type PCBP2 (which causes a distinct shift in RNA mobility in Fig. 4B, lane 4), and increasing amounts of ΔKH1-PCBP2 (Fig. 4B, lanes 5–8). When 100–500 nM ΔKH1-PCBP2 was incubated with wild-type PCBP2, a decrease in complex formation resulted; however, 1000 nM ΔKH1-PCBP2 had a reduced inhibitory effect on RNA binding (Fig. 4B). ΔKH1-PCBP2 inhibits wild-type PCBP2 binding to stem–loop IV RNA, consistent with the inhibition it causes in poliovirus translation. We propose that ΔKH1-PCBP2 is able to form multimers with wild-type PCBP2 and acts as a poison subunit in a PCBP complex by inhibiting binding to poliovirus stem–loop IV RNA and translation. The reduced inhibitory activity of ΔKH1-PCBP2 in PV translation and stem–loop IV binding observed at high concentrations may be due to the formation of tightly associated ΔKH1-PCBP2 homomultimers that cannot dissociate and interact with wild-type PCBP2. ΔKH1-PCBP2 inhibits RNA binding and translation when present at 100–250 nM, consistent with the strong interaction in the yeast two-hybrid assay (Fig. 1B). Work performed in our lab and by others has demonstrated that the concentration of PCBP2 in the cytoplasm of HeLa cells is ~100 nM, correlating with the amounts of recombinant proteins used in our in vitro assays (P. Sean, K. Bedard, and B.L. Semler, unpubl.; Silvera et al. 1999).
Mapping the PCBP2 multimerization domain
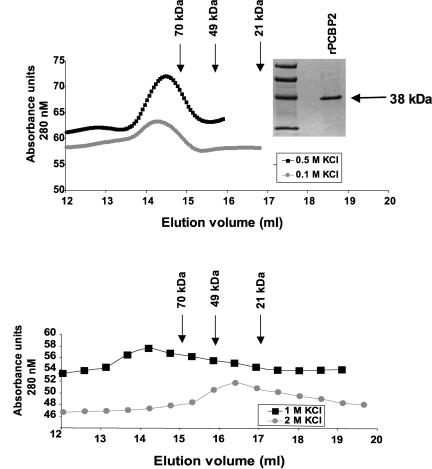
To analyze PCBP2 multimerization, recombinant His-tagged PCBP2 was subjected to gel-filtration chromatography on a Superdex S-200 column. The Coomassie-stained gel shows recombinant PCBP2 utilized in gel-filtration experiments is ~90% pure and is a single 38-kDa species (Fig. 5). In low or medium salt buffer (100 or 500 mM KCl), wild-type recombinant PCBP2 eluted from the column as a complex >70 kDa relative to standards of known molecular weight (Fig. 5), correlating with a PCBP2 dimer and confirming previously published results that PCBP exists in HeLa cytoplasmic extracts as a protein dimer (Gamarnik and Andino 1997). From all elution fractions collected (up to 18 mL), no additional protein species were observed that correlated to a PCBP2 monomer. To confirm that PCBP2 was present in elution fractions that gave high UV absorbance, we examined the fractions by visualization on a silver-stained gel. Fractions 13–15 contained detectable PCBP2 protein, and no protein was observed in fractions 16 and above (data not shown). Gel-filtration analysis with 100 and 500 μg of recombinant PCBP2 gave similar results, demonstrating that PCBP2 multimerization is not dependent on protein concentration. In the presence of 1 M KCl, PCBP2 elutes as a dimer, but the broad shoulder of the elution peak suggests that a monomer species may also exist. In 2 M KCl, the elution of recombinant PCBP2 is shifted to one peak corresponding to monomeric protein that is smaller than the 49-kDa standard (Fig. 5).
FIGURE 5.
Gel-filtration chromatography of recombinant PCBP2. Gel filtration of histidine-tagged recombinant PCBP2 was carried out using a 24-mL Superdex S-200 column. The PCBP2 protein subjected to gel filtration was bacterially expressed and purified using a nickel column and 500 μg of protein was loaded onto the S-200 column. The recombinant protein is ~90% pure as determined by visualization on a Coomassie-stained polyacrylamide gel shown at the top. (Top) The square symbols represent gel filtration in buffer containing 0.5 M KCl, and the circles represent chromatography in 0.1 M KCl. (Bottom) The squares represent gel filtration in buffer containing 1 M KCl, and the circles represent chromatography in 2 M KCl. Absorbance units at a wave length of 280 nM were measured to determine total protein eluted from the gel-filtration column. Standards of known molecular weight were used to generate a standard curve of elution volume vs. molecular weight of proteins.
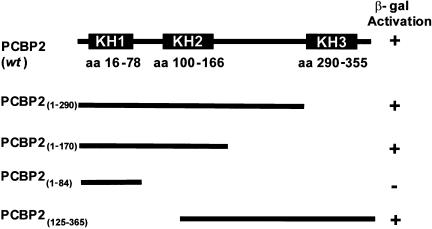
To further define the domain of PCBP2 involved in multimerization, we engineered truncations in the C-terminal portion of the protein (shown in Fig. 6) and tested them for multimerization in the yeast two-hybrid assay. Expression of the C-terminally truncated PCBP2-Gal4 binding domain fusion proteins was confirmed in yeast by Western blot analysis (data not shown), and multimerization was tested using a colorimetric β-gal assay in yeast expressing wild-type PCBP2 protein (Fig. 6). Deletion of the N-terminal domain (KH1), C-terminal domain (KH3), and the linker between KH2 and KH3 did not inhibit the interaction with wild-type PCBP2 in the yeast two-hybrid assay (Fig. 6). However, when both the KH2 and KH3 domains were deleted, β-gal activation was abolished, suggesting that a region within KH2 (amino acids 125– 170) is required for PCBP multimerization.
FIGURE 6.
The KH2 domain is required for PCBP2 multimerization. This schematic represents the C-terminal truncations engineered in PCBP2 to identify the multimerization domain. Interaction of the truncated proteins with wild-type PCBP2 was tested using the yeast two-hybrid assay. Positive β-gal activation was determined using the colorimetric assay. The numbers in subscript represent the amino acids contained in each truncated form of PCBP2.
The multimerization domain (KH2) is required for RNA binding and PV translation
We confirmed that the KH2 domain mediates PCBP2 multimerization by deleting amino acids 125–158 of PCBP2 and testing the ability of this protein to interact with GST-PCBP2. To confirm that differences in protein activity were not due to significant variability in protein concentration or purity, we examined purified wild-type PCBP2 and ΔKH2-PCBP2 on a Coomassie-stained SDS-containing, polyacrylamide gel (data not shown). In GST pull-down assays, ΔKH2-PCBP2 does not interact with either the GST control or the GST-PCBP2 fusion protein, whereas wild-type PCBP2 interacts with GST-PCBP2 above background interaction with GST alone (Fig. 7A). Moreover, yeast two-hybrid assays performed with ΔKH2-PCBP2 and wild-type PCBP2 resulted in no growth on selective medium, confirming that the KH2 domain is required for PCBP2 multimerization (data not shown). These data provide evidence that the KH2 domain is required for multimerization, but do not examine whether KH2 is sufficient for PCBP2 multimerization. To address this question, we generated a recombinant protein containing the KH2 domain (amino acids 70–270) fused to the GST protein, and tested its ability to interact with wild-type PCBP2 in GST pull-down assays. This protein (GST-KH2 PCBP2) was able to interact with His-tagged PCBP2 in pull-down assays (Fig. 7B, lanes 3,4), but with a slightly lower affinity than wild-type GST-PCBP2 (Fig. 7B, lane 2). These data suggest that the KH2 domain is sufficient for multimerization, but other protein domains may stabilize the interaction, resulting in the increased level of wild-type PCBP2 multimerization compared with KH2 alone.
FIGURE 7.
The KH2 domain of PCBP2 is necessary and sufficient for multimerization with wild-type PCBP2. (A) GST pull-down assays with recombinant GST-PCBP2 and histidine-tagged ΔKH2-PCBP2. GST-PCBP2 and wild-type His-PCBP2 were loaded in lanes 1 and 2 as size markers. The pull-down assays were carried out as described in Figure 2, using glutathione Sepharose to collect GST-PCBP2 and interacting proteins. Wild-type PCBP2 and ΔKH2-PCBP2 of equal concentration and purity were used in the pull-down assays. GST alone was used to detect nonspecific protein interactions in lanes 3 and 5. Wild-type PCBP2 was incubated with GST-PCBP2 in lane 4 as a positive control. ΔKH2-PCBP2 was incubated with GST (lane 5) and GST-PCBP2 (lane 6). Recombinant ΔKH2-PCBP2 was included in lane 7 as a size comparison. (B) GST pull-down assays with histidine-tagged wild-type PCBP2 and GST-KH2 PCBP2. The KH2 domain was fused to the GST protein and tested for its ability to interact with PCBP2. Wild-type PCBP2 (histidine-tagged) was incubated with GST alone (lane 1; negative control), full-length GST-PCBP2 (lane 2; positive control), GST-KH2 PCBP2 (lanes 3,4; referred to in the figure as GST-KH2), or beads alone (lane 5; negative control), and assays were performed as described above. The GST-KH2 PCBP2 protein used in lanes 3 and 4 is from two independent preparations of recombinant protein. To detect the interaction with histidine-tagged PCBP2, a Western blot was performed, and PCBP2 (histidine-tagged) was detected with a monoclonal antibody to histidine. Western blot analysis was used to distinguish between GST-KH2 PCBP2 and full-length His-PCBP2. Recombinant His-PCBP2 was included in lane 6 of the Western blot as a size marker.
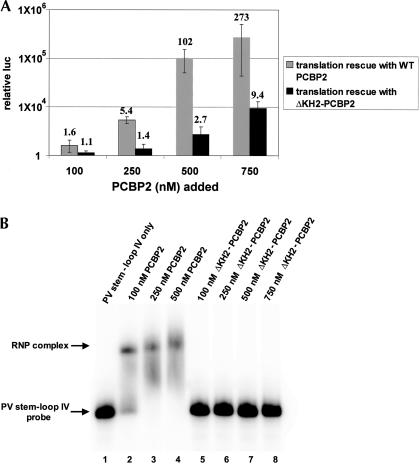
To determine the necessity of PCBP2 multimerization in poliovirus translation and stem–loop IV binding, ΔKH2-PCBP2 was used in translation experiments and electrophoretic mobility shift assays. Poliovirus translation assays were carried out in vitro using HeLa cytoplasmic S10 extracts that were depleted of PCBP using a poly(rC) RNA affinity column (Walter et al. 1999). These depleted extracts contained no detectable PCBP proteins by Western blot analysis, and were not capable of supporting efficient poliovirus translation. ΔKH2-PCBP2 was added to translation reactions in depleted S10 extracts to compare its ability to function in PV translation with that of wild-type PCBP2. Wild-type PCBP2 increased translation over 200-fold above PCBP-depleted background levels, compared with ΔKH2-PCBP2, which only rescued translation by <10-fold at the highest concentrations of protein added (Fig. 8A). These data suggest that deletions within the multimerization domain (KH2) inhibit PCBP2 function in poliovirus translation.
FIGURE 8.
ΔKH2-PCBP2 does not mediate poliovirus translation or efficiently bind to PV stem–loop IV RNA. (A) Poliovirus translation in PCBP-depleted S10 with the addition of wild-type PCBP2 or ΔKH2-PCBP2. In vitro translation assays were carried out in HeLa cell S10 extracts that were depleted of endogenous PCBP proteins. Translations were programmed with transcribed RNA containing the PV 5′ NCR and the luciferase coding region, and either wild-type or ΔKH2-PCBP2 was added. Poliovirus translation levels were determined by luciferase assays. Data is presented as relative luciferase units on a logarithmic scale. The numbers indicated above the data columns represent fold above translation in PCBP depleted S10 with no protein added. (B) Electrophoretic mobility shift assays with PV stem–loop IV RNA and ΔKH2-PCBP2. [32P]UTP PV stem–loop IV RNA was incubated with recombinant wild-type PCBP2 or ΔKH2-PCBP2 at the indicated concentrations. Wild-type PCBP2 was used as a positive control for RNA binding in lanes 2–4. ΔKH2-PCBP2 was incubated with stem–loop IV RNA under the same conditions in lanes 5–8.
To test the necessity of PCBP2 multimerization in stem–loop IV RNA binding, we utilized ΔKH2-PCBP2 in electrophoretic mobility shift assays with PV stem–loop IV RNA. Wild-type PCBP2 forms a complex with stem–loop IV RNA at 250 nM protein; however, there is no detectable complex between ΔKH2-PCBP2 and stem–loop IV at 750 nM protein in these RNA binding assays, demonstrating that ΔKH2-PCBP2 does not bind stem–loop IV (Fig. 8B). Electrophoretic mobility shift assays were repeated with 1500 nM ΔKH2-PCBP2, and no complex with stem–loop IV RNA was observed (data not shown). Previous reports have shown that the KH1 domain of PCBP2 alone can bind to PV stem–loop IV RNA at high concentrations (~10 μM), demonstrating that KH1 alone binds to stem–loop IV RNA with low efficiency (Silvera et al. 1999). Although KH1 is the primary domain of PCBP2 involved in RNA binding, our data suggest that the multimerization domain (KH2) is required for efficient binding to poliovirus stem–loop IV RNA and translation initiation.
DISCUSSION
Poly(rC) binding proteins have been shown to be involved in both picorna-virus translation and RNA replication by binding two distinct stem–loop structures, stem–loop IV and stem–loop I, (respectively) in the 5′ NCR of polio-virus RNA. Through RNA footprinting and site-directed mutagenesis experiments, the binding site of PCBP2 on poliovirus stem–loop IV RNA is thought to be a “bulge” of four C residues at the top of stem–loop IV (Gamarnik and Andino 2000). This RNA sequence has also been shown through deletion analysis to be important for PCBP2 binding and poliovirus translation (J. Nguyen, L. Leong, S. Stewart, and B. Semler, unpubl.). PCBP2 is abundantly expressed in HeLa cells and is present at high concentrations in cytoplasmic extracts that are utilized for in vitro poliovirus translation and replication studies. By utilizing a poly(rC) column, it is possible to effectively deplete PCBP (both PCBP1 and PCBP2) to nondetectable levels in our cytoplasmic extracts, resulting in a drastic decrease in viral translation. The addition of recombinant PCBP2 protein to translation reactions rescued poliovirus translation to mock-depleted levels; however, the addition of recombinant PCBP1 had no stimulatory effect on translation (Blyn et al. 1997; Walter et al. 2002). This suggests that PCBP2 independently functions in cap-independent translation, but PCBP1 alone cannot function in translation. Interestingly, our data provided the first genetic evidence for heteromul-timerization between PCBP1 and PCBP2, raising the possibility that these complexes might function in translation initiation mediated by the poliovirus IRES.
To further elucidate the role of PCBP2 in cap-independent translation of poliovirus, we have studied the protein–protein interactions of PCBP2. We carried out a yeast two-hybrid screen of a HeLa cDNA library to detect binding partners of PCBP2. In our initial screen, several clones isolated as interacting with PCBP2 contained PCBP sequence, confirming previous studies suggesting that PCBP exists as a multimer. This was not surprising, considering several proteins containing KH domains, including poly(rC) binding proteins, have been shown to multimerize (Kim et al. 2000; Git and Standart 2002). This interaction is readily detectable in the yeast two-hybrid assay and is stable in buffer containing 1 M KCl, demonstrated by our gel-filtration analysis, implying that the majority of endogenous PCBP2 is present as a multimer. Mapping the multimerization domain through mutational analysis demonstrated that the C-terminal portion of the KH2 domain (amino acids 125–158) is required. In a previous study by Walter et al. (2002), substitutions were made within the N-terminal portion of KH2 at amino acid positions 121 and 124 of PCBP2, and were shown to have little effect on binding to poliovirus stem–loop IV RNA and poliovirus translation. Recombinant PCBP2 with these amino acid substitutions was tested in GST pull-down assays and was able to multimerize similar to wild-type PCBP2 (data not shown). Therefore, the N-terminal portion of KH2 (up to amino acid 125) is not absolutely required for PCBP2 multimerization, RNA binding, or translation.
To examine whether the KH2 domain was sufficient for multimerization, we engineered a recombinant protein containing the KH2 domain fused to the GST protein and tested its ability to multimerize in a GST pull-down assay. GST-KH2 PCBP2 was shown to interact with wild-type His-PCBP2; however, the affinity of the interaction was slightly less than that of wild-type PCBP2 multimerization in the same assay. This experiment provides evidence that the KH2 domain is sufficient for PCBP2 multimerization, but other domains (KH1, KH3, or both) may increase the stability of the interaction.
The function of PCBP2 multimerization in poliovirus RNA binding and translation has yet to be examined, but may provide insights into how this cellular protein facilitates cap-independent translation. We have utilized electrophoretic mobility shift assays and in vitro translation experiments to study the function of multimerization in both RNA binding and translation. Because ΔKH1-PCBP2 was shown to multimerize, but was not able to bind stem–loop IV RNA, we predicted that this protein would not function independently in poliovirus translation, but might act as a dominant negative during PV translation through its interaction with wild-type endogenous PCBP2. To test this hypothesis, we added recombinant ΔKH1-PCBP2 to in vitro poliovirus translation assays and saw a significant decrease in translation, possibly due to the formation of nonfunctional multimer complexes. To further examine the function of multimerization in RNA binding, we added mixtures of wild-type PCBP2 and ΔKH1-PCBP2 to electrophoretic mobility shift assays with PV stem–loop IV RNA. ΔKH1-PCBP2 also inhibited the interaction of wild-type PCBP2 with stem–loop IV RNA, suggesting that PCBP2 binds to stem–loop IV as a multimer.
In both PV translation and RNA binding experiments, ΔKH1-PCBP2 inhibits the function of PCBP and acts as a poison subunit in a functional PCBP complex. Although results from experiments with this dominant-negative PCBP protein suggested that a PCBP multimer functions in RNA binding and translation, we wanted to formally test the requirement for PCBP multimerization in both RNA binding and translation. We engineered a PCBP that could not multimerize (ΔKH2-PCBP2) and tested its ability to function in RNA binding and translation. ΔKH2-PCBP2 was severely deficient in rescuing PV translation in PCBP-depleted cytoplasmic extracts and in binding to stem–loop IV RNA when compared with wild-type protein. Global misfolding of ΔKH2-PCBP2 could account for the lack of RNA binding in our RNA electrophoretic mobility shift assays; however, ΔKH2-PCBP2 maintains some ability to rescue PV translation in HeLa extracts depleted of PCBP. Therefore, we predict that ΔKH2-PCBP2 is a functional protein with a decreased ability to rescue PV translation. Previously, amino acid substitutions that disrupted a portion of the KH2 domain were made, and the resulting PCBP2 protein was functional and presumed properly folded (Walter et al. 2002). Structural studies on proteins that contain KH-domains have revealed that these domains are independently folded; therefore, we predict that disturbing one KH-domain will not affect the overall structure of the protein (Musco et al. 1996, 1997). These experiments demonstrated that PCBP multimerization is necessary for both PV stem–loop IV RNA binding and translation, and that multimerization occurs through the KH2 domain.
The role of PCBP2 multimerization in RNA binding and translation expands our understanding of picornavirus cap-independent translation initiation and suggests new mechanistic possibilities for ribosome recruitment and binding in poliovirus translation. Does a multimer complex of PCBP make multiple RNA contacts within the 5′ NCR of the viral RNA that arranges the RNA structurally for ribosome binding? This is an attractive mechanistic possibility, as mutational analysis of the 5′ NCR of several picornaviruses has demonstrated that RNA secondary structure is critical for efficient viral translation. Secondary structures within Type I and Type II IRES containing viruses may be a commonality in the mechanism of cap-independent translation of picornaviruses and cellular mRNAs. Another possibility is that the PCBP complex recruits ribosomal proteins through other protein–protein interactions. Several cellular proteins were shown to interact with PCBP2 in the yeast two-hybrid assay (data not shown); however, the significance of these interactions in picornavirus translation and/or replication is still being explored.
MATERIALS AND METHODS
Plasmid design
Synthetic oligonucleotides (primers) were as follows:
PC2del#3 (5′-GAAGTAGTCTCTGCAGATGCA-3′);
PC2del#2 (5′-CAACATGACCCTGCAGATCTGT-3′);
PC2del#1 (5′-GAGCTGCAGATGTCCTCTTCCT-3′);
PC2ER1 (5′-ACAGAATTCATTAAAGAGGAG-3′);
PCBP2-552-minus (5′-TGTACTCTCTGTATTTCCTTGA-3′);
PCBP2-652-plus (5′-CAGATCTGCGTGGTCATGTTGGA-3′);
PCBP2 KH2 5′ (5′-GGCTGGATCCACTAATGCCA-3′);
PCBP2 KH2 3′ (5′-GGAGAGCTGAATTCAATGCCA-3′).
pT220-460, which contains the cDNA for stem–loop IV RNA of the poliovirus IRES downstream of the T7 promoter, the PCBP2 wild-type bacterial expression vector, pQE30-PCBP2, and p5′PVLuc, which contains the 5′ noncoding of poliovirus followed by the luciferase coding region, have all been previously described (Dildine and Semler 1992; Blyn et al. 1996; Walter et al. 1999).
Full-length PCBP2 coding sequence from pQE30-PCBP2 was inserted into the Gal 4 binding domain fusion plasmid, pGBT-9, (Clontech) by digesting with BamHI and PstI and performing a DNA ligation. The HeLa cDNA library utilized for screening is present in the pGAD-GH Gal 4-activation domain plasmid (Clontech). The pGAD 424 DNA clone containing PCBP2125–365 sequence was isolated from yeast following the interaction screen, digested with BamHI and PstI, and cloned into the QE30 expression vector. The GST-PCBP2 plasmid was generated by cloning the PCBP2 coding sequence from pQE30-PCBP2 into the pGEX 2T vector (Pharmacia).
The PCBP2 truncations were generated by PCR amplification of pQE30-PCBP2 and were inserted into the QE30 vector. The PCR-amplified fragments generated an EcoRI site at the 5′ end of the fragment and a PstI site at the 3′ end of the fragment; therefore, all fragments were cloned into the EcoRI and PstI sites of the QE30 expression vector. The PC2del#3 primer and the PC2ERI primer were used to PCR amplify the coding sequence of PCBP21–290, PC2del#2 and PC2ERI were used to clone PCBP21–170, and PC2del#1 and PC2ERI were used to amplify PCBP21–84. Each PCR product was digested with EcoRI and PstI and cloned into the QE30 vector. PCBP2 containing a deletion in the KH2 domain between amino acids 125 and 158 was generated by PCR amplifying QE30-PCBP2 (wild-type) with PCBP2-552-minus and PCBP2-652-plus and performing a blunt-end ligation. The GST-KH2 PCBP2 expression construct was generated by PCR amplifying QE-30 PCBP2 (wild-type) with KH2 5′ and KH2 3′ oligo-nucleotide primers, digesting with BamHI and EcoRI and ligating into the pGEX-4T1 vector.
Yeast transformations
The yeast two-hybrid assay was performed as described in the Matchmaker protocol (Clontech). The bait plasmid containing the complete PCBP2 coding sequence cloned into the Gal 4 binding domain expression vector and the HeLa cDNA library were co-transformed into HF7c yeast. The yeast were grown overnight in nonselective YPD medium, diluted 1:20 in YPD (100 mL) and grown to an optical density (OD) of 0.5 at 30°C. The yeast were washed in water and then resuspended in a transformation buffer [10 mM Tris-HCl, 1 mM EDTA (pH 7.5), and 100 mM lithium acetate (pH 7.5)]. A total of 1 mg of bait plasmid DNA, 0.5 mg of the cDNA library DNA, and 20 mg of herring testes carrier DNA were added to 8 mL of competent yeast cells with 60 mL of transformation buffer containing 40% polyethylene glycol, avg. mol. wt. = 3350 (Sigma). Transformations were incubated at 30°C for 30 min with shaking. DMSO was added to a final concentration of 10%, cells were incubated at 42°C for 15 min, and then placed on ice. Cells were collected by centrifugation, resuspended in TE buffer and 200 μL of cells were plated on 150-mm plates of selective medium and incubated at 30°C until growth was seen (up to 7 d). Transformations were plated on medium lacking tryptophan and leucine to detect transformation efficiency, and additionally without histidine to detect interactions. Selective medium plates contained 6.7 g of yeast nitrogen base without amino acids, 20 g of dextrose, and 2 g of amino acid mixtures (without tryptophan and leucine or without trytophan, leucine, and histidine) per liter of medium (pH 5.8).
β-Gal activation assays
Colorimetric assays were done as described in Matchmaker (Clontech) protocol using filter lift and liquid assays. Yeast transformations were grown on selective medium, and then transferred to Whatman 1 filters. The filter papers were submerged in liquid nitrogen, thawed at room temperature, and placed on top of filters that were presoaked in Z buffer solution (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) with 3.6 mM β-mercaptoethanol (Sigma) and 0.33 mg/mL 5-bromo-4-chloro-3-in-dolyl-β-D-galactopyranoside (X-gal; Sigma). X-gal was dissolved in N,N-dimethylformamide (DMF) and added to Z buffer. Assays were incubated at 30°C until color change (blue) appears. For the liquid assays, transformed yeast were grown in selective medium overnight. Cultures were diluted 1:4 in YPD medium and grown to an OD600 = 0.5. A total of 1.5 mL of cells were collected by centrifugation, washed with water, and resuspended in 0.3 mL of Z buffer. Cells were split into 0.1-mL fractions, frozen in liquid nitrogen, and thawed at 37°C for 1 min. After 0.16 mL of ONPG solution [4 mg/mL o-nitrophenyl β-D-galactopyranoside (Sigma) in Z buffer] was added, reactions were incubated at 30°C until the reactions turned yellow, then 0.4 mL 1M Na2CO3 was added to stop the reactions. The cell debris was pelleted and the supernatant was read at an OD420 to calculate Miller units.
Transcription and RNA purification
The luciferase RNAs used in the in vitro translation experiments were generated by in vitro transcription of p5′PVLuc plasmid linearized with XbaI using the T7-based Megascript in vitro transcription kit (Ambion). Following transcription, the reactions were treated with DNase I, phenol/chloroform extracted, and ethanol precipitated in the presence of 700 mM ammonium acetate. Transcribed RNA used as a probe in the electrophoretic mobility shift assays was generated in a similar manner. The poliovirus stem–loop IV plasmid was linearized with HindIII and transcribed in the presence of 1 mM ATP, 1 mM CTP, 1 mM GTP, 50 μM unlabeled UTP, and 50 μCi of (α-32P)UTP (specific activity: 3000 Ci/mmole; Amersham Pharmacia Biotech). This RNA was purified through a gel-filtration spin column (Chromaspin DEPC-30 column), phenol/chloroform extracted, and ethanol precipitated in the presence of 700 mM ammonium acetate.
Purification of recombinant PCBP
Purification of histidine-tagged PCBP2 proteins was performed as described by Parsley et al. (1997) with the following modifications. PCBP2 present in the QE30 expression vector was induced with IPTG in Escherichia coli JM109 cells at 30°C overnight. JM109 cells expressing PCBP2 were incubated in lysozyme buffer [25 mM Tris-HCl (pH 8.0), 2.0 × 105 U/mL chicken egg white lysozyme (Sigma)] for 30 min on ice. Prior to use in functional assays, the recombinant proteins were dialyzed overnight in initiation factor buffer [5 mM Tris-HCl (pH 7.4), 100 mM KCl, 0.05 mM EDTA, 1 mM DTT, 5% glycerol (Brown and Ehrenfeld 1979)]. Concentration and purity of protein preps were determined by Bradford protein assay (Bio-Rad) and SDS-PAGE, respectively. Wild-type and truncated proteins used in these experiments were at least 90% pure. GST fusion proteins were purified from the pGex 4T vectors (Pharmacia Biotech). GST proteins were induced with IPTG in E. coli JM109 cells and lysed as described above for the histidine-tagged proteins. Cell lysates (100 mL) were incubated with glutathione Sepharose in 1× PBS for 30 min at 4°C, added to a 5-mL column, and washed with 10 mL of 1× PBS at room temperature. Bound proteins were eluted at room temperature by adding 1 mL of elution buffer [10 mM reduced glutathione, 50 mM Tris (pH 8.0)] and incubating for 10 min. Purified proteins were dialyzed overnight in initiation factor buffer and examined by Bradford assay and SDS-PAGE.
GST pull-down assay
GST pull-down assays were carried out with recombinant GST-PCBP2 and histidine-tagged proteins. Glutathione Sepharose was washed and resuspended in TENN buffer [50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 0.5% NP-40 and 50 mM NaCl]. Glutathione Sepharose was preincubated with recombinant histidine-tagged proteins for 15 min at 4°C to eliminate nonspecific interactions. The histidine-tagged proteins (10 μg) were then incubated with the GST fusion protein (2 μg) at 4°C for 1 h. A total of 200 μL of glutathione Sepharose was added to the reactions and incubated for 1 h at 4°C. The Sepharose was collected by centrifugation 1000g and washed three times with TENN buffer. Sepharose was collected and bound proteins were eluted by adding 30 μL of 1× Laemmli sample buffer and boiling for 3 min. Proteins that bound to the GST-tagged protein were subjected to electrophoretic separation on an SDS-polyacrylamide gel and examined by silver staining.
RNA electrophoretic mobility shift assays
Electrophoretic mobility shift assays were performed as described by Dildine and Semler (1992). Purified PCBP2 at the indicated concentration was preincubated for 10 min at 30°C in RNA binding buffer [5 mM HEPES-KOH (pH 7.8), 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 3.8% glycerol, 2 mM DTT, 1 mg/mL E. coli tRNA (Sigma), 0.5 mg/Ll BSA] in a total volume of 10 μL. 32P-labeled RNA probe was added to a final concentration of 10 nM and incubated at 30°C for 10 min. A total of 2.5 μL of 50% glycerol was added to each reaction and the complexes were resolved on a 5% native polyacrylamide (40:1 acrylamide-bisacrylamide)-5% glycerol-0.5× TBE (45 mM Tris-borate, 1 mM EDTA) gel and visualized by autoradiography.
HeLa cell cytoplasmic extract preparation
Cytoplasmic extracts were prepared as described in Barton and Flanegan (1993) as modified by the original protocol from Brown and Ehrenfeld (1979). The slight differences are as follows: HeLa cell pellets were resuspended in an equal volume of hypotonic buffer, the cells were lysed with 12–15 strokes of the glass dounce homogenizer to obtain around 95% lysis as determined by trypan blue exclusion, and the extract was treated with 120 U micrococcal nuclease (Worthington Biochemical) per milliliter of S10 supernatant at 13°C for 15 min. The nuclease was inactivated by adding EGTA to a final concentration of 4 mM. The HeLa S10 extracts are stored under liquid nitrogen.
In vitro translation
Translation experiments were carried out as described by Walter et al. (1999). Briefly, the reactions (20 μL) contained 68% HeLa S10, 50 fmole PV luciferase reporter RNA, 16 mM HEPES-KOH (pH 7.4), 60 mM K(CH3CO2), 1 mM ATP, 250 μM GTP, 250 μM CTP, 250 μM UTP, 30 mM creatine phosphate, 400 μg of creatine-kinase (Boehringer Mannheim) per milliliter, and 15% initiation factor buffer that contained wild-type PCBP2, mutated PCBP2 proteins, or no protein. Reactions were incubated at 30°C for 75 min, and then translations were terminated and placed on ice.
Luciferase assay
Luciferase assays were performed in three or four independent experiments. Samples (5 μL) from each reaction were added to 100 μL of reconstituted Luciferase Assay Substrate (Promega), followed by measuring the relative light units for 10 sec to quantitate luciferase activity using a Berthold luminometer.
Acknowledgments
We thank Dr. Rushika Perera for her assistance with the gel-filtration experiments and critical review of the manuscript, Sarah Daijogo for her help in cloning ΔKH2-PCBP2, and Santos Rojas for work on constructing the GST-PCBP2 clone. We are also thank Gwendolyn Jang for her helpful suggestions on the manuscript. K.M.B. was a predoctoral trainee of Public Health Service training grant AI 07319. B.L.W. was supported by a research fellowship from the American Heart Association, Western States Affiliate. This work was supported by Public Health Service grant AI 26765 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7070304.
REFERENCES
- Andino, R., Rieckhof, G.E., and Baltimore, D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63: 369–380. [DOI] [PubMed] [Google Scholar]
- Andino, R., Rieckhof, G.E., Achacoso, P.L., and Baltimore, D. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12: 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D.J. and Flanegan, J.B. 1993. Coupled translation and replication of poliovirus RNA in vitro: Synthesis of functional 3D polymerase and infectious virus. J. Virol. 67: 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn, L.B., Swiderek, K.M., Richards, O., Stahl, D.C., Semler, B.L., and Ehrenfeld, E. 1996. Poly(rC) binding protein 2 binds to stem–loop IV of the poliovirus RNA 5′ noncoding region: Identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. 93: 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn, L.B., Towner, J.S., Semler, B.L., and Ehrenfeld, E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71: 6243–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, B.A. and Ehrenfeld, E. 1979. Translation of poliovirus RNA in vitro: Changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97: 396–405. [DOI] [PubMed] [Google Scholar]
- Dildine, S.L. and Semler, B.L. 1992. Conservation of RNA-protein interactions among picornaviruses. J. Virol. 66: 4364–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke, B., Zuleger, B., Benavente, R., Schuster, T., Goller, M., Stevenin, J., and Horak, I. 1996. The mouse poly(C)-binding protein exists in multiple isoforms and interacts with several RNA-binding proteins. Nucleic Acids Res. 24: 3821–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik, A.V. and Andino, R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3: 882–892. [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74: 2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Git, A. and Standart, N. 2002. The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA 8: 1319–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Holcik, M. and Liebhaber, S.A. 1997. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. 94: 2410–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. 2002. Proteins involved in the function of Picornavirus internal ribosome entry sites. In Molecular biology of picornaviruses (eds. B.L. Semler and E. Wimmer), pp. 171–183. ASM Press, Washington, DC.
- Jang, S.K., Krausslich, H.G., Nicklin, M.J., Duke, G.M., Palmenberg, A.C., and Wimmer, E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian, M., Wang, X., and Liebhaber, S.A. 1995. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 14: 4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., Hahm, B., Kim, Y.K., Choi, M., and Jang, S.K. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298: 395–405. [DOI] [PubMed] [Google Scholar]
- Lindquist, J.N., Kauschke, S.G., Stefanovic, B., Burchardt, E.R., and Brenner, D.A. 2000. Characterization of the interaction between alphaCP(2) and the 3′-untranslated region of collagen alpha1(I) mRNA. Nucleic Acids Res. 28: 4306–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev, A.V. and Liebhaber, S.A. 2000. Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics 67: 301–316. [DOI] [PubMed] [Google Scholar]
- ———. 2002. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 8: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musco, G., Stier, G., Joseph, C., Castiglione Morelli, M.A., Nilges, M., Gibson, T.J., and Pastore, A. 1996. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 85: 237–245. [DOI] [PubMed] [Google Scholar]
- Musco, G., Kharrat, A., Stier, G., Fraternali, F., Gibson, T.J., Nilges, M., and Pastore, A. 1997. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 4: 712–716. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., Standart, N., and Thiele, B.J. 1994. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 13: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsley, T.B., Towner, J.S., Blyn, L.B., Ehrenfeld, E., and Semler, B.L. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3: 1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J., Kaplan, G., Racaniello, V.R., and Sonenberg, N. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell Biol. 8: 1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyreva, M., Schullery, D.S., Suzuki, H., Higaki, Y., and Bomsztyk, K. 2000. Interaction of two multifunctional proteins, heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein 1. J. Biol. Chem. 275: 15498–15503. [DOI] [PubMed] [Google Scholar]
- Silvera, D., Gamarnik, A.V., and Andino, R. 1999. The N-terminal K homology domain of the Poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 274: 38163–38170. [DOI] [PubMed] [Google Scholar]
- Siomi, H., Matunis, M.J., Michael, W.M., and Dreyfuss, G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic. Acids. Res. 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic, B., Hellerbrand, C., Holcik, M., Briendl, M., Liebhaber, S., and Brenner, D.A. 1997. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol. Cell Biol. 17: 5201–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommerup, N. and Leffers, H. 1996. Assignment of the human genes encoding 14,3-3 Eta (YWHAH) to 22q12, 14-3-3 zeta (YWHAZ) to 2p25.1–p25.2, and 14-3-3 β (YWHAB) to 20q13.1 by in situ hybridization. Genomics 33: 149–150. [DOI] [PubMed] [Google Scholar]
- Walter, B.L., Nguyen, J.H., Ehrenfeld, E., and Semler, B.L. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5: 1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, B.L., Parsley, T.B., Ehrenfeld, E., and Semler, B.L. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76: 12008–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, I.M. and Liebhaber, S.A. 1994. Erythroid cell-specific determinants of α-globin mRNA stability. Mol. Cell Biol. 14: 8123–8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1995. Erythroid cell-specific mRNA stability elements in the α 2-globin 3′ nontranslated region. Mol. Cell Biol. 15: 2457–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]