Abstract
The emergence of viral escape mutants is usually a highly undesirable phenomenon. This phenomenon is frequently observed in antiviral drug applications for the treatment of viral infections and can undermine long-term therapeutic success. Here, we propose a strategy for evaluating a given antiviral approach in terms of its potential to provoke the appearance of resistant virus mutants. By use of Qβ RNA phage as a model system, the effect of an antiviral gene therapy, i.e., a virus-specific repressor protein expressed by a recombinant Escherichia coli host, was studied over the course of more than 100 generations. In 13 experiments carried out in parallel, 12 phage populations became resistant and 1 became extinct. Sequence analysis revealed that only two distinct phage mutants emerged in the 12 surviving phage populations. For both escape mutants, sequence variations located in the repressor binding site of the viral genomic RNA, which decrease affinity for the repressor protein, conferred resistance to translational repression. The results clearly suggest the feasibility of the proposed strategy for the evaluation of antiviral approaches in terms of their potential to allow resistant mutants to appear. In addition, the strategy proved to be a valuable tool for observing virus-specific molecular targets under the impact of antiviral drugs.
Ideally, antiviral strategies should be highly virus specific and should not affect the host organism. In addition, they should exhibit a minimal potential to allow viral escape mutants to appear. In this paper, we propose a general strategy to evaluate a given antiviral approach with regard to resistance phenomena that may occur due to the emergence of escape mutants. Utilizing RNA phage Qβ as a model system, we investigated the potential of virus-specific translational repressor proteins to suppress viral propagation. Among the mechanisms that control gene expression, translational regulation is certainly one of the most specific and therefore should be a valuable target mechanism for antiviral strategies.
First observed in the RNA phages (29), translational repression has since been discovered in many bacteriophages (47), prokaryotes (11, 16, 45), and eukaryotes (46) as well as in several mammalian viruses (14, 17, 35, 44). Within the closely related RNA phage species R17, MS2, F2, fr, and Qβ, two proteins, namely, the coat protein and the replicase, are translational regulators of each other. Shortly after single-stranded viral RNA enters the host cell, replicase protein is translated from the proximal end of the phage genome, leading to high levels of replicated phage RNA.
However, it has been shown that replication cannot occur when phage RNA is being translated (25, 47). Therefore, strict translational regulation of the remaining cistrons, namely, A1/coat protein and A2, from the distal portion of the Qβ RNA genome is essential for the replication. Hence, blocking the translation of the coat protein, which is produced abundantly during the late phase of the viral infection cycle, suppresses translation of the entire RNA (51). During late infection, repression of replicase translation by growing numbers of coat proteins occurs naturally. This phenomenon is regarded as a means by which coat protein synthesis is increased in order to package all the RNA that has been produced.
The mechanism of repression by the coat protein has been studied in detail. In the case of Qβ, the coat protein binds a 27-base hairpin structure of the phage RNA covering the replicase start codon (50). Witherell and Uhlenbeck synthesized a 29-base RNA that contains the same sequence as that of the hairpin structure (53). Coat protein bound to this short RNA has an association constant (Ka) of 570 μM−1, which is comparable to the association constant value for coat protein when intact genomic Qβ RNA is complexed with it (Ka = 156 μM−1). Mutation analysis revealed that all eight base pairs of the hairpin structure are required for binding. The stem sequence, however, is not important for coat protein binding. In the 3-base loop, only the A residue at position +8 relative to the A (+1) of the replicase start codon is essential for coat protein binding. When A at +8 is replaced by G or U, the Ka is reduced by more than 100-fold. The same result was observed for loop sizes extended to 4 or more nucleotides. In summary, coat protein binding to Qβ RNA is a highly specific interaction. In addition to its regulatory function, the coat protein binding site of the viral RNA is also required for specific genome encapsidation. In fact, coat protein even at low concentrations will aggregate around phage RNA containing the binding site (9), leading to intracellular self-assembly of new phage particles.
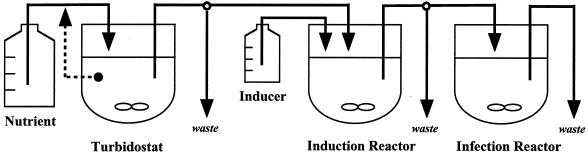
Translational suppression has been known for quite some time to be a natural strategy by which host cells defend themselves against virus infection (24). In addition, plasmid-encoded repressor proteins have been shown to down-regulate expression (31, 34). Berkhout and Jeang found that a prokaryotic RNA-binding protein is able to down-regulate human immunodeficiency virus type 1 gene expression (7). Thus, translational repression may have the potential to suppress viral propagation and may therefore serve as a novel antiviral strategy. In this paper, we make use of the RNA phage Qβ as a model system to show that its coat protein, a natural repressor of replicase translation, inhibits bacteriophage propagation if it is expressed by the host bacterium during early infection. To investigate the long-term efficacy of such an antiviral strategy, we grew phage continuously in a three-step bioreactor setup (Fig. 1). This system is based on the one described by Husimi and coworkers (22). In our system, F-piliated Escherichia coli host bacteria were grown continuously in a turbidostat under physiologically constant conditions. These host bacteria, which were preinduced to express large amounts of coat protein, served as a limiting “nutrient” which was continuously supplied to a number of phage infection reactors at dilution rates that largely exceeded the bacterial growth rate. By keeping host bacteria and phage culture(s) separate, coevolution of the host and the parasite, which may lead to the takeover of phage-resistant host cells (19, 30, 49), was prevented. In the case of normal host bacteria, bacteriophage particles are usually not diluted out under these conditions because their growth rate is much higher than that of the host cell. The system has already been used to serve different purposes: comparisons of the growth kinetics of phage with theoretical models (20, 26, 41), analysis of reversion kinetics starting with recombinant phage (21), production of deletion mutants by the growing of phage in a complementing host (26), and selection of RNase-resistant lytic phages (5). Furthermore, the system has been optimized to serve as a device for automated phage display experiments (41).
FIG. 1.
Three-stage continuous culture system for bacteriophages. Host cells were grown in a turbidostat. Host cell suspension and inducer stock solution were transferred into a stirred flow reactor called the induction reactor. The dilution rate was adjusted to reveal a mean residence time of approximately 40 min for host bacteria in this reactor. The suspension of induced host bacteria was finally transferred into as many as six stirred flow reactors called infection reactors. In these infection reactors, host bacteria became infected by bacteriophages.
In our study, a modified version of the original two-stage system was employed to continuously cultivate Qβ bacteriophage by use of recombinant E. coli host cells that produce plasmid-encoded phage coat protein. Phage mutants resistant to coat protein repression could be isolated after several days of culture. In 12 out of 13 cultures, bacteriophage managed to escape from the selection pressure of the translational repressor. In only one culture did bacteriophage become extinct. Sequence analysis of reverse-transcribed phage RNA revealed two distinct variants, each containing nucleotide base substitutions within the coat protein binding site.
MATERIALS AND METHODS
Bacterial and viral strains.
E. coli strain JM105 [thi rpsL endA sbcBC hsdR4 Δ(lac-proAB) (F′ traD36 proAB LacIqZΔM15)] (54) and vector pKK223-3 (8) were obtained from Stratagene (Heidelberg, Germany). Wild-type phage Qβ was kindly provided by C. K. Biebricher.
Fermentation medium.
The turbidostat fermentation medium contained 100 mM Tris-HCl (pH 7.4), 27 mM KCl, 37 mM NH4Cl, 1 mM K2HPO4, 2 mM trisodium citrate, 0.1 mM CaCl2, 2.5 mM MgSO4, 1 mg of FeCl2 · 4H2O, 2 g of glucose, 1 g of Casamino Acids (Oxoid), and 2 ml of vitamin solution (as described by Schlegel [39]) per liter. To induce gene expression prior to infection, the turbidostat suspension (2 × 108 cells/ml, corresponding to 12 nephelometric turbidity units) was transferred to a stirred flow reactor with a 50 mM stock solution of IPTG (isopropyl-β-d-thiogalactopyranoside), resulting in a 10:1 mixture of turbidostat culture to IPTG stock solution.
Phage titer measurements.
Phage titers were determined by counting the plaques of dilution series by the standard double-agar-layer plating method with E. coli JM105, with or without pBL410 as an indicator bacterium (2). All phage titers reported in this paper are mean values of at least two measurements.
Reverse transcription of Qβ RNA.
Qβ RNA was isolated from phage suspension by proteinase K digestion, phenol-chloroform extraction, subsequent ethanol precipitation, and resuspension in double-distilled water according to standard protocols (38). This phage RNA solution was used for subsequent reverse transcription reactions. Twenty-five microliters of a 20 μM solution of one of the coat protein primers (COAT2N, coat protein cistron, 5′-TCGACTTGGGTCGC-ATAACTTCGAAGCTCAGTATAG-3′; QCRT coat protein binding site, 5′-CTATGGCTTCGACAACGGAC-3′) was added to a 0.5-ml Eppendorf tube before being lyophilized and resuspended in 6.1 μl of phage RNA solution. After this solution was heated for 1 min at 90°C and then incubated for 2 min at room temperature, 45 μl of avian myeloblastosis virus reaction mixture (250 mM HEPES-Na [pH 7.9], 40% glycerol, 30 mM MgCl2, 200 mM KCl, 5 mM dithiothreitol [DTT], 500 μM [each] dATP, dGTP, dCTP, and dTTP) was added. The reaction was started by the addition of 2.5 μl of avian myeloblastosis virus reverse transcriptase (55 U). After 1 h at 42°C, the reaction was stopped by the addition of 50 μl of 100 mM EDTA (pH 8.0). Nucleic acids were purified by phenol extraction and ethanol precipitation, followed by lyophilization and resuspension in 10 μl of double-distilled water.
Construction of the expression vector for coat protein production.
The coat protein gene was PCR amplified from Qβ cDNA with the primers COAT1 (5′-CCGAATTCCATGGCAAAATTAGAGACTG-3′) and COAT2N and 1 μl of the purified reverse transcription product as the template in a PCR mixture with a total volume of 100 μl. The PCR product was cleaved with EcoRI and HindIII and ligated into the EcoRI- and HindIII-double-digested vector pKK223-3 by standard protocols (38). The identity of the ligation product, construct pBL410, was confirmed by DNA sequencing with a Genesis 2000 DNA analysis system (DuPont, Bad Homburg, Germany).
Expression of the coat protein.
E. coli JM105 transformed with pBL410 was grown in Trypticase yeast extract medium containing 100 μg of ampicillin per ml. At an A600 of 0.7, IPTG was added to a final concentration of 5 mM, and the bacterial suspension was further incubated at 37°C for 2 h. Cells were collected by centrifugation, and the pellets were frozen in liquid nitrogen. After thawing on ice, the cells were resuspended in 10 mM Tris-HCl (pH 8.0)-100 mM NaCl-1 mM EDTA, disrupted by lysozyme treatment (25 mg/ml for 30 min at room temperature), and sonicated (five bursts, 30 s and 80 W each, on ice). Cellular debris was removed by centrifugation at 10,000 rpm (SS34 rotor). Proteins were precipitated from the supernatant with 60% (wt/vol) ammonium sulfate and collected by centrifugation. The ammonium sulfate pellet was resuspended in 100 mM NaCl-10 mM Tris-HCl (pH 7.5)-0.1 mM MgSO4-0.01 mM EDTA. The suspension was then applied to a 1.6- by 40-cm column of Sepharose CL-4B (exclusion limit, 2 × 107 Da; Pharmacia-LKB, Freiburg, Germany) and eluted in the same buffer. The position of the elution of whole authentic Qβ virus had been previously determined for comparison purposes. Fractions of 5 ml each were collected. Aliquots of proteins present in each fraction were separated by denaturing polyacrylamide gel electrophoresis (28). Coat protein was visualized by Western blotting and Coomassie brilliant blue staining. Peak fraction 6 appeared to contain highly pure coat protein and was therefore chosen for further concentration with Centricon-30 columns (Amicon, Witten, Germany).
Electron microscopic analysis of virus-like particles formed from plasmid-produced coat protein.
A 7-μl drop (5 μg/μl) of purified fraction 6 obtained from Sepharose CL-4B chromatography of the coat protein was adsorbed onto a prepared carbon grid for 1 min. The grid was then immersed in 250 μl of 1% uranyl acetate solution for 2 min. Excess liquid was removed with filter paper, and the grid was allowed to air dry for 1 h. Images were recorded at 75 kV by a transmission electron microscope (Phillips Medical Systems).
Translational repression in batch culture.
E. coli strain JM105/pBL410 was cultivated in turbidostat fermentation medium. At an A600 of 0.7, the culture was distributed into two flasks and IPTG was added to one culture to a final concentration of 5 mM. Both cultures were then vigorously shaken at 37°C. After 1 h, bacteriophage Qβ was added to both cultures (multiplicity of infection, 1,000), followed by further incubation at 37°C. The efficacy of infection was analyzed by monitoring the A600.
Construction of a three-stage flow reactor for continuous cultivation of phage.
To investigate bacteriophage evolution under the selection pressure of host translational-repressor expression, we constructed a three-stage culture system (Fig. 1) based on the previously described two-stage system (41). Briefly, the system consists of a 2.5-liter turbidostat serving as a constant supply of receptive host cells that were grown at 30°C. The turbidity was adjusted to 12 nephelometric turbidity units (corresponding to approximately 2 × 108 E. coli cells ml−1) by means of an external turbidometer (Hach, Namur, Belgium) with a flow cell. In contrast to the previously described system (41), an additional, smaller flow reactor was positioned between the turbidostat and the phage infection reactors and served to induce coat protein expression by the host cells on addition of IPTG and to increase the incubation temperature to 37°C. This configuration accelerated the bacterial growth rate prior to their transfer into the infection reactors. The induction reactor medium consisted of a 10:1 mixture of the turbidostat culture and the 50 mM IPTG stock solution. To achieve sufficient induction of coat protein expression, the flow rate and volume of the induction flow reactor were adjusted to allow a mean residence time of approximately 40 min for host bacteria inside the induction reactor. The analysis of batch cultures described above had indicated that expression reached approximately 50% of the maximal level after 40 min of induction (data not shown). Induced host cells were transferred via a manifold to a maximum of six stirred flow reactors (infection reactors) operated in parallel for the propagation of bacteriophage. The infection reactor volumes were adjusted to 20 ml each. The flow rates through the infection reactors were regulated to yield a dilution rate of 1.5 h−1. Samples (100 μl) were periodically withdrawn from the infection reactors. One aliquot of each sample was immediately used to determine the phage titer, while another aliquot was frozen in liquid nitrogen and stored at −20°C for subsequent (sequence) analysis.
Sequence analysis of bacteriophage populations from infection reactors.
An aliquot (100 μl) of the infection reactor suspension was centrifuged at 10,000 rpm for 15 min at 4°C (SS34 rotor). Bacteriophage RNA was then isolated from the supernatant, reverse transcribed, and PCR amplified with the primers QCRT and QBCOAT3B (5′-CAAGCCGTGATAGTCGTTCC-3′), as described above. Sequence analysis was performed with a Genesis 2000 DNA analysis system (DuPont) according to standard protocols (6).
Filter binding assays.
Qβ coat protein was prepared by a modification of the cold acetic acid method (53). Qβ phage (2.53 mg) in 6 ml of cold 66% acetic acid was vortexed for 30 s every 10 min for 1 h. Qβ RNA was precipitated by centrifugation at 10,000 rpm for 20 min at 4°C (SS34 rotor). The supernatant was removed and dialyzed overnight at 4°C against three 1-liter changes of 1 mM acetic acid-10 mM DTT (pH 3.2). The coat protein was stored at 4°C at a concentration of 2.8 μM in 1 mM acetic acid-10 mM DTT (pH 3.2).
Wild-type, cbsmut 1, and cbsmut 2 RNAs were prepared by in vitro transcription with the T7 RNA polymerase system. Double-stranded DNA templates for in vitro transcription were generated by cloning double-stranded DNA cassettes into the EcoRI-digested pUC19 vector. Each cassette comprised the T7 promoter and the sequence from position −11 to +66 relative to the A of the replicase start codon of the respective Qβ RNA variant. Each cassette also contained EcoRI-compatible overhangs at the 5′ ends, so ligation resulted in only one functional EcoRI site at the end of the viral sequences, which then could be used for vector linearization. In vitro transcription of the EcoRI-linearized vector was carried out according to standard methods (32). Equilibrium binding constants were determined by the two-step procedure described previously (53). To calculate the fraction of active coat protein, binding of excess RNA was analyzed at concentrations of 50 nM for the coat protein dimer and 10 to 60 nM for RNA. Binding of excess protein was examined at concentrations of 10 pM for RNA and 0.5 μM to 0.16 nM for coat protein. The protein concentration at half saturation is equal to the inverse of the equilibrium binding constant for the reaction if the fraction of protein active in RNA binding is known (53).
RESULTS
Construction of the expression vector.
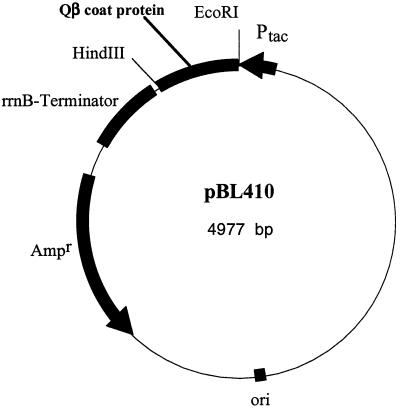
The construct which was used in this study, pBL410, is depicted in Fig. 2. It was constructed by ligation of the entire PCR-amplified coat protein gene from bacteriophage Qβ between the tac promoter and the strong terminator rrnBT1 T2 of plasmid pKK223-3. Coat protein expression from the new construct could therefore be induced by the addition of IPTG to the fermentation medium.
FIG. 2.
Plasmid map of construct pBL410. The gene of Qβ coat protein was amplified by PCR and cloned into the EcoRI and HindIII sites of vector pKK223-3 (8). The tac promoter and the concomitant rrnB terminator are designated Ptac and rrnB, respectively. ori, origin of replication
Purification and analysis of plasmid-produced coat protein.
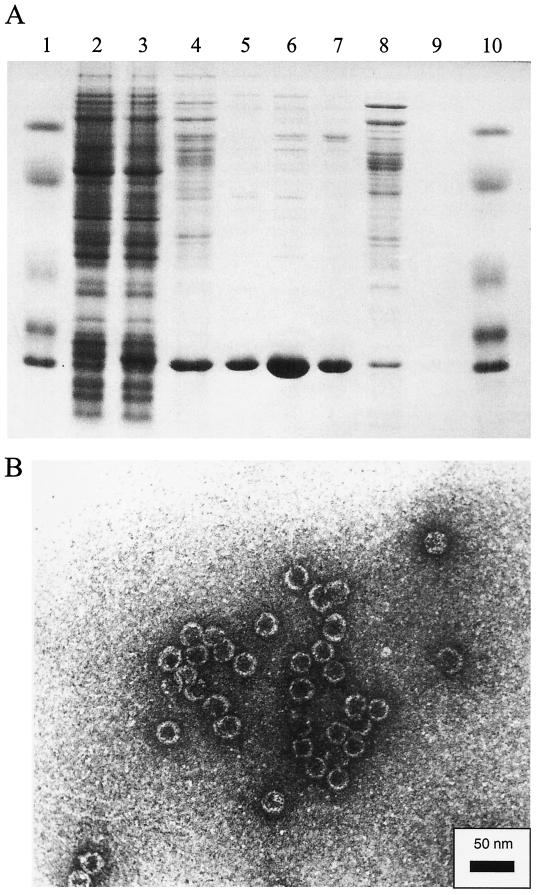
The construct pBL410 permits expression of soluble Qβ phage coat protein. To prove that biologically active coat protein is produced in bacteria harboring plasmid pBL410, we purified the coat protein from a batch culture of JM105/pBL410 that had been induced with IPTG by following a modified version of the protocol of Peabody (34). Briefly, the ability of the plasmid-encoded coat protein molecules to form capsids of a size and appearance similar to those of authentic Qβ phages provides a basis for both purification of the protein by gel filtration on Sepharose CL-4B and identification of biological activity by electron microscopy. Qβ coat protein eluted at approximately the same position as that of whole Qβ phage (data not shown). When subjected to denaturing polyacrylamide gel electrophoresis, however, the coat protein showed up as a strong band of the expected size of the monomer. Figure 3A indicates the purity of the coat protein at each stage of the purification procedure. To demonstrate that plasmid-produced coat protein is in its native, biologically active conformation, we analyzed purified coat protein preparations by electron microscopy. Figure 3B shows an electron micrograph of virus-like particles from purified coat protein. It seems reasonable to assume that only properly folded coat protein molecules are capable of forming virus-like capsids, as the formation of a capsid requires multiple intersubunit contacts. Hence, we conclude that plasmid-produced coat protein is in its native state.
FIG. 3.
(A) Results of denaturing polyacrylamide gel electrophoresis of the purified coat protein from host cells harboring plasmid pBL410. Lanes: 1 and 10, protein markers (66, 45, 24, 18.4, and 14.3 kDa); 2, cell extract of JM105/pBL410 without induction by IPTG; 3, cell extract of JM105/pBL410 with induction by IPTG; 4 and 8, protein fraction after precipitation with ammonium sulfate; 5, 6, and 7, fractions 5, 6, and 7 from size exclusion chromatography on Sepharose CL-4B, respectively. The coat protein monomer has a molecular mass of 16.9 kDa (52). (B) Electron micrograph of virus-like particles from purified coat protein. Coat protein isolated from E. coli JM105 cells harboring plasmid pBL410 spontaneously formed virus-like capsids that were purified by size exclusion chromatography. The virus-like particles are similar in size and shape to Qβ virions and capsids synthesized in a previous work (27).
Analysis of the effect of coat protein expression on bacteriophage infection.
The observation that plasmid-encoded coat protein is in its native conformation suggests that it should act as a translational repressor in vivo, as has been previously shown with the MS2 system (34). We therefore compared the levels of growth of E. coli JM105/pBL410 with and without induction of coat protein expression by IPTG after infection with Qβ phage. A bacterial culture was grown until it reached an A600 of 0.7. The culture was then equally divided, and after addition of IPTG to one of the cultures, both cultures were incubated at 37°C for 1 h. Finally, Qβ phage was added to both cultures and the optical density was monitored. While the noninduced culture showed a dramatic decrease in the A600 after about 20 min postinfection, the A600 of the induced culture further increased, indicating that bacteria continued to divide and did not lyse (data not shown). Plating of different concentrations of wild-type Qβ phage on both induced and noninduced JM105/pBL410 as the host bacterium also showed a significant difference. The amount of phage that produced 5.6 × 109 plaques on noninduced host bacteria generated only one plaque on induced host bacteria. The differences were even more pronounced when induced E. coli JM105/pBL410 was compared with plasmid-free JM105 host cells. In this case, the ratio of the number of PFU obtained from the former to that obtained from the latter was smaller than 1:1011.
Evolution of bacteriophage in continuous culture.
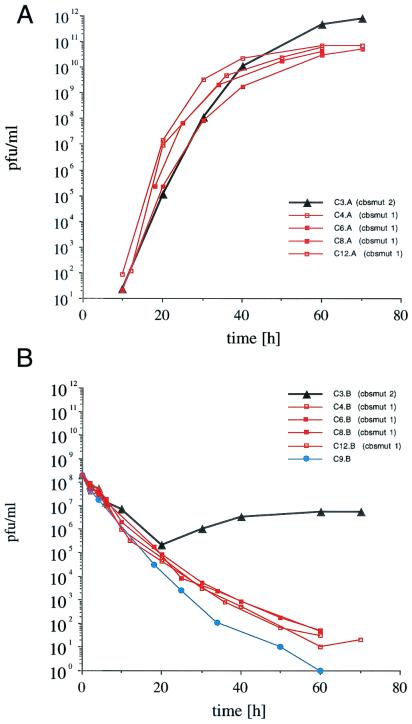
Three experiments were carried out to investigate the evolution of phage Qβ under the selection pressure of viral coat protein expression by the host bacteria. In each experiment, one infection reactor was operated without phage infection to monitor cross-contamination. Figure 4 shows the time course of phage titers in six representative infection reactors. Phage titers were determined with induced E. coli JM105/pBL410 and plasmid-free JM105. During the first 20 h, the bacteriophage titers in all infection reactors decreased considerably. However, in only 1 out of 13 populations did this trend continue until bacteriophage had been diluted out completely. The remaining 12 bacteriophage populations overcame the selection pressure of the translational repressor protein and grew to steady-state titers of 5 × 1010 to 7 × 1010 PFU/ml (11 populations; CBSMUT1) or 8.2 × 1011 PFU/ml (1 population; CBSMUT2), as determined with induced indicator bacteria. Phage titers determined with plasmid-free indicator bacteria were as low as 21 PFU/ml, except in the case of the latter culture (cbsmut 2), where a titer of 5.6 × 106 PFU/ml was observed.
FIG. 4.
Time courses of phage titers in six representative infection reactors. Phage titers were determined with induced E. coli JM105/pBL410 (A) and plasmid-free JM105 (B). Depicted are the time courses for one population that revealed a variant of the major class (cbsmut 1) of escape mutants (squares), four populations that yielded the cbsmut 2 mutant (triangles), and one population that died out during the time course of the experiment (circles).
Sequence analysis of mutant populations evolved in continuous culture.
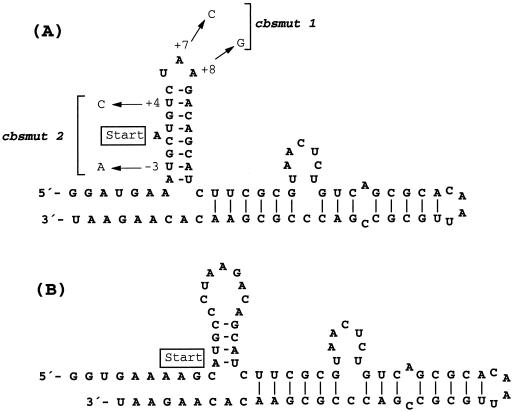
To determine whether differences (or similarities) in steady-state titers reflected differences (or similarities) at the sequence level, phage RNAs from all 12 escape populations were sequenced. Only two types of coat protein binding site mutants were isolated. As shown in Fig. 5A, the mutations were superimposed on the secondary structure of the coat protein binding site. The CPSMUT1 mutant (containing the cbsmut 1 gene) appeared 11 times. It showed a double mutation (C at position +7 and G at position +8 relative to the replicase start codon) in the 3-base loop of the hairpin structure essential for coat protein binding (53). In contrast, the unique CPSMUT2 mutant (containing the cbsmut 2 gene) revealed two more distant mutations (A at position −3 and C at position +4) that were both in the stem region of the hairpin. As the mutations could also affect the local secondary structure of the coat protein binding site, an RNA secondary-structure calculation based on a genetic algorithm (48) implemented into the STAR program package (1) was carried out. In the case of the CBSMUT1 mutant, a secondary structure that was identical to that of the wild type was predicted. For the CBSMUT2 mutant, however, an alternative structure (Fig. 5B) was suggested by the algorithm. In comparison with the wild-type secondary structure, this alternative structure had a calculated difference in free energy of 7.9 kcal/mol. Possible non-Watson-Crick pairs, e.g., G-A pairs, were not considered. At the level of the amino acid sequence, the cbsmut 1 mutations are conservative: the replicase lysine 3 is changed to arginine. In the CBSMUT2 mutant, the replicase proline 2 is changed to a serine. The second mutation in CBSMUT2 is located in the untranslated region 5′ of the replicase start codon. To investigate whether second-site mutations in the coat protein occurred in the CBSMUT1 and CBSMUT2 mutants, the coat protein genes of several isolates were also sequenced. However, no mutations were detected (data not shown).
FIG. 5.
Putative secondary structure of the coat protein binding site. (A) The local secondary structure of Qβ wild-type RNA is depicted. The sequence shown corresponds to the transcripts used for binding studies in vitro, except that an additional U is present at the 3′ terminus. Arrows indicate the point mutations found in the cbsmut 1 and cbsmut 2 variants; the start codon of the replicase gene is also indicated. (B) For the cbsmut 2 variant, an alternative fold can be calculated by standard methods (1).
Binding of Qβ coat protein to mutant RNAs.
To investigate the effect of mutations in CBSMUT1 and CBSMUT2 on Qβ coat protein binding, we determined the equilibrium association constants of mutant RNAs as described previously (53). In contrast to the previous work (53), we used a 77-base fragment transcribed from synthetic DNA for coat protein binding assays. This fragment was chosen to cover the sequence portion necessary to form the alternative structure shown in Fig. 5B. If we assume, as other researchers have done (9, 15, 36, 37), that each Qβ coat protein dimer binds a single RNA hairpin, then the Ka of the wild-type 77-base fragment is 510 μmol−1. This corresponds well to the Ka of 570 μmol−1 previously reported for a 29-base RNA fragment (53). The introduction of mutations C at position +7 and G at position +8 (cbsmut 1) or A at position −3 and C at position +4 (cbsmut 2) reduced the Ka below the detection limit of the assay (<12 μmol−1).
DISCUSSION
The evolution of viral escape mutants is frequently observed as a consequence of the application of antiviral therapies with a single virus-specific drug or even combinations of such drugs. This phenomenon severely constrains therapeutic success. It is therefore important to assess the potential for such mutants to evolve and proliferate under a novel antiviral strategy. In addition, it would be highly desirable to understand the mechanisms by which resistance is acquired. In this paper, we report a strategy for evaluating the propensity of a given antiviral agent to promote viral escape mutants. We demonstrate how a population of the RNA phage Qβ evolves under the selection pressure of a virus-specific translational repressor protein expressed by host bacteria.
The expression of biologically active coat protein in vivo was demonstrated by electron microscopic analysis of capsids forming spontaneously by use of purified bacterially expressed coat protein. When coat protein expression was induced, bacteria suppressed phage Qβ propagation, as deduced by (i) optical density measurements of bacterial suspension infected with phage and (ii) the comparison of plating efficiencies between induced and noninduced host bacteria. To study the evolutionary escape of bacteriophage from the selection pressure of the host-produced antiviral translational repressor, wild-type phage Qβ was cultivated in a three-stage continuous culture reactor with recombinant host bacteria expressing viral coat protein. Among 13 different phage populations analyzed, only 1 was not able to overcome the novel selection pressure. Twelve populations succeeded in adapting to the selection pressure. PFU counts with coat protein-expressing strain E. coli JM105/pBL410 (Fig. 4A) were significantly higher than those determined with plasmid-free JM105 (Fig. 4B). In the case of the CBSMUT2 mutant, plaque counts differed by approximately 5 orders of magnitude; for the CBSMUT1 mutant, plaque counts differed by as much as 9 orders of magnitude. This result may reflect the differences in plating efficiencies, i.e., different levels of dependence on the coat protein-expressing host bacteria. The observation of plaque count differences with two different host bacteria, however, is also consistent with the observation of differences in the population structures between the CBSMUT1 and CBSMUT2 mutants, e.g., different fractions of well-adapted clones. In this case, however, a future detailed analysis of single phage clones is necessary to clarify this point beyond speculation.
The molecular basis for viral adaptation appears to stem from mutations within the coat protein binding site of the viral genomic RNA. No mutations were found in the coat protein genes themselves, and we did not expect to find mutations or deletions such as those observed, e.g., in T7 phages growing on a complementing host (26). The read-through protein A1 and therefore the intact viral coat protein sequence are essential for assembly and viability (3, 18). In addition, translation of the viral coat/A1 protein is a prerequisite to translational initiation of viral replicase (4, 23, 43). The observed mutations do not seem to affect the replicase gene product. In the first place, only conservative changes in the mutated codons could be traced. In addition, previous work has shown that the amino terminus is much more permissive of amino acid insertions and substitutions than is the replicase core region (33).
One mutation in the cbsmut 2 variant of Qβ phage is located in the 5′ untranslated region upstream of the replicase start codon. This change, however, is unlikely to interfere with the ribosomal entry site (42) of the replicase gene. However, mutations may alter RNA structure, in particular the coat protein binding site. The CBSMUT1 mutant contains an A→C mutation at position +8 relative to the A of the replicase start codon. Witherell and Uhlenbeck have previously shown that an A residue at this position is the most important sequence constraint for coat protein binding in the context of the Qβ system (53). As anticipated, coat protein binding is greatly reduced in the CBSMUT1 mutant. The equilibrium association constant Ka measured for the 77-base fragment of the cbsmut 1 coat protein binding site was reduced more than 42-fold compared to that for the corresponding wild-type fragment. Assuming a comparable reduction in the affinity of the full-length genomic RNA for the coat protein, it may be surmised that translational repression is less effective in the case of the CBSMUT1 mutant.
The interpretation of sequence variations in the CBSMUT2 mutant is somewhat less obvious. Here, mispairings are introduced in the hairpin at positions −3 and +4. Previous work has revealed that full binding of the coat protein requires the intact 8-bp hairpin from nucleotides −4 to +16 but that a detectable level of binding can be achieved with nucleotides −2 to +14, encompassing the upper 6-bp hairpin (53). Mutations in the CBSMUT2 mutant may therefore destabilize the hairpin and hence decrease the affinity for the coat protein. To further examine this possibility, secondary-structure predictions of wild-type and mutant RNAs were carried out. Whereas wild-type and cbsmut 1 RNAs were predicted to fold into the same structure (Fig. 5A), cbsmut 2 RNA revealed an alternative structure (Fig. 5B). In this alternative structure, the central hairpin lacks two features that have proved to be important for Qβ coat protein binding sites. A loop size of more than 3 nucleotides and a shortened stem region should prevent the coat protein from binding to this structure. As expected, coat protein binding is greatly reduced in the CBSMUT2 mutant. The Ka measured for the 77-base fragment of the cbsmut 2 coat protein binding site was reduced below the detection limit of the assay. Hence, translational repression may also be less effective in vivo. This fact would, however, have consequences for the regulation of replicase translation. Either regulation of replicase translation at late stages of the viral life cycle is not absolutely vital for virus propagation or coat protein-mediated shutoff of replicase translation in the CBSMUT1 and CBSMUT2 mutants appears at the significantly higher intracellular coat protein concentrations provided by both host-born and virus-born coat protein synthesis. The latter idea is supported by two findings. First, efficient virus propagation became dependent on a coat protein-producing host, as indicated by the different steady-state titers for different host bacteria (Fig. 4). Second, as already mentioned, CBSMUT1 and CBSMUT2 mutants kept an intact coat/A1 protein gene sequence, i.e., no mutations or deletions occurred.
Evolutionary processes are inherently stochastic (12). It is thus not surprising that this attribute is illustrated by our observation that two different molecular adaptations evolved that solved the problem of translational repression but that one population did not adapt and thus died out over the course of the experiment. It is, however, notable that one molecular adaptation like that in the CBSMUT1 mutants is realized much more frequently than the other. Cross-contamination can be excluded as a possible cause, since control populations that were not infected remained uninfected throughout the experiments.
In both CBSMUT1 and CBSMUT2 mutants, two mutations were observed. It would be very interesting to determine whether the paired mutations occurred simultaneously or sequentially and in different orders in the different experiments. Preliminary sequence analysis of populations at 30 h, however, did not reveal any indication of single-point mutants at that time point. This finding therefore supports the idea of simultaneous mutations. Alternatively, one may argue in favor of a selection process rather than evolution. In this case, one has to assume that a small number of well-adapted mutants was already present in the initial phage population. Two arguments support this idea. First, virus populations are quasispecies that contain a vast fraction of sequences that differ from the master sequence (10, 13). Second, plating efficiencies determined for the initial phage population indicate that for the 5.6 × 109 plaques that were obtained with noninduced JM105/pBL410 host bacteria, the same amount of phage produced at least one plaque with an induced host. Both scenarios could in principle be tested in future experiments, as selection and evolution processes exhibit different dependencies on initial population size. While smaller populations would slow down adaptation in evolution, in a pure selection procedure, smaller populations would reduce the probability of any survival at all. This means that within the scope of the experiments described herein, fewer populations would be expected to overcome the selection pressure.
Our results indicate that viral populations evolve to incorporate adaptations that enable them to overcome antiviral strategies. This phenomenon can be analyzed over the course of multiple viral generations. Wild-type viruses, which are unable to replicate in hosts that express a virus-specific translational repressor, mutated to variants that replicated with high efficiency and became dependent on the altered host conditions. The mechanism of resistance acquisition can be analyzed at the molecular level. In the context of evolution, such experiments therefore may help to define the Achilles heel of a virus, i.e., conservative genomic regions that need to remain constant to yield infectious viral particles. Preliminary testing of the replicator strategy (40) indicates that, e.g., targeting the replication machinery of the virus may indeed prevent resistance acquisition. In summary, the strategy of generating escape mutants by continuous cultivation of viruses is a useful tool for designing antiviral strategies and thus accelerates the development of more efficient therapies.
Acknowledgments
We thank M. Eigen for many fruitful discussions and in particular for providing a very stimulating working atmosphere. We also very much appreciate the discussions with C. K. Biebricher, who provided us with the Qβ phage, and M. Gebinoga. M. Friedlein, R. Weise, and W. Simm assisted in building the system for continuous bacteriophage cultivation. M. Sjödin assisted in writing the control software of the bioreactor system. K. Weber and M. Osborn introduced us to electron microscopy.
This work was in part supported by grants 031 6420 A-7.A, BEO 0310665, and BioFuture 0311852 from the Bundesministerium für Forschung und Technologie.
REFERENCES
- 1.Abrahams, J. P., M. van den Berg, E. van Batenburg, and C. Pleij. 1990. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 18:3035-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, M. H. 1959. Bacteriophages. Wiley Interscience, New York, N.Y.
- 3.Arora, R., C. Priano, A. B. Jacobson, and D. R. Mills. 1996. cis-acting elements within an RNA coliphage genome: fold as you please, but fold you must. J. Mol. Biol. 258:433-446. [DOI] [PubMed] [Google Scholar]
- 4.Ball, L. A., and P. Kaesberg. 1973. A polarity gradient in the expression of the replicase gene of RNA bacteriophage Qβ. J. Mol. Biol. 74:547-562. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, G., J. S. McCaskill, H. Otten, and A. Schwienhorst. 1989. Evolution im Laboratorium. Nachrbl. Chem. Tech. Lab. 37:484-488. [Google Scholar]
- 6.Bauer, G. J. 1990. RNA sequencing using fluorescent-labeled dideoxynucleotides and automated fluorescence detection. Nucleic Acids Res. 18:879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout, B., and K. T. Jeang. 1990. Down modulation of HIV-1 gene expression using a prokaryotic RNA-binding protein. Nucleic Acids Res. 18:6903-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey, J., V. Cameron, P. L. deHaseth, and O. C. Uhlenbeck. 1983. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry 22:2601-2610. [DOI] [PubMed] [Google Scholar]
- 10.Domingo, E., E. Martinez-Salas, F. Sobrino, J. C. de la Torre, A. Portela, J. Ortin, C. López-Galindez, P. Pérez-Brena, N. Villanueva, R. Nájera, S. van de Pol, D. Steinhauer, N. De Polo, and J. Holland. 1985. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene 40:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Draper, D. E. 1987. Translational regulation of ribosomal proteins in Escherichia coli: molecular mechanisms, p. 1-26. In J. Ilan (ed.), Translational regulation of gene expression. Plenum Press, New York, N.Y.
- 12.Eigen, M. 1971. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58:465-523. [DOI] [PubMed] [Google Scholar]
- 13.Eigen, M., and C. K. Biebricher. 1988. Sequence space and quasispecies distribution, p. 211-245. In E. Domingo, J. J. Holland, and P. Alquist (ed.), RNA genetics, vol. III. CRC Press, Boca Raton, Fla. [Google Scholar]
- 14.Gingras, A. C., Y. Svitkin, G. J. Belsham, A. Pause, and N. Sonenberg. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. USA 93:5578-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gralla, J., J. A. Steitz, and D. M. Crothers. 1974. Direct physical evidence for secondary structure in an isolated fragment of R17 bacteriophage mRNA. Nature 248:204-208. [DOI] [PubMed] [Google Scholar]
- 16.Hatfield, G. W., and J. A. Sharp. 1987. Translational control of transcription termination in prokaryotes, p. 447-472. In J. Ilan (ed.), Translational regulation of gene expression. Plenum Press, New York, N.Y.
- 17.Henry, G. L., S. J. McCormack, D. C. Thomis, and C. E. Samuel. 1994. Translational control and the RNA-dependent protein kinase (PKR): antagonists of PKR enhance the translational activity of mRNAs that include a 161 nucleotide region from reovirus S1 mRNA. J. Biol. Regul. Homeost. Agents 8:15-24. [PubMed] [Google Scholar]
- 18.Hofstetter, H., H. J. Monstein, and C. Weissmann. 1974. The readthrough protein A1 is essential for the formation of viable Qβ particles. Biochim. Biophys. Acta 374:238-251. [DOI] [PubMed] [Google Scholar]
- 19.Horne, M. 1970. Coevolution of Escherichia coli and bacteriophages in chemostat culture. Science 168:992-993. [DOI] [PubMed] [Google Scholar]
- 20.Husimi, Y. 1989. Selection and evolution of bacteriophages in cellstat. Adv. Biophys. 25:1-43. [DOI] [PubMed] [Google Scholar]
- 21.Husimi, Y., and H.-C. Keweloh. 1987. Continuous culture of bacteriophage Qβ using a cellstat with a bubble wall-growth scraper. Rev. Sci. Instrum. 58:1109-1111. [Google Scholar]
- 22.Husimi, Y., K. Nishigaki, Y. Kinoshita, and T. Tanaka. 1982. Cellstat—a continuous culture system of a bacteriophage for the study of the mutation rate and the selection process at the DNA level. Rev. Sci. Instrum. 53:517-522. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson, A. B., R. Arora, M. Zuker, C. Priano, C. H. Lin, and D. R. Mills. 1998. Structural plasticity in RNA and its role in the regulation of protein translation in coliphage Qβ. J. Mol. Biol. 275:589-600. [DOI] [PubMed] [Google Scholar]
- 24.Jay, G., and R. Kaempfer. 1975. Translational repression of a viral messenger RNA by a host protein. J. Biol. Chem. 250:5749-5755. [PubMed] [Google Scholar]
- 25.Kolakofsky, D., and C. Weissmann. 1971. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat. New Biol. 231:42-46. [DOI] [PubMed] [Google Scholar]
- 26.Kong, D., and J. Yin. 1995. Whole-virus vaccine development by continuous culture on a complementing host. Bio/Technology 13:583-586. [DOI] [PubMed] [Google Scholar]
- 27.Kozlovska, T. M., I. Cielens, D. Dreilinna, A. Dislers, V. Baumanis, V. Ose, and P. Pumpens. 1993. Recombinant RNA phage Qβ capsid particles synthesized and self-assembled in Escherichia coli. Gene 137:133-137. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lodish, H. F., and N. D. Zinder. 1966. Mutants of the bacteriophage f2. J. Mol. Biol. 19:333-348. [DOI] [PubMed] [Google Scholar]
- 30.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, E. S., J. Karam, M. Dawson, M. Trojanowska, P. Gauss, and L. Gold. 1987. Translational repression: biological activity of plasmid-endoded bacteriophage T4 RegA protein. J. Mol. Biol. 194:397-410. [DOI] [PubMed] [Google Scholar]
- 32.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills, D. R., C. Priano, P. DiMauro, and B. D. Binderow. 1988. Qβ replicase: mapping the functional domains of an RNA-dependent RNA polymerase. J. Mol. Biol. 205:751-764. [DOI] [PubMed] [Google Scholar]
- 34.Peabody, D. S. 1990. Translational repression by bacteriophage MS2 coat protein expressed from a plasmid. A system for genetic analysis of a protein-RNA interaction. J. Biol. Chem. 265:5684-5689. [PubMed] [Google Scholar]
- 35.Poulin, L., M. Fauchon, A. Darveau, and J. A. Levy. 1994. Inhibition of protein synthesis by the human immunodeficiency virus type 1 nef gene product. J. Gen. Virol. 75:2977-2984. [DOI] [PubMed] [Google Scholar]
- 36.Romaniuk, P. J., P. Lowary, H.-N. Wu, G. Stormo, and O. C. Uhlenbeck. 1987. RNA binding site of R17 coat protein. Biochemistry 26:1563-1568. [DOI] [PubMed] [Google Scholar]
- 37.Rossmann, M. G. 1984. Constraints on the assembly of spherical virus particles. Virology 134:1-11. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schlegel, H. G. 1985. Allgemeine Mikrobiologie. Thieme Verlag, Stuttgart, Germany.
- 40.Schwienhorst, A., and B. F. Lindemann. 1998. A novel vector for generating RNAs with defined 3′ ends and its use in antiviral strategies. BioTechniques 24:116-126. [DOI] [PubMed] [Google Scholar]
- 41.Schwienhorst, A., B. F. Lindemann, and M. Eigen. 1996. Growth kinetics of a bacteriophage in continuous culture. Biotechnol. Bioeng. 50:217-221. [DOI] [PubMed] [Google Scholar]
- 42.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skripkin, E. A., and A. B. Jacobson. 1993. A two-dimensional model at the nucleotide level for the central hairpin of coliphage Qβ RNA. J. Mol. Biol. 233:245-260. [DOI] [PubMed] [Google Scholar]
- 44.Sonstegard, T. S., and P. B. Hackett. 1996. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 70:6642-6652. [PMC free article] [PubMed] [Google Scholar]
- 45.Springer, M., and M. Grunberg-Manago. 1987. Escherichia coli threonyl-transfer RNA synthetase as a model system to study translational autoregulation in prokaryotes, p. 51-62. In J. Ilan (ed.), Translational regulation of gene expression. Plenum Press, New York, N.Y.
- 46.Standart, N., and R. J. Jackson. 1994. Regulation of translation by specific protein/mRNA interactions. Biochimie 76:867-879. [DOI] [PubMed] [Google Scholar]
- 47.Stormo, G. D. 1987. Translational regulation in bacteriophages, p. 27-49. In J. Ilan (ed.), Translational regulation of gene expression. Plenum Press, New York, N.Y.
- 48.van Batenburg, F. H. D., A. P. Gultyaev, and C. W. A. Pleij. 1995. An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J. Theor. Biol. 174:269-280. [DOI] [PubMed] [Google Scholar]
- 49.Varon, M. 1979. Selection of predation-resistant bacteria in continuous culture. Nature 277:386-388. [Google Scholar]
- 50.Weber, H. 1976. The binding site for coat protein on bacteriophage Qβ RNA. Biochim. Biophys. Acta 418:175-183. [DOI] [PubMed] [Google Scholar]
- 51.Weber, H., M. A. Billeter, S. Kahane, C. Weissmann, J. Hindley, and A. Porter. 1972. Molecular basis for repressor activity of Q replicase. Nat. New Biol. 237:166-170. [DOI] [PubMed] [Google Scholar]
- 52.Weber, K., and W. Konigsberg. 1974. Proteins of the RNA phages, p. 51-84. In N. D. Zinder (ed.), RNA phages. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Witherell, G. W., and O. C. Uhlenbeck. 1989. Specific RNA binding by Qβ coat protein. Biochemistry 28:71-76. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]