Abstract
An important step in mRNA biogenesis is the export of mRNA from the nucleus to the cytoplasm. In this work, we provide evidence that the previously uncharacterized gene APQ12 functions in nucleocytoplasmic mRNA transport in Saccharomyces cerevisiae. First, apq12Δ strains manifest 3′ hyperadenylated mRNA similar to other previously characterized RNA export mutants. Second, bulk poly(A)+ RNA is retained in the nucleus in apq12Δ cells. Third, an Apq12p–GFP chimeric protein is localized to the nuclear periphery. Fourth, mRNA in apq12Δ cells is stabilized, consistent with a defect in the rate of nuclear export. Interestingly, apq12Δ mutants are severely compromised for growth and display atypical cell morphology. Because this aberrant cell morphology is not seen with other viable export mutants, Apq12p must have either an additional cellular function, or preferentially impinge on the export of mRNAs regulating cell growth. Together, these findings support a role for APQ12 in nucleocytoplasmic transport of mRNA.
Keywords: mRNA transport, hyperadenylation, RNA metabolism, cell morphology
INTRODUCTION
An important feature of eukaryotic gene expression is the compartmentalization of mRNA metabolism. Transcription, pre-mRNA splicing, and 3′ end formation are nuclear events that, once completed, yield an mRNA capable of export to the cytoplasm. Transport of mRNA from the nuclear to the cytoplasmic environment signals a transition in the fate of an mRNA, wherein it becomes a template for protein synthesis and ultimately is degraded by the mRNA turnover machinery.
A fertile area of research for understanding mRNA transport and its relationship to other aspects of mRNA metabolism has come from the isolation of numerous mutants that affect mRNA export in Saccharomyces cerevisiae. These mRNA export mutants were identified by several criteria including screening for lesions that result in the nuclear accumulation of poly(A)+ RNA based on in situ hybridization with labeled oligo(dT). These screens led to the identification of ribonucleic acid trafficking (RAT), mRNA transport (MTR), mRNA export (MEX), and GLFG-lethal (GLE) genes (Amberg et al. 1992; Kadowaki et al. 1994; Murphy et al. 1996; Segref et al. 1997). One interesting aspect of many export mutants is the production and nuclear retention of hyperadenylated mRNAs (Hilleren and Parker 2001; Hilleren et al. 2001; Jensen et al. 2001). Given this, a promising screen for mutations affecting mRNA export is the identification of yeast strains with hyperadenylated mRNAs.
To distinguish novel genes affecting mRNA export, we screened the collection of viable yeast deletion mutants for strains that led to the production of hyperadenylated mRNAs. This investigation led to the identification of apq12Δ as a lesion leading to hyperadenylated poly(A) tails, suggesting that Apq12p, a protein of previously unknown function, might affect mRNA nucleocytoplasmic transport. Consistent with Apq12p functioning in mRNA export, apq12Δ strains accumulate poly(A)+ in the nucleus and show additional defects in the metabolism of mRNAs. In addition, Apq12p localizes to the nuclear periphery. In summation, this work defines Apq12p as a novel factor contributing to mRNA export.
RESULTS AND DISCUSSION
Identification of APQ12 as a gene influencing mRNA poly(A) tail length
In an endeavor to identify novel factors involved in mRNA metabolism, we screened the EUROSCARF collection of ~5000 yeast nonessential gene deletion strains (Winzeler et al. 1999) for alterations in mRNA poly(A) tail length or distribution. The effect of each deletion mutant on poly(A) tail metabolism was determined by analyzing endogenous MFA2 mRNA by high-resolution Northern blot analysis of total cellular RNA isolated from each single-gene knockout strain. Inspection of MFA2 mRNA by this procedure resulted in the identification of several strains that displayed an increase in poly(A) tail length. These included ccr4Δ and pop2Δ, strains that manifest increased average poly(A) tail lengths owing to defects in the major cytoplasmic deadenylase (Tucker et al. 2001). In addition, hyperadenylated poly(A) tails were observed for sac3Δ, hpr1Δ, plc1Δ, and apq12Δ strains. Strains lacking Sac3p, Hpr1p, and Plc1p have previously been described as being defective in nuclear mRNA export (Schneiter et al. 1999; York et al. 1999; Fischer et al. 2002; Strasser et al. 2002; Lei et al. 2003). In contrast, APQ12 encodes a protein of unknown function.
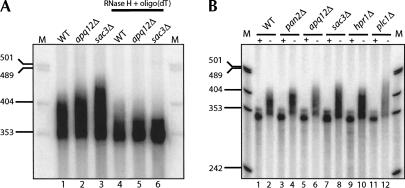
In an apq12Δ strain, the distribution of MFA2 mRNA extended 20–40 residues longer than in a wild-type strain (Fig. 1A, lane 1 vs. 2). An enhanced mRNA size distribution was also observed for endogenously expressed PGK1 mRNA (data not shown). To confirm that the more slowly migrating MFA2 mRNA from the apq12Δ strain is caused by heterogeneity of the poly(A) tail at the mRNA 3′ terminus, RNA samples were treated with RNase H in the presence of oligo(dT) to remove mRNA poly(A) tails. As shown in Figure 1A (lanes 4,5), treatment of the RNA results in a homogeneous MFA2 mRNA population that is similar in size for wild-type and apq12Δ samples. These data demonstrate that MFA2 mRNA is hyperadenylated in apq12Δ cells.
FIGURE 1.
APQ12 is required for normal mRNA poly(A) tail distribution. High-resolution Northern blots of endogenously expressed MFA2 mRNA from (A) wild-type, apq12Δ, and sac3Δ cells. Poly(A) tail lengths were determined by comparing the size of the heterogeneous population in untreated RNA samples (lanes 1–3) as compared with samples in which the poly(A) tail has been removed by treatment with oligo(dT) and RNase H (lanes 4–6). Molecular size markers (M) are labeled at the left of the panel. (B) The mRNA hyperadenylation observed for apq12Δ cells is intermediate to that seen for other transport mutants. Shown is MFA2 mRNA from pan2Δ, apq12Δ, sac3Δ, hpr1Δ, and plc1Δ strains either untreated (−) or treated with oligo(dT)/RNase H to remove poly(A) tails (+).
The hyperadenylated mRNA poly(A) tail phenotype observed for apq12Δ cells is similar to that previously observed for lesions in SAC3, HPR1, RIP1, and PLC1, genes implicated in mRNA transport (Schneiter et al. 1999; York et al. 1999; Jones et al. 2000; Fischer et al. 2002; Strasser et al. 2002). In addition, the temperature-sensitive mex67-5, rat7-1, rat8-2, and gle1-37 mutations all produce hyperadenylated mRNA at the restrictive temperature (Hilleren and Parker 2001; Jensen et al. 2001). Hyperadenylated mRNAs can also be observed in yeast strains lacking the Pab1p-associated poly(A) nuclease, Pan2p/Pan3p, which appears to function, in part, to trim a nascent poly(A) tail to a shorter species after export of the RNA to the cytoplasm (Boeck et al. 1996; Brown et al. 1996). The hyperadenylation observed in apq12Δ cells is more extensive than that observed in cells void of Pan2p and Hpr1p, a member of the transcription/RNA export complex (Strasser et al. 2002; Fig. 1B, cf. lanes 4,6,10), and slightly shorter than the hyperadenylation observed in other viable mRNA export mutants such as plc1Δ or sac3Δ (Fig. 1B, cf. lanes 6,8,12). Given that apq12Δ manifests hyperadenylated mRNA reminiscent of mRNA from various RNA transport mutants, we hypothesized that the Apq12 protein also affects mRNA export.
Apq12p is encoded by the YIL040w locus and represents an uncharacterized gene in the Saccharomyces Genome Data Base. We confirmed that deletion of APQ12 was necessary for the hyperadenylated mRNA phenotype by complementing an apq12Δ strain with a single-copy plasmid encoding wild-type APQ12. The complemented strain demonstrated a wild-type poly(A) tail distribution of MFA2 mRNA by Northern blot analysis (data not shown). In addition, the plasmid also complemented the severe growth phenotype observed for apq12Δ strains (see below; data not shown).
APQ12 encodes a protein of ~16.5 kDa that demonstrates no strong structural relatedness to any other protein in S. cerevisiae. Whereas Apq12p homologs with high identity (~80%) exist in several Saccharomyces genomes (Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevii, and Saccharomyces bayanus, e.g.), no obvious homolog can be identified in Schizosaccharomyces pombe or within genomes outside of fungi. Interestingly, apq12Δ is synthetically lethal with cdc73Δ, a protein also involved in mRNA biosynthesis, and suggests a connection between Apq12p and proteins involved in mRNA metabolism and export (Tong et al. 2004). Two-hybrid analysis determined that Apq12p can interact with either Pea2p or Ssa2p (Ito et al. 2001); however, the relevance of these putative protein–protein interactions in RNA metabolism remains unclear.
Loss of Apq12p results in the accumulation of poly(A)+ RNA in the nucleus
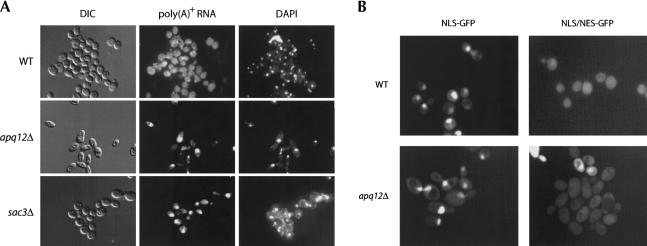
To evaluate if Apq12p influences mRNA nucleocytoplasmic transport, we examined the subcellular localization of poly(A)+ mRNA in apq12Δ, sac3Δ (as a positive control), and wild-type cells by in situ hybridization of a digoxigenin-conjugated oligo(dT) probe and indirect immunofluorescence. In wild-type yeast, poly(A)+ staining is observed as a diffuse, uniform signal throughout the cell, including the nucleus and the cytoplasm (Fig. 2A). In contrast, a majority of apq12Δ cells demonstrate poly(A)+ RNA as a single focus that is coincident with nuclear DNA as visualized by DAPI staining (Fig. 2A). Cells that do not demonstrate poly(A)+ foci also fail to stain with DAPI, suggesting that nuclear division and/or segregation many be compromised in the apq12Δ strain. Similar to apq12Δ cells, bulk poly(A)+ RNA in an sac3Δ is nuclear in its localization (Fig. 2A), consistent with the characterized role of SAC3 in nucleocytoplasmic mRNA transport (Jones et al. 2000; Fischer et al. 2002).
FIGURE 2.
APQ12 affects bulk poly(A)+ RNA accumulation but not protein trafficking. (A) Distribution of bulk poly(A)+ RNA within wild-type and apq12Δ cells. Poly(A)+ RNA was visualized by oligo(dT) hybridization and indirect immunofluorescence. Cell nuclei are revealed by staining of nuclear DNA with DAPI. Bulk poly(A)+ RNA distribution is also shown for an sac3Δ mutant that has been previously shown to demonstrate strong nuclear retention of poly(A)+ RNA. (B) Subcellular localization patterns of NLS(GFP)3 and NLS/NES(GFP)2 protein reporters in wild-type and apq12Δ cells. The foci observed for NLSp–GFP in both wild-type and apq12Δ cells colocalize with nuclear DNA as confirmed by DAPI staining (data not shown).
Our in situ hybridization results suggested a role for APQ12 in mRNA export; to assess whether APQ12 also functions in protein trafficking, we examined the localization of GFP-tagged protein transport reporters in living wild-type and apq12Δ cells. These reporters included the nondiffusible SV40 T antigen nuclear localization signal construct [NLS(GFP)3] and an NLS/NES–GFP reporter [NLS/NES(GFP)2] containing the SV40 NLS and a protein kinase inhibitor (PKI) NES sequence (Stade et al. 1997; de Bruyn Kops and Guthrie 2001). In wild-type cells, the NLS–GFP reporter revealed the expected nuclear distribution (Fig. 2B). Similarly, in apq12Δ cells, the NLS–GFP reporter protein is nuclear (Fig. 2B; data not shown). The equivalent distribution of the NLS/NES–GFP reporter within the nucleus and cytoplasm is indistinguishable between wild-type and apq12Δ cells (Fig. 2B). These data indicate that protein trafficking is generally unaffected in apq12Δ cells.
The cellular morphology of apq12Δ mutants was observed to be strikingly aberrant as compared with wild-type cells (see Fig. 2A, DIC images). In addition to the presence within the population of cells that appear to lack a nucleus, apq12Δ cells demonstrated an elongated cell shape morphology. This latter observation is consistent with a role for APQ12 in apical growth. Accordant with this, the YIL040w/APQ12 locus was previously identified in a screen for enhanced APical growth as detected with Quantitative analysis (Y. Ohya and D. Watanabe, pers. comm.). Interestingly, an apical cell morphology phenotype is not observed for sac3Δ mutants (Fig. 2A), nor, to our knowledge, has it been reported for other characterized mRNA transport mutants. It is unclear whether the aberrant cell morphology observed for apq12Δ cells is related to the defect in mRNA export or represents an additional cellular function of Apq12p linked to cell growth.
APQ12p localizes to the nuclear periphery
The observation that apq12Δ mutants retain bulk poly(A)+ RNA in the nucleus is similar to that seen for several mutants defective in mRNA transport factors that are localized to the nucleus or to the nuclear pore complex (NPC) itself (Amberg et al. 1992; Kadowaki et al. 1994; Murphy et al. 1996; Segref et al. 1997; Lei et al. 2003). We therefore evaluated the subcellular localization of the Apq12p protein. In wild-type cells, the APQ12 locus was engineered by yeast recombination to express a C-terminally tagged Apg12p–green fluorescent protein (Apq12p–GFP; Longtine et al. 1998). Expression of the Apq12p–GFP chimera was confirmed by Western blot analysis and determined to be the anticipated size (data not shown). Importantly, the fusion protein was fully functional because neither bulk poly(A)+ RNA localization nor MFA2 mRNA poly(A) tail distribution was affected in strains harboring the APQ12–GFP chromosomal fusion as compared with wild-type cells (data not shown).
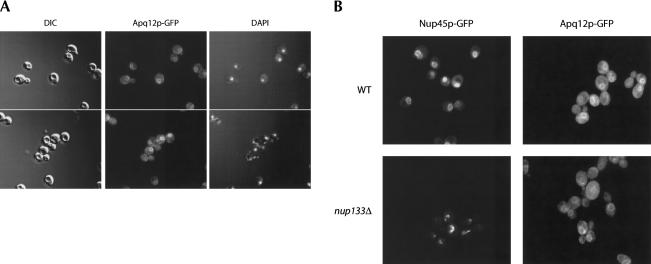
The Apq12p–GFP fusion protein was visualized by direct fluorescence microscopy and appears as a ring structure localized to the entire nuclear rim (Fig. 3A). Interestingly, this ring-like staining was not punctate but smooth. Furthermore, a discontinuous labeling close to the plasma membrane was observed that may represent the underlying ER (Preuss et al. 1991). These results suggest Apq12p is localized both to the ER and to the specialized ER that makes up the nuclear envelope. Consistent with being localized to the NE/ER, Apq12p has predicted membrane-spanning domains. The distribution of Apq12p–GFP around the nuclear periphery is consistent with APQ12 playing a role in nucleocytoplasmic transport of mRNA.
FIGURE 3.
Apq12p localizes to the nuclear periphery. (A) The subcellular distribution of Apq12p is shown by direct visualization of a C-terminally tagged Apq12p–GFP protein chimera in living cells. The position of the nucleus is visible by DAPI staining of nuclear DNA. (B) The subcellular localization patterns for Nup45p–GFP and Apq12p–GFP in wild-type and nup133Δ cells. Nup45p–GFP, a known component of the nuclear pore complex, displays protein clustering in nup133Δ cells. In contrast, Apq12p–GFP fails to display clustering in the nup133Δ mutant.
To determine whether Apq12p is associated with nuclear pore complexes, an assay for clustering of NPC components in cells lacking the nucleoporin Nup133p was undertaken (Doye et al. 1994; Heath et al. 1995; Belgareh and Doye 1997). In wild-type cells, GFP-labeled nucleoporin Nup45p appeared as a ring structure surrounding the nuclear envelope (Fig. 3B; data not shown). In contrast, in nup133Δ cells, Nup45p–GFP clusters into a single bright focus adjacent to nuclear DNA (Fig. 3B; data not shown). When the subcellular distribution of Apq12p–GFP was evaluated in nup133Δ cells, the staining pattern remained ring-shaped and was indistinguishable from that observed in wild-type cells (Fig. 3B). These data suggest that although Apq12p is localized to the nuclear envelope/ER membrane, it is not strictly associated with nuclear pore complexes embedded within the nuclear membrane. Despite this observation, these findings are similar to those observed for Brr6p, a nuclear-envelope integral membrane protein implicated in nucleocytoplasmic transport (de Bruyn Kops and Guthrie 2001).
Apq12p is required for appropriate mRNA metabolism
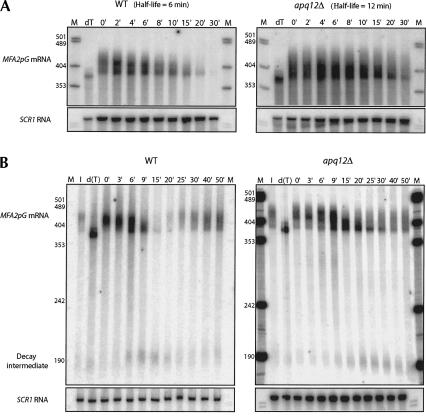
Several transport mutants that demonstrate hyperadenylated mRNA have been shown also to influence RNA metabolism and manifest retention at the site of transcription (Hilleren et al. 2001; Jensen et al. 2001). We therefore evaluated a possible role for APQ12 in RNA catabolism. In yeast, mRNA turnover is initiated by deadenylation of the RNA poly(A) tail, followed by decapping, and exonucleolytic decay of the transcript body in a 5′ →3′ direction (for review, see Coller and Parker 2004). We used both a transcriptional shut-off and pulse-chase experimental protocol to measure mRNA half-life and the mode of decay of a reporter mRNA in apq12Δ cells. Wild-type and apq12Δ strains were transformed with plasmid DNA encoding MFA2 under control of an inducible galactose (GAL1) promoter and harboring an insertion within the MFA2 3′ UTR that encodes 18 guanosine residues [poly(G) tract], which facilitates determination of the directionality of mRNA degradation (Decker and Parker 1993). For the transcriptional shut-off experiment, cells were grown in the presence of galactose to mid-log phase, at which time glucose was added to achieve transcriptional repression of GAL1–MFA2pG expression, and cell aliquots were removed at various time points. Northern blot analysis of the reporter RNA demonstrated that MFA2pG mRNA was stabilized approximately twofold in the apq12Δ mutants (Fig. 4A). The half-life of MFA2pG mRNA from wild-type cells was 6 min, similar to that observed previously (Decker and Parker 1993). In contrast, MFA2pG mRNA half-life in apq12Δ cells was 12 min. The increase in mRNA half-life in the apq12Δ strain is consistent with a kinetic delay in the export of mRNA from the nucleus (Hilleren and Parker 2001).
FIGURE 4.
APQ12 is required for appropriate mRNA metabolism. (A) Half-life determination and (B) transcriptional pulse-chase analysis of MFA2pG mRNA from either wild-type (WT) or apq12Δ cells. Steady-state mRNA (t = 0) subjected to oligo(dT)/RNase H cleavage (dT) represents a fully deadenylated mRNA. RNA isolated from cell aliquots removed prior to induction of transcription of the MFA2pG reporter mRNA is indicated (I). Time points are specified above each lane in minutes. The relative positions of the full-length polyadenylated mRNA, nonadenylated species, and the decay intermediate are indicated at the left in B. Half-life determinations in A were made by quantitating MFA2pG mRNA levels standardized to an SCR1 RNA loading control.
To determine the mode of decay of MFA2pG mRNA in apq12Δ strains, we examined its metabolism by a transcriptional pulse-chase experiment, wherein carbon source regulation of the GAL1 promoter is used to create a synchronous pulse of transcripts whose fate can be evaluated throughout its lifetime (see Materials and Methods). Analysis of the plasmid-encoded MFA2pG mRNA by the transcriptional pulse-chase protocol revealed a temporal lag in mRNA metabolism in apq12Δ stains as compared with wild type. Specifically, the MFA2pG mRNA from apq12Δ cells deadenylated more slowly (cf. 9-min time points, e.g.), and the decay intermediate of mRNA degradation required more time to accumulate. Interestingly, the delay in RNA degradation is less severe than observed for the temperature-dependent lethal mutations, rat7-1, rat8-1, and mex67, where a hyperadenylated mRNA pool is produced that persists for prolonged periods of time (>60 min; Hilleren and Parker 2001). The intermediate mRNA metabolism phenotype could explain why apq12Δ cells are viable.
Pre-mRNAs that are not fully spliced do not exit the nucleus (Legrain and Rosbash 1989; Rain and Legrain 1997) and may, presumably, influence the export of all mRNA, including, non-intron-containing mRNA such as MFA2 mRNA. We evaluated whether deletion of APQ12 elicits an influence on pre-mRNA splicing through assessing the accumulation of pre-U3 snoRNA. We observed elevated levels of pre-U3 snoRNA in the splicing-defective strains, lsm6Δ or lsm7Δ, as previously demonstrated (Mayes et al. 1999). However, we were unable to observe an accumulation of pre-U3 snoRNA in apq12Δ cells (data not shown), suggesting that the efficiency of splicing is not affected in this strain. The absence of a splicing deficiency in the apq12Δ strain argues that the observed mRNA export defect is not caused by inhibition of pre-mRNA splicing.
Conclusion
Several observations illustrate that the Apq12 protein affects mRNA export in yeast. First, apq12Δ strains accumulate poly(A)+ mRNA in the nucleus as assessed by in situ hybridization (Fig. 2). Second, an apq12Δ strain manifests hyperadenylated mRNAs, similar to other mRNA export factor mutants (Fig. 1A,B). Third, the subcellular distribution of Apq12p is perinuclear, a location consistent with a role in mRNA export (Fig. 3). Finally, apq12Δ strains show defects in mRNA metabolism consistent with a delay in export of the RNA to the cytoplasm. Interestingly, apq12Δ mutants are also severely compromised for growth and display atypical cell morphology. Because this aberrant cell morphology is not seen with other viable export mutants, Apq12p must have either an additional cellular function, or preferentially impinge on the export of mRNAs regulating cell growth; future work can be directed at addressing these hypotheses.
MATERIALS AND METHODS
Yeast strains and RNA analyses
Total cellular mRNA was isolated from the ~5000 EUROSCARF nonessential gene knockout strain collection. All cells carry the genotype MATa, his3Δ, leu2Δ, met15Δ, ura3Δ, and, for these experiments, a single gene replacement of APQ12, SAC3, PAN2, HPR1, PLC1, or NUP133 (Winzeler et al. 1999). Cells were cultured in YPD at 24°C and harvested at mid-log phase (OD600 = 0.3–0.4). Isolation of total cellular RNA was performed as previously described (Decker and Parker 1993). Alterations in size distribution for each RNA sample were determined by Northern blot analysis of 40 μg of total RNA electrophoresed through 6% polyacylamide gels. Endogenous MFA2 mRNA was visualized by PhosphorImager analysis (Typhoon 9410; Amersham Biosciences) of nylon membranes (Schleicher and Schuell) probed with uniformly [32P]-radiolabeled DNA complementary to the entire MFA2 coding region (Decker and Parker 1993).
Transcriptional shut-off and pulse-chase experiments were performed as previously described (Caponigro et al. 1993; Decker and Parker 1993). Briefly, yeast were transformed with pRP485, harboring MFA2pG under control of the inducible GAL1 UAS. For the transcriptional pulse-chase protocol, cells were grown in minimal media supplemented with 2% raffinose/1% sucrose (pH 6.5) to OD600 = 0.4, placed in 2% galactose for an 8-min “induction period,” and harvested, and cell aliquots were taken at time points after the immediate addition of minimal media supplemented with 4% glucose. RNA was extracted and monitored by PAGE and Northern blot analysis.
Fluorescence in situ hybridization (FISH) of poly(A)+ RNA
The subcellular distribution of poly(A)+ RNA was examined by in situ hybridization using a digoxigenin-conjugated oligo(dT) probe and indirect immunofluorescence microscopy (Corbett et al. 1997). Briefly, oligo(dT)50 (MWG Biotech) was 3′ end-labeled with digoxigenin (Roche Applied Science), and rhodamine-conjugated anti-digoxigenin Ab (1:200 dilution; Roche Applied Science) was used to visualize poly(A)+ RNA in cells grown to mid-log phase at 24°C.
Microscopic techniques
To examine the localization of Apq12p when expressed at endogenous levels, the chromosomal APQ12 reading frame in wild-type and nup133Δ cells was fused in-frame to GFP to generate APQ12–GFP using published recombinant methods (Longtine et al. 1998). The C-terminally tagged Apq12p–GFP chimera was localized by directly viewing the fluorescence signal in log-phase cells through a GFP-optimized filter (Chromas) using a Leica DM-RXA microscope equipped with a mercury xenon light source. Images were captured by a Retiga EX (Q Imaging) digital camera and manipulated using Metamorph 6.0 software. Analysis of poly(A)+ RNA by FISH was achieved using a rhodamine-optimized filter (Chromas). Cells were stained with 4′,6-diamidino-2-phenylindole-dehydro-chloride (DAPI) to detect chromatin and visualized using a DAPI-optimized filter (Chromas).
The C-terminally tagged Nup45p–GFP plasmid construct (pAC342; Jones et al. 2000) was obtained from the laboratory of Anita Corbett. Constructs for the evaluation of protein trafficking [NLS(GFP)3 and NLS/NES(GFP)2] were obtained from Christine Guthrie (Stade et al. 1997; de Bruyn Kops and Guthrie 2001).
Acknowledgments
We thank Yoshikazu Ohya and Daisuke Watanabe for communicating results prior to publication and the laboratories of Christine Guthrie and Anita Corbett for generously providing plasmids. We also thank the following individuals for expertise and assistance during the course of this work: Denise Muhlrad, Robin Staples, Ujwal Sheth, Anne Webb, Bettsy Garcia, and Pat Hilleren. In addition, we thank Meenakshi Kshirsagar for assistance with database analysis, and the entire Parker lab for their contributions to this work. Lastly, we thank Paul Muhlrad, Nicole Washington, and Sam Ward for their assistance with various microscopic techniques. This work was supported by funds from the Howard Hughes Medical Institute and the National Institutes of Health (NIH grant GM45443).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7420504.
REFERENCES
- Amberg, D.C., Goldstein, A.L., and Cole, C.N. 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 6: 1173–1189. [DOI] [PubMed] [Google Scholar]
- Belgareh, N. and Doye, V. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck, R., Tarun Jr., S., Rieger, M., Deardorff, J.A., Muller-Auer, S., and Sachs, A.B. 1996. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271: 432–438. [DOI] [PubMed] [Google Scholar]
- Brown, C.E., Tarun Jr., S.Z., Boeck, R., and Sachs, A.B. 1996. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5744–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., Muhlrad, D., and Parker, R. 1993. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Mol. Cell. Biol. 13: 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M. and Paker, R. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73: 861–890. [DOI] [PubMed] [Google Scholar]
- Corbett, A.H., Koepp, D.M., and Silver, P.A. 1997. Indirect immunofluorescence in Saccharomyces cerevisiae. In Cell biology: A laboratory manual (ed. David Spector), pp. 369–382. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- de Bruyn Kops, A. and Guthrie, C. 2001. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 20: 4183–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- Doye, V., Wepf, R., and Hurt, E.C. 1994. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13: 6062–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, T., Strasser, K., Racz, A., Rodriguez-Navarro, S., Oppizzi, M., Ihrig, P., Lechner, J., and Hurt, E. 2002. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21: 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, C.V., Copeland, C.S., Amberg, D.C., Del Priore, V., Snyder, M., and Cole, C.N. 1995. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol. 131: 1677–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P. and Parker, R. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7: 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898. [DOI] [PubMed] [Google Scholar]
- Jones, A.L., Quimby, B.B., Hood, J.K., Ferrigno, P., Keshava, P.H., Silver, P.A., and Corbett, A.H. 2000. SAC3 may link nuclear protein export to cell cycle progression. Proc. Natl. Acad. Sci. 97: 3224–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Hitomi, M., Chen, S., and Tartakoff, A.M. 1994. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell 5: 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain, P. and Rosbash, M. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583. [DOI] [PubMed] [Google Scholar]
- Lei, E.P., Stern, C.A., Fahrenkrog, B., Krebber, H., Moy, T.I., Aebi, U., and Silver, P.A. 2003. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell 14: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Mayes, A., Verdone, L., Legrain, P., and Beggs, J. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18: 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, R., Watkins, J.L., and Wente, S.R. 1996. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell 7: 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., Mulholland, J., Kaiser, C.A., Orlean, P., Albright, C., Rose, M.D., Robbins, P.W., and Botstein, D. 1991. Structure of the yeast endoplasmic reticulum: Localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast 7: 891–911. [DOI] [PubMed] [Google Scholar]
- Rain, J. and Legrain, P. 1997. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 16: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., Guerra, C.E., Lampl, M., Gogg, G., Kohlwein, S.D., and Klein, H.L. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19: 3415–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Luhrmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., Ford, C.S., Guthrie, C., and Weis, K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050. [DOI] [PubMed] [Google Scholar]
- Strasser, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Lesage, G., Bader, G.D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G.F., Brost, R.L., Chang, M., et al. 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.I., and Parker, R. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- York, J.D., Odom, A.R., Murphy, R., Ives, E.B., and Wente, S.R. 1999. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285: 96–100. [DOI] [PubMed] [Google Scholar]