Abstract
Discrete RNA secondary and higher-order structures, typically local in extent, play a fundamental role in RNA virus replication. Using new bioinformatics analysis methods, we have identified genome-scale ordered RNA structure (GORS) in many genera and families of positive-strand animal and plant RNA viruses. There was remarkably variability between genera that possess this characteristic; for example, hepaciviruses in the family Flaviviridae show evidence for extensive internal base-pairing throughout their coding sequences that was absent in both the related pestivirus and flavivirus genera. Similar genus-associated variability was observed in the Picornaviridae, the Caliciviridae, and many plant virus families. The similarity in replication strategies between genera in each of these families rules out a role for GORS in a fundamentally conserved aspect of this aspect of the virus life cycle. However, in the Picornaviridae, Flaviviridae, and Caliciviridae, the existence of GORS correlated strongly with the ability of each genus to persist in their natural hosts. This raises the intriguing possibility of a role for GORS in the modulation of innate intracellular defense mechanisms (and secondarily, the acquired immune system) triggered by double-stranded RNA, analogous in function to the expression of structured RNA transcripts by large DNA viruses. Irrespective of function, the observed evolutionary conservation of GORS in many viruses imposes a considerable constraint on genome plasticity and the consequent narrowing of sequence space in which neutral drift can occur. These findings potentially reconcile the rapid evolution of RNA viruses over short periods with the documented examples of extreme conservatism evident from their intimate coevolution with their hosts.
Keywords: virus, persistence, evolution, innate immune response, molecular clock, RNA structure
INTRODUCTION
The genomes of many important animal and plant virus pathogens, such as hepatitis C virus (HCV), foot and mouth disease virus (FMDV), and potyviruses consist of single-stranded positive-sense ribonucleic acid. The inherent error-prone replication of these and other RNA viruses provides a distinct evolutionary advantage, allowing them to escape from innate defense mechanisms and acquired immune surveillance of the host, and to rapidly adapt to new cell types, tissues, or species (Moya et al. 2000; Baranowski et al. 2001; Weiss 2002).
Many events in virus replication involve structured RNA elements. For example, several viruses have evolved internal ribosome entry sites (IRESs), consisting of extensive regions of structured RNA, to recruit ribosomes for translation (Pelletier and Sonenberg 1988; Tsukiyama Kohara et al. 1992; Belsham and Sonenberg 1996). Subsequent genome replication may involve interaction of the viral polymerase with structured RNA elements, often referred to as cis-acting replication elements (CREs), for transcription initiation (Xiang et al. 1997; Goodfellow et al. 2000; Joost Haasnoot et al. 2002; Mason et al. 2002). In the latter stages of the replication cycle, RNA structures form packaging signals used during encapsidation of the genome into progeny virus particles (Schlesinger et al. 1994; Huthoff and Berkhout 2002).
The application of increasingly advanced computational techniques has resulted in the prediction of similar structured RNA elements in these and other viruses for which no function has yet been defined (Simmonds and Smith 1999; Witwer et al. 2001; Tuplin et al. 2002). Reverse genetic approaches to mapping structure and function of such elements can therefore provide important insights into the life cycle of RNA viruses (Evans 1999). Similar structural prediction analyses suggest that many RNA viruses possess more extensive RNA structure. For example, Palmenberg has suggested that picornavirus genomes exhibit extensive folding resulting in the juxtaposition of the 5′- and 3′-termini (Palmenberg and Sgro 1997). Our own studies of HCV and the distantly related hepatitis G virus or GB virus C (HGV/GBV-C) have provided evidence for a wide range of evolutionarily conserved defined RNA structures of unknown function (Simmonds and Smith 1999; Tuplin et al. 2002).
In this study we have used large-scale thermodynamic prediction methods to investigate the occurrence of more extensive RNA structure in the genomes of the Flaviviridae and Picornaviridae. We discovered significant differences in the extent of structure between closely related genera in these families, and thus extended this analysis to include a wide range of animal and plant RNA viruses to determine what compositional or biological factors underlie these differences in RNA structure. Finally, we investigated the sensitivity of the predicted RNA structures to introduced neutral nucleotide substitutions, and thus the effect of sequence drift on the maintenance of large-scale internal RNA folding. Our results have fundamental implications for the understanding of virus replication and evolution.
RESULTS
Large-scale thermodynamic prediction of minimal free energy (MFE) on folding
MFOLD thermodynamic analysis was used to estimate MFEs of virus genomes and control sequences (Zuker 1989,Zuker 2003). This method predicts the free energy of the most stable RNA secondary structure for a given sequence, the significance of the predicted folding being ascertained by comparison with the null expectation, that is, the MFE of sequence-order randomized controls. MFEs of 372 complete genome sequences of viruses from four genera of Flaviviridae and Picornaviridae (flaviviruses, hepaciviruses [HCV and related viruses], HGV/GBV-C-like viruses, and pestiviruses; enteroviruses, aphthoviruses, teschoviruses, and hepatoviruses, respectively; listed in Materials and Methods) were compared with corresponding MFEs of 50 separate sequence order randomizations of each sequence segment using an algorithm that preserved dinucleotide frequencies (NDR; see below). MFE results were expressed either as MFE differences (MFEDs), that is, the percentage difference between the MFE of the native sequence from that of the mean value of the 50 sequence order randomized controls, or as a Z score, which is the position of the MFE of the native sequence within the distribution of MFEs of the randomized sequences, expressed as the number of standard deviations from the mean value; thus, values between −2 and +2 fall within the range of 95% of their MFE values (Workman and Krogh 1999). For analysis of RNA folding in different virus genera, Z scores of each 498-base fragment from each available complete genome sequence of viruses within the genus were computed, and combined to produce a composite distribution of Z scores. These were similarly calculated for control sequences: 57 human mRNA sequences, and complete genomes of bacteria (Escherichia coli, Archaeoglobus fulgidus) and large DNA viruses (eight herpesviruses and 13 poxvirus).
The mean Z score of the set of mammalian mRNA sequence fragments approximated to zero (0.027; MFED −0.2%; Fig. 1). The distribution of Z-score values corresponded closely to the null expectation, where the 95% percentile range of Z scores of −2.21–+1.8 was close to the expected values −2.0–+2.0 of unstructured sequences. The minor discrepancy was the result of a slightly skewed leftward distribution (S) of some Z scores toward lower values (S = −0.51), and a kurtosis (K) value of 0.9 (flattening of distribution). For the other control sequences (E. coli, A. fulgidus, large DNA viruses), mean MFEDs ranged from 1.1% to 2.7% (Z scores: −0.32–−0.72). In each case, distributions again were slightly skewed leftward (S values of −0.79, −0.40, −1.32, respectively) and showed flattened distributions (K = 2.0, 0.54, 8.1). The nonnormal distributions of MFEDs and Z scores from the large DNA viruses were caused by the presence of highly structured outlier sequences, which on inspection were found to originate from a self-complementary noncoding repeat region of HHV-8, and similar repetitive sequences in other herpesviruses (data not shown). It seems likely that structured nucleotide elements involved in transcriptional or translational control and genes for structured RNAs (see Discussion) contributed to the other slight deviations from normality in the bacterial and DNA virus sequence data sets.
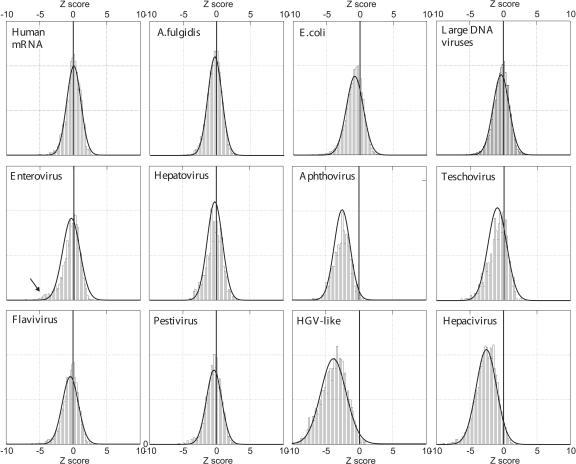
FIGURE 1.
Distribution of Z scores for RNA viruses and controls. Distribution of Z scores of the set of 498-base fragments of control sequences (row 1), and different genera within the Picornaviridae and Flaviviridae (rows 2,3). MFEDs were calculated using NDR for sequence randomization. Each observed distribution (bars) was overlaid with a symmetrical best fit normal distribution (solid black line); the Z-score = 0 value was highlighted by vertical bar. The arrow in the enterovirus panel indicates fragments containing highly structured RNA elements of known function (see text).
The distribution of MFEDs in the control sequences was markedly different from those of certain genera of the two families of positive-stranded viruses analyzed (Fig. 1). Remarkably, the entire distributions of Z scores (and MFEDs) from aphthoviruses, teschoviruses, hepacivirus, and HGV/GBV-C-like viruses were shifted away from zero (mean Z scores: −2.58 [−4.98–−0.40], −0.90 [−3.98–+1.42], −2.5 [−5.77–+0.15], and −3.80 [−7.70–−0.52], respectively). Examination of MFEDs for aligned genome fragments showed that predicted RNA structure was distributed throughout most of the genomes of these virus groups (Fig. 2). It was notable that MFEDs of regions with known, functional RNA structures, such as the 5′- and 3′-UTRs, were generally little different from or frequently less than those of coding regions of these viruses. For example, the highly structured aphthovirus 5′-UTR and IRES had MFEDs of around 5%, lower than the 9.7% mean of the entire genome.
FIGURE 2.
Distribution of MFEDs across the genomes of the Picornaviridae and Flaviviridae. Mean MFEDs for individual 498-base sequence fragments across the genomes of Picornaviridae (top row) and Flaviviridae (bottom row) genera that possess (Aphthovirus, Teschovirus, HGV/GBV-C, HCV) or lack (Hepatovirus, Enterovirus, Flavivirus, Pestivirus) GORS are shown. The mean value and variability between members of each genus (standard deviation) are shown as bars and lines, respectively.
In stark contrast, distributions of MFEDs in the flavivirus, pestivirus, hepatovirus, and enterovirus genera centered around zero (mean Z scores: 0.39 [−3.09–+1.55] and −0.45 [−3.25–+1.53]). Each of these distributions showed a slight leftward skew (S values ranging from −0.35 to −1.00). This was particularly marked in the enteroviruses, whose sets of MFED Z-score values included a second distribution of highly structured fragments corresponding to sequences from the 5′-UTR, 3′-UTR, and the CRE (arrowed in Fig. 1).
In this study, we use the term Genome-scale Ordered RNA structure (GORS) to indicate the existence of widely distributed predicted RNA folding throughout the genomes of virus groups, such as the hepaciviruses and aphthoviruses (Figs. 1, 2). Other genera within the Flaviviridae and Picornaviridae, and the large DNA viruses, and bacterial and mammalian coding sequences analyzed were considered to lack GORS because their distributions of Z scores centered around zero.
Sequence randomization strategies
The use of thermodynamic methods, such as MFOLD, to predict RNA folding requires that the folding free energy (MFE) of test sequences are compared with sequence order randomized controls (Zuker 2003). These controls, however, are not valid if they destroy certain nonrandom features of the native sequence. For example, disruption of naturally occurring biases in dinucleotide frequencies and positional differences in base composition in sequences of many eukaryotic organisms have been common sources of erroneous conclusions in previous studies (discussed in Workman and Krogh 1999; Rivas and Eddy 2000). This is illustrated for sequences analyzed in the present study. Although sequence randomization of the human mRNA sequences using NDR produced a mean MFED of approximately zero (−0.02%), a mean MFED value of 5.0% was obtained when MFEDs were based on sequences scrambled using a method (NOR; Tuplin et al. 2002) that failed to preserve dinucleotide frequencies (data not shown). To further investigate this and other potential artifacts associated with different sequence order randomization strategies, we used a range of alternative sequence scrambling methods that preserved other nonrandom characteristics of the sequences (Fig. 3). One method we developed limits nucleotide exchange only to adjacent matched dinucleotide triplets (NDS) and thus retains any regional differences in base composition, as well as preserving dinucleotide composition. The use of NDS to scramble sequences of eight virus genera and control sequences produced MFEDs similar to those obtained using NDR (Fig. 3). We also developed a new method (CDLR) for protein-coding sequences, in which codon composition and codon order were preserved, and which also retains dinucleotide frequencies, equivalent in effect to DicodonShuffle (Katz and Burge 2003). CDLR, therefore, combines the attributes of two previously described methods, CLR and CDR, that could only maintain codon order and dinucleotide frequencies, respectively (Tuplin et al. 2002). Despite the fact that CDLR is based on a quite different strategy from NDR, it produced equivalent results when used for sequence randomization of the coding sequences of the eight virus genera in the Picornaviridae and Flaviviridae (Fig. 3).
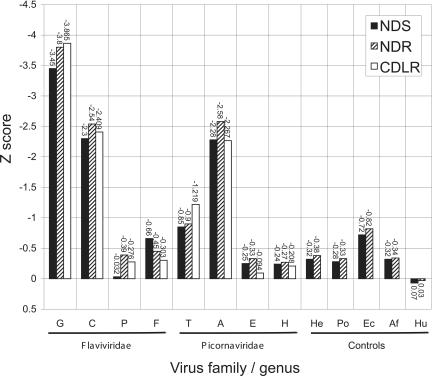
FIGURE 3.
Comparison of sequence randomization methods used for calculation of MFEDs. Mean Z scores for individual 498-base sequence fragments of RNA viruses and control sequences using different sequence randomization methods. These preserve different nonrandom sequence ordering features, such as dinucleotide frequencies (NDR, NDS) or both the dinucleotide frequency and codon order (CDLR) in the open reading frame. Abbreviations for virus and control groups on the X-axis (from left) are Flaviviridae (G) HGV-like, (C) hepaci-viruses, (P) pestiviruses, (F) flaviviruses; Picornaviridae (T) teschoviruses, (A) aphthoviruses, (E) enteroviruses, (H) hepatoviruses; Controls (He) herpesviruses, (Po) poxviruses, (Ec) Escherichia coli, (Af) Archaeoglobus fulgidus, (Hu) human mRNA.
Among other possible artifacts associated with MFED prediction, it is possible that the frequencies of higher-order combinations of bases were biased and influenced folding. To investigate this, we developed a new scrambling method that preserved tri- and tetranucleotide frequencies (NRR, NTR) through randomization or neighbor exchange of the central base, N, in the following 5- or 7-base contexts: abNxy or abcNxyz, where a–c and x–z represent matched bases surrounding the nucleotide to be exchanged (analogous to the exchange of N between matched aNx triplets for dinucleotide randomization). In sequences of 6000 nt (the maximum analyzable by MFOLD), the limited number of abcNxyz sites restricted possible divergence to 13% using the NTR method. Mean MFEDs of 6000-base, overlapping sequences spanning the HGV/GBV-C genome (genotypes 1–4) shuffled by NTR were 7.9% for the 5′-fragment and 10.9% for the 3′-fragment. These MFEDs were comparable to those obtained for the two genomic fragments using NDR (8.5%, 10.0%) or NRR (8.5%, 10.9%), where the extent of shuffling was limited to reproduce the degree of divergence achieved by NTR (13%). Therefore, MFEDs observed in the structured genera were not artifacts of triplet or higher-order sequence constraints that influenced RNA folding.
RNA structure in other viruses
To determine whether genus-specific differences in GORS were also found in other virus families, we carried out a large-scale survey of MFEDs of 512 complete genome sequences from a range of animal and plant viruses (Fig. 4). Virus groups included each of the positive-stranded RNA virus families (including retroviruses and the Nidovirales), and negative-stranded or ambi-sense RNA viruses infecting vertebrates (listed in Materials and Methods). Genomic RNA structure was also investigated in the main groups of positive-stranded RNA viruses infecting plants, divided into groups depending on the virion structure.
FIGURE 4.
Extended survey of MFEDs in animal and plant RNA viruses. MFEDs of sequence fragments of positive-stranded RNA viruses infecting animals and plants (top two panels), and negative-stranded RNA viruses (lower panel), divided into orders (or virion structure for plant viruses), families, and where applicable, genera. MFEDs for sequences in their native genomic configuration are shown as filled boxes; MFEDs for reverse complementary sequences are shaded (for clarity, smaller values are stacked in front).
Adding to our previous results, considerable variability was observed between the other genera in the Picornaviridae, and within the calicivirus family. Other positive-stranded viruses also showed evidence for GORS, particularly the Astroviridae, and members of the order Nidovirales. However, mean MFEDs were close to zero for all families within the Mononegavirales, and in the segmented bunya-viruses. Evidence for RNA structure in many families of plant viruses was also obtained, particularly those groups with icosahedral or rod-like morphology. For almost all virus families or genera showing evidence for GORS, MFEDs of the positive strand (corresponding to genomic sequences for the majority of viruses analyzed) were greater than those predicted for the sequence in reverse complementary orientation (corresponding to the replication intermediate; Fig. 4). For example, mean MFEDs of genomic RNA sequences for hepaci- and HGV-like genera in the Flaviviridae were two to three times greater than the reverse complementary sequences (8.5% and 12.9% compared with 4.6% and 4.0%, respectively). Similar discrepancies in MFEDs between sense (coding) and antisense strands were found in structured viruses within the Picornaviridae (particularly koboviruses and aphthoviruses), vesiviruses, and several genera and/or families of icosahedral plant viruses.
Correlates of GORS in positive-strand RNA viruses
The thermodynamic analysis carried out on this large number of viruses provided an extensive data set with which to investigate correlations between MFED and sequence composition parameters that varied between viruses (for comparison of mean values of complete genome sequences, >650 paired values for each parameter were available for analysis). The variables considered included MFE and MFE differences between sense and antisense sequences, frequency of each base, G+C composition, purine and pyrimidine composition, dinucleotide frequencies, and composition mismatches between complementary bases.
Consistent with previous observations (Rima and McFerran 1997), all of the animal and plant RNA virus groups analyzed showed variable degrees of underrepresentation of the CG and UA dinucleotides and marked overrepresentation of CA and UG (all generically pyrimidine/purine; Y/R), comparable to those observed in the set of human mRNA sequences (R = 0.95; p = 5 × 10−7). The degree of each YR bias was generally specific to individual virus genera or families, and showed no correlation with MFEDs, either when compared by fragment, by complete virus genome, by family, or by any other higher-order grouping of viruses (data not shown). The use of a scrambling method that preserves dinucleotide frequencies (see Materials and Methods) in the calculation of MFEDs further guaranteed that this variable did not influence our analysis of GORS differences between viruses.
As expected, MFEs showed an inverse correlation with G+C content (R = −0.82 [animal viruses] and −0.59 [plants]; p < 10−8), with outliers being those with marked MFEDs (such as some members of the Flaviviridae and Picornaviridae that have greater MFEs than expected from their G+C contents), and those with gross differences in frequencies of complementary bases, leading to lower MFEs than expected from G+C content (such as tymoviruses and marifiviruses, many of which showed genomic compositions of 41%–42% C and <20% G residues). No other associations between MFEDs or other composition or thermodynamic folding variables were detected on analysis of the RNA virus data set taken as a whole, specifically within animal or plant viruses, or within individual orders (data not shown).
RNA structure and virus persistence
Because GORS was not universally present in all genera of, for example, the Picornaviridae and Flaviviridae, we considered that it was unlikely to function in fundamentally conserved aspects of the virus replication strategy. For example, all picornaviruses exhibit IRES-mediated translation and share a common replication strategy involving known structured elements in the 5′ and 3′ noncoding regions and a variably located CRE (Rohll et al. 1995; Xiang et al. 1997; Goodfellow et al. 2000). RNA structures involved in these translation and replication functions, therefore, cannot account for the observed differences in GORS between the aphthoviruses and enteroviruses. Instead, we considered that GORS may be associated with virus phenotypes that differ between genera. Preliminary analysis revealed certain general correlations; GORS was absent from many mammalian or plant virus families with helical nucleocapsids, suggesting that the association with nucleocapsid proteins limits or restricts the need for this type of RNA structure. The apparent exceptions, the coronaviruses, criniviruses, and tobacco mosaic virus, which possess both helical nucleocapsids and GORS (Fig. 4), are notable in that they all expose single-stranded RNA during replication. This suggests that a helical nucleocapsid per se is not incompatible with the extensive RNA structure we propose, but that genomic exposure to the cellular environment may be a determining feature.
To investigate if there was a correlation between MFEDs and the ability of different virus genera to persist in their natural hosts, we classified different genera from the flavi-virus, picornavirus, togavirus, HEV-like, and calicivirus families into those capable of establishing persistence and those in which infection was acute and self-limiting in an immunocompetent host (Fig. 5). We accept that a simple, two-way division of viruses into persistent and nonpersistent is overly simplistic, and that outcomes of infection can vary depending on the maturity and functional capacity of the immune system of the affected host. However, following the criteria described in Materials and Methods, there was a clear association between MFEDs and their classification as persistent or nonpersistent. Without exception, mean MFEDs (NDR) of genera of nonpersistent viruses were lower than those of persistent viruses (Fig. 5); mean MFED values of 2.2% (range 1.0%–3.9%) for the 10 nonpersistent genera were significantly lower than those of the five persistent viruses (8.5% [4.9%–12.9%]; p = 0.022 [Mann-Whitney U test]). Weaker, independent segregation between persistent and nonpersistent viruses was also apparent on ranking genera by mean CG dinucleotide under-representation (Fig. 5; mean values for nonpersistent virus genera 0.51 [range 0.26–0.82] compared with 0.69 [0.56–0.84] for persistent viruses; p = 0.066). In other comparisons, no other composition variables were significantly associated with infection outcome, such as MFE (p = 0.14), G+C content (p = 0.086), and other dinucleotide frequency biases, such as those of UA (p = 0.46) and UU (p = 0.62).
FIGURE 5.
Relationship between host persistence with MFED and CG underrepresentation. Virus genera were ranked by MFED (left panel) or CG underrepresentation (right panel); viruses were scored as persistent (filled boxes) or nonpersistent (gray). Histograms show mean values for the genus; the error bar corresponds to one standard deviation from the mean within the virus genus/family. First letter (virus family) abbreviations are (P) Picornaviridae; (second-letter, genus abbreviations are [A] aphthovirus; [C] cardiovirus; [P] parechovirus; [H] hepatovirus; [E] enterovirus); (F) Flaviviridae ([P] pestivirus; [F] flavivirus; [C] hepacivirus; [G] HGV-like virus); (C) Caliciviridae ([S] sapovirus; [N] norovirus; [V] vesivirus; [L] lagovirus); (TA) Togaviridae, alphavirus genus; (HE) HEV-like viruses.
The influence of GORS on virus evolution
Irrespective of the biological function of GORS, or whether indeed the primary purpose of the sequence ordering we have observed is to promote RNA folding in the way that is conventionally modeled, the maintenance of GORS must be a significant factor that limits the diversification of structured RNA viruses.
To investigate the extent of these limitations, we artificially mutated coding sequences of each genotype of HCV, HGV/GBV-C, and aphthovirus (n = 6, 4, and 4, respectively), using an algorithm that introduced random changes into the sequence but preserved specific characteristics of naturally occurring virus diversity within each group (including the ratio of synonymous to nonsynonymous substitutions, transition/transversion ratios, and base composition at first, second, and third codon positions; Kimura 1983). Sets of 10 independently mutated sequences were generated, each differing from the original by a range of sequence distances. For example, HCV sequence sets differing from the starting sequence by 2%, 5%, 8.8%, 12%, 15%, 25%, and 33% were generated. The sequence distances of 8.8%, 25%, and 33% corresponded to the mean differences within genotype 1b, between subtypes, and between genotypes, respectively.
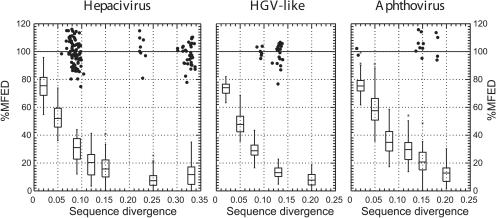
Despite the close match of the introduced sequence changes to expected neutral evolutionary drift, the introduced sequence changes dramatically reduced MFEDs in each virus group (Fig. 6). This reduction was apparent even in sequences differing by only 2% from the original sequence (mean MFEDs were 76%, 74%, and 75% of those of the original native sequences of HCV, HGV/GBV-C, and FMDV, respectively), whereas much greater reductions were observed for each virus at 5% (53%, 48%, and 57% of original values). At divergences of >10%, MFEDs approached those of unstructured RNA viruses, DNA viruses, or bacterial genomes (Fig. 6). The initial gradient of the reduction in MFEDs with divergence provides an indirect indication of the contribution of individual or specific bases to RNA structure; the very rapid decline even at the 2% divergence level suggests a large proportion of individual nucleotides are involved in RNA structure formation.
FIGURE 6.
Effect of simulated neutral sequence drift on MFEDs of viruses with predicted RNA structure. Coding regions of complete genome sequences of HCV genotype 1b (GenBank accession number HPCJ491), HGV/GBV-B (genotype 2; HGU94695), and FMDV (PIFMDV2) were each mutated independently 10 times to produce sequences showing a range of sequence divergence from the original (X-axis). MFEDs, using the NDR algorithm to generate 50 sequence order randomized sequences, were calculated for each and expressed as a percentage of the MFED of the starting sequence (Y-axis). The distributions of MFED values for the mutated sequences are represented as box and whisker plots (showing 95% percentile [line], standard deviation [upper and lower box], and mean values [line within box], with outliers indicated by the symbol *). For comparison, MFEDs for naturally occurring variants in each virus group expressed as a percentage of the MFED of the starting sequences were plotted using the symbol •.
DISCUSSION
Detection of GORS in RNA virus genomes
Defined RNA secondary and higher-order structures have fundamental roles in cell biology, including transcription initiation, elongation, and termination; translation (both in the formation and control of the ribosomal machinery); RNA localization; splicing; transport; stability; and catalytic activity. Where defined, these functions are usually ascribed to structured RNA elements essentially local in extent. RNA viruses generally have small genomes and must control genome replication and protein expression and, as obligate intracellular parasites, need to interact with the cellular bio-synthetic machinery for these processes. To date, the structured RNA elements identified as being involved in these processes are usually discrete, usually involving short-range Watson-Crick base-pairing in an otherwise essentially unstructured genomic environment.
In contrast, we present evidence for the existence of a very distinct biological phenomenon in the genome of some positive-sense, single-stranded RNA viruses. This characteristic, the presence of extensive tracts of RNA structure as defined by the application of whole-genome thermodynamic analysis, we term genome-wide predicted RNA structure (GORS). In viruses that possess GORS, the thermodynamic stability—and hence distribution of structured RNA—is significantly greater than the discrete, highly ordered structures that function during genome replication and translation. For example, prior to this analysis, the most extensive characterized region of secondary RNA structure in the Picornaviridae was a 300–450-nt region of the 5′-UTR responsible for the recruitment of ribosomes for translation. In those genera that lack GORS, but use an IRES (e.g., enteroviruses and pestiviruses), the genome segments that span the IRES are clearly visible in the Z score and MFED distributions (Fig. 1) of RNA folding. In contrast, the known structured IRES elements of GORS-possessing genera (e.g., aphthoviruses and hepaciviruses) are completely obscured in the Z-score analysis, reflecting the significantly greater level of RNA structure throughout the genome. This is clearly evident from comparison of the genomic MFED distribution of enteroviruses with aphthoviruses or pestiviruses with HCV in Figure 2. In the majority of the genera analyzed that possess GORS, the MFED levels were broadly similar throughout the genome. However, the teschoviruses (Fig. 2) display an intriguing switch from low to high MFEDs located near the junction of the capsid-encoding region of the genome, an area associated with recombination events that are common in the Picornaviridae. Further detailed analysis of the distribution of MFEDs in virus genomes may uncover unsuspected relationships that reflect the evolutionary history or aspects of the replication strategy of the virus.
The apparent differences in GORS between virus genera and families is consistent with previous analyses of these groups using other RNA prediction methods. For example, phylogenetic covariance analyses of discrete stem–loop structures in members of different picornavirus genera revealed a greater number of predicted structures in the coding regions of aphthoviruses, cardioviruses, and teschoviruses than in other genera of picornaviruses such as enteroviruses, hepatoviruses, and parechoviruses (Witwer et al. 2001). Similarly, several stem–loops have been predicted in the coding regions of the HCV and HGV/GBV-C genomes by covariance analysis (Simmonds and Smith 1999; Tuplin et al. 2002), but none was apparent in an alignment of pestiviruses (Tuplin et al. 2002), concordant with an absence of GORS in these viruses (Fig. 1).
The existence of GORS is not a consequence of the application of an inappropriate scrambling strategy that distorts any of the known fundamental organizational principles of the nucleic acid sequence. We were careful to use a range of sequence-order scrambling approaches that avoided disturbing di-, tri- and tetranucleotide frequencies, coding sequence order, or—in the CDLR method—retained both dinucleotide frequencies and coding sequence order. When present, GORS was clearly apparent, irrespective of the scrambling strategy used (Fig. 3). To facilitate comparison of coding and noncoding regions, or comparative analysis of complete genomes with varying amounts of coding capacity, we standardized on a randomization strategy (NDR) that was applicable to all sequences. The absence, in an extensive data set of RNA viruses (Fig. 4), of any correlation between the presence of GORS and a wide range of composition variables including G+C content, mismatches in base frequencies, and biases in dinucleotide constitution is further evidence that the significant differences we observe in MFEDs were not artifacts of the scrambling methods used to generate the sequences (Workman and Krogh 1999; Rivas and Eddy 2000; Tuplin et al. 2002).
Possible function of GORS in virus persistence
Eukaryotic cells possess a formidable array of innate defense mechanisms, operating at a more fundamental level than the acquired immune system of vertebrates. Most antiviral response pathways in animals and plants recognize viruses by the presence of dsRNA sequences in the cytoplasm, frequently through interaction with a family of structurally related dsRNA-binding proteins (DRBPs; Levy and Garcia-Sastre 2001; Girardin et al. 2002; Saunders and Barber 2003). Although the mechanism of substrate recognition is shared (typically to A-form double-helix RNA of minimum length of 11–16 bp), DRBPs are coupled to a wide range of antiviral, effector pathways. In vertebrates these include the protein kinase dsRNA-dependent (PKR)-mediated induction of apoptosis and modulation of the interferon response pathways, and activation of oligoadenylate synthetase resulting in RNAse L production and consequent cytoplasmic RNA cleavage (Player and Torrence 1998). Perhaps more fundamental, being shared by plant and animal cells, is the production of small interfering RNA (siRNA) by Dicer-mediated cleavage of dsRNA and the resulting targeting and destruction of complementary RNA sequences by an siRNA-armed RNA-induced silencing complex (Bernstein et al. 2001; Martinez et al. 2002). The efficacy and importance of these innate defense strategies is emphasized by the range, ubiquity, and sophistication of the mechanisms viruses have evolved to evade or subvert their action, reflecting the intensity of past evolutionary battles between viruses and the organisms they infect (Levy and Garcia-Sastre 2001; Katze et al. 2002). Evasion mechanisms may prevent recognition of dsRNA (Imani and Jacobs 1988) or inhibit steps in response or effector pathways. Typically, even the small RNA viruses described in this study may contain a battery of evasion measures, variously inhibiting the activation of PKR, inhibition of IFN response pathways (IRF-3, Grb-2, and JAK-STAT; Goodbourn et al. 2000; Tan and Katze 2001; Foy et al. 2003).
The ability of certain viruses to persist in the host demonstrates the capacity to defeat or circumvent both innate defenses and immune system responses. The association of GORS with virus persistence suggests that the formation of extensive RNA secondary structure plays a role in the evasion of cell defenses, potentially by facilitating escape from innate responses induced by certain structured RNAs. It is not clear why cellular structured RNAs such as ribosomal RNA and tRNA do not trigger dsRNA-induced cellular responses, and it is possible that GORS has evolved as a mechanism to conformationally mimic cellular structured RNAs, for example, by the formation of stem–loop configurations that are not recognized by DRBPs. An alternative scenario is that GORS is functionally analogous to the abundant, short, structured viral RNA transcripts that competitively inhibit induction of the PKR-mediated antiviral state (for review, see Goodbourn et al. 2000). The adeno-virus VAI, Epstein-Barr virus EBER, and HIV-TAR transcripts (Gunnery et al. 1990; Sharp et al. 1993; Mathews 1995) are thought to bind avidly to PKR, so preventing dimerization and consequent translational suppression and stimulation of cytokine production and apoptosis (Clemens and Elia 1997). Understanding the molecular basis of this evasion will require more information on mechanisms of dsRNA recognition, and the differentiation of viral RNA sequences from other structured elements in the cytoplasm.
Interestingly, persistent and nonpersistent viruses also differ in the extent of their dinucleotide frequency biases, with those causing acute infections showing patterns of under- and overrepresentation comparable to those of host cells (Fig. 5), similar to that in other nonpersistent virus families and orders (such as the Mononegavirales). Selection for a different pattern of dinucleotide bias in persistent viruses may reflect further interactions with the host cell. Although there are clearly several factors contributing to the development of large-scale RNA structure in virus genomes, the strong association between MFEDs of mammalian RNA viruses with persistence provides a new insight into the complex field of virus/host interactions, and allows several experimentally testable hypotheses to be made (such as the effect on IFN induction and RNA interference following transfection of structured and unstructured RNA sequences in animal and plant cells).
We are presently investigating the nature of the RNA structures that form in GORS-rich genera to determine whether they form static, ordered structural elements or, as we favor, a dynamic structured environment that can readily accommodate the unwinding, elongation, and exposure of different regions of the genome during the replication cycle. Of particular interest is the marked differences in strand-specificity of MFEDs (Fig. 4); HCV, HGV/GBV-C, and aphthoviruses exhibit two to three times greater MFEDs in the genomic (sense) strand than the antisense orientation, whereas the coronaviruses (Fig. 4) and bacteriophage alleloviruses (data not shown) have high levels of MFEDs in both sense and antisense orientation. This may reflect differences in the replication strategies of these viruses.
Evolutionary implications of GORS
The results from the simulated evolution experiments contrast strikingly with the preservation of MFEDs during the evolution of HCV in vivo. For example, MFEDs are conserved between genotypes (and between subtypes of HCV), and between epidemiologically unlinked variants with genotypes of each virus family (Fig. 6). The retention and narrow range of MFEDs in individual virus genera (including viruses as diverse as GBV-B in the hepaciviruses, and GBV-A in the HGV-like genus) indicate that GORS is an evolutionarily conserved feature, and therefore likely to be a significant factor in fitness optimization in these virus groups. The extreme sensitivity of MFEDs to experimental drift indicates that the fitness space around naturally occurring structured viruses is therefore correspondingly narrow and rugged. This has several important implications for the evolution of structured RNA viruses. Firstly, the pathways followed on natural sequence drift must be extremely constrained, and lead to substantial homoplasy and sequence convergence, which can be studied experimentally. For example, HCV variants infecting different individuals 20 yr after exposure to a shared source of genotype 1b infection (Power et al. 1994) each provides independent examples of how HCV could have achieved their current divergence (4%) while maintaining RNA structure. Indeed, specific comparison of these sequences with those derived through computer-simulated neutral drift will provide novel insights into constraints operating in vivo, and, more mechanistically, how RNA structure is maintained during virus diversification.
Secondly, the GORS restriction on sequence drift greatly complicates the use of a molecular clock to predict times of divergence, as no assumptions can be made about the number of neutral sites or the limitations on sequence change at variable sites. Similarly, the existence of paired and unpaired bases in a sequence, as well as variable restrictions on the introduction of covariant and semicovariant substitutions in RNA structures, adds several new dimensions of complexity to distance calculations that allow for rate differences in individual base changes, and at different codon positions (Yang 1993). The constriction of sequence space of viruses with GORS implies that many of the branches evident on phylogenetic analysis of contemporary sequences that define virus species, genotypes, or genera occurred at remote times in the past. GORS may thus underlie the extremely limited sequence divergence of HGV/GBV-C variants infecting different human racial groups, whose geographical distribution suggests that they coevolved in modern human populations after their emergence from Africa 100,000–150,000 years ago (Tanaka et al. 1998). The similar sensitivity of GORS to sequence drift is consistent with the hypothesis that genotypes of HCV and FMDV arose over an equivalent or even greater timescale.
MATERIALS AND METHODS
Data sets
Nucleotide sequences (n = 916) were downloaded from GenBank and bear the following accession numbers:
Sequences used for the comparison of different genera of the Flaviviridae and Picornaviridae, and control DNA sequences (Figs. 1, 2, 3)
Flaviviridae
Hepaciviruses: AB030907, AB031663, AB047639, AB047640, AB047641, AB047642, AB047643, AB047644, AB047645, AF169002, AF169003, AF169004, AF169005, AF177036, AF238481, AF238482, AF238483, AF238484, AF238485, AF238486, HPCJ8G, HPCPOLP, AF046866, HCVCENS1, HP-CEGS, HPCFG, HPCJK046E2, HPCK3A, HCV4APOLY, HCV1480, D84262, D84263, D84264, D84265, HCV12083, HPCJK049E1, AF011751, AF064490, AF511948, AF511950, AY051292, D50409, HEC278830, HPCCGS, HPCHCJ1, HPC-PLYPRE, AB016785, AB049087, AB049088, AB049089, AB049090, AB049091, AB049092, AB049093, AB049094, AB049095, AB049096, AB049097, AB049098, AB049099, AB049100, AB049101, AB080299, AF139594, AF165045, AF165047, AF165049, AF165051, AF165053, AF165055, AF165057, AF165059, AF165061, AF165063, AF176573, AF207752, AF207753, AF207754, AF207756, AF207757, AF207758, AF207759, AF207760, AF207761, AF207762, AF207763, AF207764, AF207765, AF207766, AF207767, AF207768, AF207769, AF207770, AF207771, AF207772, AF207773, AF207774, AF208024, AF313916, AF333324, AF356827, AF483269, AF511949, AY045702, D85516, D89815, D89872, HCJ238799, HCU01214, HCU45476, HCV132996, HCVJK1G, HCVPOLYP, HPCCGENOM, HPCGENANTI, HPCHUMR, HPCJ491, HPCJCG, HPCJRNA, HPCJTA, HPCK1R1, HPCK1R2, HPCK1R3, HPCPP, HPCRNA, HPCUNKCDS, HPVHCVN, NC_001655.
HGV-like viruses: AB003288, AB003289, AB003290, AB003291, AB003292, AB003293, AB008342, AB013500, AB013501, AB018667, AB021287, AF006500, AF031827, AF104403, AF121950, D87255, D87262, D87708, D87709, D87710, D87711, D87712, D87713, D87714, D87715, D90600, D90601, GBV-tro, HGU36380, HGU44402, HGU45966, HGU63715, HGU75356, HGU94695.
Pestiviruses: AB078950, AB078951, AF091605, AF220247, BVDPOLYPRO, BVDPP, NC_001461, PTU86600, AF002227, AF145967, AF502399, BVU18059, NC_002032, NC_002657, BDU70263, NC_003679, AF091507, AF092448, AF407339, AF531433, AY072924, AY259122, HCU45478, HCVCG3PE, HCVCGSA, HCVPOLYPR, HCVSEQB, NC_003677, NC_003678.
Flaviviruses: AB051292, AB062063, AB062064, AF022438, AF038402, AF045551, AF100462, AF100465, AF100466, AF100468, AF202541, AF206518, AF217620, AF226685, AF289029, AF298807, AF309641, AF315119, AF317203, AF326573, AF350498, AF404757, AF489932, AF527415, AY037116, AY149904, AY182009, AY217093, AY262283, AY277251, DEN2JAMCG, DENCME, DENT1SEQ, JEVLINGCG, KUNCG, L40361, NC_000943, NC_001474, NC_001563, NC_001564, NC_001672, NC_001809, NC_002031, NC_003218, NC_003635, NC_003675, NC_003676, NC_003687, NC_003690, NC_003996, NC_004119, NC_004355, YFU54798.
Picornaviridae
Enterovirus: AF039205, AF081485, AF083069, AF085363, AF114383, AF114384, AF119795, AF162711, AF176044, AF177911, AF241359, AF302996, AF304459, AF311938, AF311939, AF316321, AF317694, AF328683, AF405666, AF405669, AF405682, AF462418, AF504533, AF524866, AF524867, AF541919, AY036578, AY036579, AY167103, AY167104, AY167105, AY167106, AY167107, AY184220, CXA24CG, CXA3CG, CXA3G, CXA9CG, CXAB3CG, CXB5CGA, CXU57056, E11276224, E6U16283, EC12TCG, ECHOV9XX, ETU22521, ETU22522, EV11VPCD, EV9GENOME, HEC295172, HPO132960, HPO132961, HPO293918, NC_001428, NC_001430, NC_001472, NC_001612, NC_002058, NC_003986, NC_003988, PICOXB4, PIP03XX, POL2CG1, POL2LAN, POL3L37.
Aphthoviruses: AB079061, AF026168, AF154271, AF189157, AF377945, AF506822, AF511039, APHA12CDR, FDI320488, FM-DVALF, FMV7572, FOO539136, FOO539139, NC_002554, NC_003992, NC_004004, PIFMDV1, PIFMDV2.
Teschoviruses: AB038528, AF231767, AF231768, AF231769, AF296087, AF296088, AF296089, AF296090, AF296091, AF296092, AF296093, AF296094, AF296096, AF296100, AF296102, AF296104, AF296107, AF296108, AF296109, AF296111, AF296112, AF296113, AF296115, AF296117, AF296118, NC_003985.
Hepatoviruses: AB020564, AB020565, AB020566, AB020567, AB020568, AF268396, AF314208, AF485328, AR141321, AR219495, AY032861, HAVCOMPL, HAVRNAGBM, HAVR-NAGWT, HEA299464, HPA18F, HPA24A, HPAACG, HPACG, NC_001489.
Human mRNA controls
AB089957, ADAMTS20, ADPRTL1, AF345347, AF389420, AF533230, AF533875, AF536753, AKAP13, APRIN, ATP2A3, AY184206, AY194287, AY196326, CDC42BPA, CDK5RAP2, CECR2, CNTN5, DISPA, DLG5, DRPLA, DUOX2, ELD/OSA1, FBN3, GPR124, HSA011972, HSA132429, HSA275213, HSA310567, HSA310931, HSA318888, HSA491324, HSA544537, HUMCO, HUMGCP372, HUMHSNF2B, HUMHT3I, HUMITS, HUMKIAAJ, HUMPTPB3, KIAA0847, KIAA1244, KIAA1404, M160, MYH11, NFAT5, NFBD1, PCDH1, PTK7, TIAF1, TJP1, TRALPUSH, TRPM3, TRPM7, UBR1, USH2A, XLHSRF-1.
Large DNA viruses
Herpesviridae: NC_001806, NC_001798, NC_001348, NC_001345, NC_001347, NC_001664, NC_001716; NC_003409.
Poxviridae: NC_001559, NC_004003, NC_003391, NC_002188, NC_004002, NC_001731, NC_003663, NC_003389, NC_001132, NC_003310, NC_001611, NC_000913, NC_000917.
Bacterial sequences
Escherichia coli: NC_004431.
Archaeoglobus fulgidis: NC_000917.
Sequences used for the extended comparison of animal and plant RNA viruses (Fig. 4)
RNA viruses
Animal viruses.
Filoviridae: NC_004161, NC_001608.
Paramyxoviridae: AF371337, NC_001498, NC_001552, NC_001803, NC_001906, NC_001921, NC_002199, NC_002617, NC_002728, NC_003044, NC_003461.
Rhabdoviridae: NC_002803, NC_000855, NC_001542, NC_001560, NC_001652, NC_002526, NC_003243, NC_001615, NC_002251, NC_003746, NC_000903.
Arteriviridae: AY150312, LDU15146, NC_001961, NC_002532, NC_002534, NC_003092.
Coronaviridae: NC_001451, NC_001846, NC_002306, NC_002645, NC_003045, NC_003436.
Arenaviridae: AF485264, AY129248, NC_004291, NC_004292, NC_004293, NC_004294, NC_004296, NC_004297.
Bunyaviridae:AF288297, AF288298, HANC12GP, HANC12RDRP, HPSGPGO, HPSNUPR, HPSVRPLA, NC_003467, NC_003468, TUVL5302, TUVM5302, NC_004158, NC_004159, NC_001925, NC_001926, NC_004108, NC_004109, NC_002043, NC_002044, NC_002050, NC_002051, NC_002052, NC_003614, NC_003616, NC_003619, NC_003620, NC_003624, NC_003625, NC_003832, NC_003841, NC_003843.
Orthomyxoviridae: NC_002021, NC_002022, NC_002023, NC_002204, NC_002205, NC_002206, FLCP3A, FLCPB1A, FLCPB2A.
Astroviridae: NC_001943, NC_002469, NC_002470, NC_003790.
Caliciviridae: NC_004064, AF258618, AF295785, NC_001543, NC_002615, RHDVCGS, RHU54983, AB039774, AB039775, AB039777, AB039779, AB039780, AB039781, AB039782, AB042808, AB081723, AF093797, AF145896, AY032605, AY134748, CRNAORFS, HCU07611, NC_001959, SOUCAPPRO, HCA249939, HECGENRA, NC_000940, AB070225, AF091736, AF109465, AF321298, AF479590, BCA011099, FCLF4, FCU13992, NC_001481, NC_002551, NC_004542, SMU15301, AY228235.
Flaviviridae: KUNCG, NC_000943, NC_001437, NC_001474, NC_001475, NC_001477, NC_001563, NC_002031, NC_002640, NC_003687, NC_001564, NC_003635, NC_003675, NC_003676, NC_003996, NC_004119, AF331718, NC_001672, NC_001809, NC_003218, NC_003690, AF011751, AF177036, HCV12083, HCV1480, HCV4APOLY, HPCEGS, NC_001655, AB018667, AF023424, AF023425, D90601, HGU22303, HGU36380, HGU44402, NC_001837, AF037405, BVDCG, BVU18059, NC_003677, NC_003678, PTU90951.
Hepatitis E-like viruses: AB074915, AB074917, AB074918, AB074920, AY115488, NC_001434.
Picornaviridae: AF154271, FMDVALF, NC_002554, NC_003982, NC_003992, NC_004004, NC_002527, MNGPOLY, NC_001366, NC_001479, NC_001479, NC_001428, NC_001430, NC_001472, NC_001490, NC_001612, NC_001617, NC_001752, NC_001859, NC_002058, NC_003986, NC_003988, POL3L37, SVDMPS, NC_003983, NC_001489, NC_003990, SHVAGM27, NC_001918, NC_004421, NC_001897, NC_003976, NC_003077, AB038528, AF231769, AF296087, AF296091, AF296093, AF296115, AF296119, NC_003985, ERVPOLY.
Togaviridae: NC_001449, NC_001512, NC_001544, NC_001547, NC_001786, NC_001924, NC_003215, NC_003899, NC_003908, SINOCK82, SPA316244, NC_001545.
Hepadnaviridae: NC_001344, NC_001486, AF242585, HBV131568, NC_001484, NC_001719, NC_001896, NC_002168, NC_003977, NC_004107.
Retroviridae: NC_001407, NC_001408, NC_001503, NC_001550, NC_000858, NC_001414, NC_001436, NC_001488, NC_001815, NC_003323, NC_001724, NC_001867, NC_000863, NC_001501, NC_001502, NC_001514, NC_001885, NC_001940, HIVMNCG, NC_001413, NC_001450, NC_001452, NC_001463, NC_001482, NC_001511, NC_001654, NC_001722, NC_001362, NC_001363, NC_001499, NC_001500, NC_001702, NC_001364, NC_001736, NC_001795, NC_001831, NC_001871, NC_002201, NC_001819
Plant viruses.
Barnaviridae: NC_001633.
Carlavirus: NC_001361, NC_002552, NC_002795, NC_003499, NC_003557.
Caulimoviridae: NC_001343, NC_001574, NC_003031, NC_003381, NC_003382, NC_001648, NC_001497, NC_001725, NC_003138, NC_003498, NC_003554, NC_004036, NC_004324, NC_001839, NC_001914, NC_001634, NC_001739, NC_003378.
Closteroviridae: NC_001598, NC_001661, NC_001836, NC_003617, NC_003618, NC_004123, NC_004124.
Foveavirus: NC_001946, NC_001948, NC_002468, NC_002729, NC_003462.
Pomovirus: NC_003510, NC_003518, NC_003519, NC_003520, NC_003723, NC_003724, NC_004423.
Potexvirus: NC_001441, NC_001455, NC_001483, NC_001642, NC_001658, NC_001748, NC_001753, NC_001812, NC_002815, NC_003400, NC_003632, NC_003794, NC_003820, NC_003849, NC_004067, NC_004322.
Potyviridae: NC_002350, NC_002990, NC_003483, NC_004016, NC_003797, AY149118, NC_000947, NC_001445, NC_001517, NC_001555, NC_001616, NC_001671, NC_001768, NC_001785, NC_001841, NC_002509, NC_002600, NC_002634, NC_003224, NC_003377, NC_003397, NC_003398, NC_003492, NC_003536, NC_003537, NC_003605, NC_003606, NC_003742, NC_004010, NC_004011, NC_004013, NC_004035, NC_004039, NC_004047, NC_004426, NC_001814, NC_001886, NC_003501, NC_003399.
Trichovirus: NC_001409, NC_002500.
Vitivirus: NC_003602, NC_003604.
Bromoviridae: NC_003543, NC_004008, NC_004120.
Comoviridae: AB054689, ADEPP, AF059532, AF059533, AF394607, NC_003495, NC_003496, NC_003544, NC_003545, NC_003549, NC_003550, NC_003738, NC_003741, NC_003741, NC_003799, AB011007, AB013616, AB018698, AB023484, AB032403, AB051386, AF104335, AF144234, AF149425, AF225955, AF228423, BBE132844, BBU65985, NC_003004, NC_003003, NC_003975, AF016626, AMVRNA2U, AY017338, AY017339, AY157994, BLU20621, GFLRNA1, NC_003502, NC_003509, NC_003621, NC_003622, NC_003623, NC_003693, NC_003694, NC_003791, NC_003792, NC_003840, NC_004439, OLA277435, S46011, TBRVEDRN2, TOSRNA2, TRU46022, TRU50869, AB030941, NC_003445, NC_003446, NC_003785, NC_003786, NC_003787, AY122330.
Luteoviridae: NC_003629, NC_002160, NC_003056, NC_003369, NC_003680, AY138970, NC_001747, NC_002198, NC_002766, NC_003688, NC_003743, NC_000874.
Marafivirus: NC_001793, NC_002164, NC_002786.
Tetraviridae: NC_001990, NC_001981.
Tombusviridae: NC_000939, NC_003633, NC_001265, NC_001504, NC_001600, NC_002187, NC_003535, NC_003821, NC_003627, NC_002598, NC_001339, NC_001469, NC_001554, NC_003500, NC_003532.
Tymovirus: NC_001480, NC_001513, NC_001746, NC_001977, NC_002588, NC_003634, NC_004063.
Hordeivirus: NC_003469, NC_003478, NC_003481, NC_001367, NC_001556, NC_001728, NC_001801, NC_001873, NC_002633, NC_002692, NC_002792, NC_003355, NC_003610, NC_003630, NC_003852, NC_003878, NC_004106, NC_004422.
Bacteriophages
Leviviridae: AY099114, NC_001890, NC_001891, NC_004301, NC_004304.
Sequence divergence between each included member of the data set was >5%, and typically included at least 10 sequences for clustering and phylogenetic analysis. As at least 95% of the genomic sequences of large DNA viruses and bacteria are transcribed, thermodynamic prediction of RNA folding in large DNA viruses and bacteria was based on their complete genomic sequences as their calculated MFEs would be predominantly determined by secondary structure in RNA transcripts.
Sequence order randomization and compositional analysis
All sequence order randomization methods were carried out using the SIMMONIC sequence editor package (Simmonds and Smith 1999). Methods used included those that preserved the dinucleotide frequencies (NDR) and CDLR, which combined the features of NDR and a method previously developed that preserves codon order (CLR; Tuplin et al. 2002). In addition, we have implemented algorithms that randomize nucleotide sequence order, while maintaining tri- or tetranucleotide base frequencies (NRR, NTR). To generate the required information on variance required for calculation of Z scores in finite time (Workman and Krogh 1999), computation time was reduced by following a strategy of dividing genomes into sequential segments of 498 bases overlapping its neighbors by 249 bases. Mean MFED values and Z scores could be calculated for the whole genome; alternatively, mean values for each 498-base segment across the genomes of several viruses could be computed. Base and dinucleotide frequencies for sequences analyzed for MFE were determined using programs within the SIMMONIC editor package (Simmonds and Smith 1999).
Calculation of minimal free energy differences (MFEDs)
All folding free energy (MFE) calculations were determined using MFOLD version 3.1 (http://www.bioinfo.rpi.edu/~zukerm/), using the implementation available in the program ZIPFOLD, which allows MFEs of large numbers of RNA sequences to be determined rapidly. Sequence submission and result retrieval were automated using perl scripts with a backend MySQL database to facilitate handling the large data sets for Z-score statistics (~25 million individual sequence submissions to the ZIPFOLD file server). Statistical analysis was carried out using SYSTAT (http://www.systat.com/).
Simulation of random sequence drift
The introduction of defined numbers of nucleotide substitutions into a viral sequence that reproduced characteristics of naturally occurring variants of the virus under a neutral model (Kimura 1983) was carried out using the Mutate program in the SIM-MONIC package (Simmonds and Smith 1999). This program allows the degree of sequence divergence, the relative numbers of synonymous and nonsynonymous substitutions, and of transitions and transversions to be specified, thereby reproducing frequencies observed naturally and maintaining the base composition of the native sequence.
Classification of viruses as persistent or nonpersistent
Genera from the flavivirus, picornavirus, alphavirus, HEV-like, and calicivirus virus families were chosen for this analysis because they contained both sufficient complete genome sequences from several genera and information on outcome of infection. Virus genera were collectively classified as persistent (capable of establishing long-term, chronic infection in their natural hosts), and nonpersistent (acute, self-limiting infections in immunocompetent hosts, i.e., excluding neonatal infection).
In the flaviviruses, the hepacivirus and HGV-like genera are classified as persistent, whereas infections with the pestiviruses and flaviviruses were classified as nonpersistent. In the four genera of Caliciviridae, vesiviruses are known to establish systemic and persistent infections, whereas the noroviruses, lagoviruses, and sapoviruses do not. The Picornaviridae can be similarly classified into genera that cause persistent disease in the appropriate host (aphthoviruses, cardioviruses) and those that are nonpersistent (enteroviruses, hepatoviruses, parechoviruses). The ability to establish persistent infections in the recently identified and classified kobuand teschoviruses is not known. The other virus groups included in the analysis were the nonpersistent viruses, HEV and the alphavirus genus within the Togaviridae. Viruses classified as Coronaviridae or Ateriviridae vary in their persistence in their natural hosts and in the extent of their predicted RNA folding. However, few complete genome sequences are available from members of these groups, and information on the specifics of their virus/host interactions is not available in most cases. For these reasons, we have not included MFED results from this virus family in the comparison of GORS with persistence, although the observation of substantial MFEDs in coronaviruses such as murine hepatitis virus is consistent with the association found in other virus families.
Acknowledgments
The authors are grateful to Michael Zuker for access and intensive use of the MFOLD server required for the free energy calculations. We thank Tony Nash and Mark Woolhouse, University of Edinburgh, and Eddie Holmes, University of Oxford, for helpful review and discussion of the paper prior to submission.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7640104.
REFERENCES
- Baranowski, E., Ruiz-Jarabo, C.M., and Domingo, E. 2001. Evolution of cell recognition by viruses. Science 292: 1102–1105. [DOI] [PubMed] [Google Scholar]
- Belsham, G.J. and Sonenberg, N. 1996. RNA–protein interactions in regulation of Picornavirus RNA translation. Microbiol. Rev. 60: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, E., Denli, A.M., and Hannon, G.J. 2001. The rest is silence. RNA 7: 1509–1521. [PMC free article] [PubMed] [Google Scholar]
- Clemens, M.J. and Elia, A. 1997. The double-stranded RNA-dependent protein kinase PKR: Structure and function. J. Interferon Cytokine Res. 17: 503–524. [DOI] [PubMed] [Google Scholar]
- Evans, D.J. 1999. Reverse genetics of picornaviruses. Advances Virus Res. 53: 209–228. [DOI] [PubMed] [Google Scholar]
- Foy, E., Li, K., Wang, C., Sumpter Jr., R., Ikeda, M., Lemon, S.M., and Gale Jr., M. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300: 1145–1148. [DOI] [PubMed] [Google Scholar]
- Girardin, S.E., Sansonetti, P.J., and Philpott, D.J. 2002. Intracellular vs extracellular recognition of pathogens—Common concepts in mammals and flies. Trends Microbiol. 10: 193–199. [DOI] [PubMed] [Google Scholar]
- Goodbourn, S., Didcock, L., and Randall, R.E. 2000. Interferons: Cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81: 2341–2364. [DOI] [PubMed] [Google Scholar]
- Goodfellow, I.G., Chaudhry, Y., Richardson, A., Meredith, J.M., Almond, J.W., Barclay, W.S., and Evans, D.J. 2000. Identification of a cis-acting replication element (CRE) within the poliovirus coding region. J. Virology 74: 4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery, S., Rice, A., Robertson, H., and Mathews, M. 1990. TAT-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded-RNA-activated protein kinase. Proc. Natl. Acad. Sci. 87: 8687–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff, H. and Berkhout, B. 2002. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 41: 10439–10445. [DOI] [PubMed] [Google Scholar]
- Imani, F. and Jacobs, B.L. 1988. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 σ 3 protein. Proc. Natl. Acad. Sci. 85: 7887–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost Haasnoot, P.C., Olsthoorn, R.C., and Bol, J.F. 2002. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem–loop C. RNA 8: 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, L. and Burge, C.B. 2003. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 13: 2042–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze, M.G., He, Y., and Gale Jr., M. 2002. Viruses and interferon: A fight for supremacy. Nat. Rev. Immunol. 2: 675–687. [DOI] [PubMed] [Google Scholar]
- Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
- Levy, D.E. and Garcia-Sastre, A. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12: 143–156. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Mason, P.W., Bezborodova, S.V., and Henry, T.M. 2002. Identification and characterization of a cis-acting replication element (CRE) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76: 9686–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, M.B. 1995. Structure, function, and evolution of adenovirus virus-associated RNAs. Curr. Top. Microbiol. Immunol. 199: 173–187. [DOI] [PubMed] [Google Scholar]
- Moya, A., Elena, S.F., Bracho, A., Miralles, R., and Barrio, E. 2000. The evolution of RNA viruses: A population genetics view. Proc. Natl. Acad. Sci. 97: 6967–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg, A.C. and Sgro, J. 1997. Topological organisation of Picornaviral genomes: Statistical prediction of RNA structural signals. Semin. Virol. 8: 231–241. [Google Scholar]
- Pelletier, J. and Sonenberg, N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from polio-virus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Player, M.R. and Torrence, P.F. 1998. The 2–5A system: Modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol.Ther. 78: 55–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J.P., Lawlor, E., Davidson, F., Yap, P.L., Kenny-Walsh, E., Whelton, M.J., and Walsh, T.J. 1994. Hepatitis C viraemia in recipients of Irish intravenous anti-D immunoglobulin. Lancet 344: 1166–1167. [DOI] [PubMed] [Google Scholar]
- Rima, B.K. and McFerran, N.V. 1997. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J. Gen. Virol. 78: 2859–2870. [DOI] [PubMed] [Google Scholar]
- Rivas, E. and Eddy, S.R. 2000. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics 16: 583–605. [DOI] [PubMed] [Google Scholar]
- Rohll, J.B., Moon, D.H., Evans, D.J., and Almond, J.W. 1995. The 3′-untranslated region of picornavirus RNA—Features required for efficient genome replication. J. Virol. 69: 7835–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, L.R. and Barber, G.N. 2003. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 17: 961–983. [DOI] [PubMed] [Google Scholar]
- Schlesinger, S., Makino, S., and Linial, M.L. 1994. cis-Acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin. Virol. 5: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds, P. and Smith, D.B. 1999. Structural constraints on RNA virus evolution. J. Virol. 73: 5787–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, S.L. and Katze, M.G. 2001. How hepatitis C virus counteracts the interferon response: The jury is still out on NS5A. Virology 284: 1–12. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Mizokami, M., Orito, E., Ohba, K., Kato, T., Kondo, Y., Mboudjeka, I., Zekeng, L., Kaptue, L., Bikandou, B., et al. 1998. African origin of GB virus C hepatitis G virus. FEBS Lett. 423: 143–148. [DOI] [PubMed] [Google Scholar]
- Tsukiyama Kohara, K., Iizuka, N., Kohara, M., and Nomoto, A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virology 66: 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin, A., Wood, J., Evans, D.J., Patel, A.H., and Simmonds, P. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8: 824–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, R.A. 2002. Virulence and pathogenesis. Trends Microbiol. 10: 314–317. [DOI] [PubMed] [Google Scholar]
- Witwer, C., Rauscher, S., Hofacker, I.L., and Stadler, P.F. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29: 5079–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman, C. and Krogh, A. 1999. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids. Res. 27: 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, W., Paul, A.V., and Wimmer, E. 1997. RNA signals in Entero-and Rhinovirus genome replication. Semin. Virology 8: 256–273. [Google Scholar]
- Yang, Z. 1993. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10: 1396–1401. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]