Abstract
Seventeen years after the discovery of tissue-specific apoB mRNA editing, only three nucleus-encoded mRNAs have been shown to undergo C-to-U editing. All three mRNAs occur in mammals. apoB mRNA editing is tissue-specific and occurs normally, whereas NF1 and NAT1 mRNA editing is found largely in tumors. Here we report the first example of C-to-U RNA editing in Caenorhabditis elegans. The gld-2 gene encodes an atypical poly(A) polymerase that governs the mitosis/meiosis decision in the germ line as well as progression through meiosis and early embryogenesis. At least two of its alternatively spliced transcripts are germline-specific. We find that most and perhaps all germline-specific transcripts generated by the gld-2 gene undergo C-to-U editing, but that somatic transcripts show no detectable editing. The gld-2 C-to-U editing event changes the codon from CCG to CUG, which is predicted to cause a proline to leucine substitution in the protein sequence. Our findings suggest the presence of a sequence- and tissue-specific cytidine deaminase acting on RNA, or CDAR. This CDAR modifies a specific base in gld-2 mRNA, and acts only in the germline.
Keywords: RNA editing, C-to-U, C. elegans, GLD-2, poly(A) polymerase
INTRODUCTION
RNA editing is a mode of regulating gene expression in which the coding capacity of an mRNA is altered by covalent modification, including the insertion or deletion of nucleotides, or base modification. In the first documented example, uridine residues were shown to be added and removed from cytochrome oxidase subunit II mRNA in trypanosome mitochondria (Benne et al. 1986). One year later, the deamination of cytidine to uridine (C-to-U editing) of mammalian apoB mRNA was identified (Powell et al. 1987). These classic examples exemplify the two broad categories of RNA editing: insertion/deletion editing and modification editing (for reviews, see Bass 1993; Simpson 1999).
Numerous examples of both types of editing have been discovered. However, only three cases of C-to-U modification editing of nuclear mRNAs have been documented: apoB, NF1, and NAT1. Both NF1 and NAT1 editing were found in tumors. In wild-type cells, NF1 editing was less than 2%, whereas NAT1 editing was not detected (Skuse et al. 1996; Yamanaka et al. 1997). NAT1 mRNA is edited at many positions (Yamanaka et al. 1997), and apoB mRNA appears to be edited at both a major and minor site (Navaratnam et al. 1991). All three mRNAs share a “mooring-like” sequence in the vicinity of the edited site. In addition, the tissues in which editing occurs all express the cytidine deaminase APOBEC-1 (Apolipoprotein B mRNA-Editing enzyme, Catalytic polypeptide 1; Navaratnam et al. 1993; Teng et al. 1993; Mukhopadhyay et al. 2002). These data suggest mechanistic links among the three editing events.
Proteins related to APOBEC-1 have been identified in mammals (for review, see Gerber and Keller 2001; Blanc and Davidson 2003). AID (Activation-Induced cytidine Deaminase) is required for antibody maturation processes encompassing class-switch recombination, somatic hyper-mutation, and gene conversion (for review, see Durandy 2003; Okazaki et al. 2003). Scores of C-to-U changes have been found in mitochondrial and chloroplast RNAs in plants, where CDAR (cytidine deaminase acting on RNA)-like protein sequences have been identified (Gerber and Keller 2001). Outside mammalian species, overexpressed yeast Cdd1 protein can edit apoB RNAs ectopically expressed in yeast, though natural mRNA targets of Cdd1 have not been identified (Dance et al. 2001).
In this study, we report the discovery of tissue-specific C-to-U editing of the gld-2 mRNA in Caenorhabditis elegans. To our knowledge, this is the first example of natural C-to-U editing of a nucleus-encoded mRNA in a nonmammalian species.
RESULTS
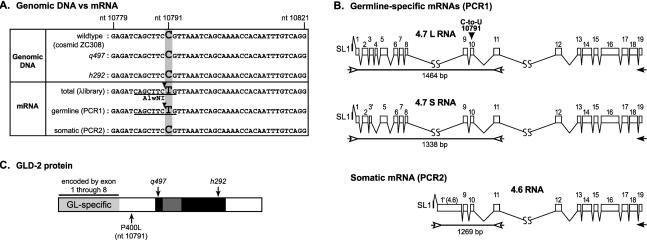
The C. elegans gld-2 gene encodes an atypical poly(A) polymerase that controls the mitosis/meiosis decision in the germ line as well as progression through meiosis and early embryogenesis (Kadyk and Kimble 1998; Wang et al. 2002). It directs the production of multiple alternatively spliced transcripts that correspond to four distinct mRNAs—two of ~4.7 kb (designated 4.7L and 4.7S), and one each of 4.6 kb and 4.0 kb (Wang et al. 2002). During the analysis of gld-2 cDNA and transcripts, we obtained a series of gld-2 RT-PCR clones and cDNAs from phage libraries. We noticed a discrepancy between the genomic sequence and that of the mRNAs it produces, in which a single position undergoes a change from C in the genome to U in the mRNA.
The GenBank genomic DNA sequence of gld-2 indicates a cytidine at position 10791 (Fig. 1A). To determine its identity independently, we analyzed genomic DNA fragments containing position 10791 from the wild-type C. elegans strain N2 and from two gld-2 mutant strains, q497 and h292 (each has a point mutation elsewhere in the coding region of gld-2; Wang et al. 2002). Genomic fragments were amplified using Hi-Fidelity PCRs and sequenced (Fig. 1A). All sequences match the one recorded in GenBank.
FIGURE 1.
Germline-specific gld-2 transcripts undergo C-to-U editing. (A) Comparison of sequences derived from genomic DNA and cDNA. Genomic DNA: The sequences of genomic DNA were derived from three sources. The sequence of cosmid ZC308 was determined by the C. elegans Sequencing Consortium. ZC308 carries a 19,559-bp insert of wild-type C. elegans genomic DNA, including the region that encodes all the gld-2 transcripts. It possesses a C at position 10791. The other two sequences were derived from PCR products (designed to include the region that contains nucleotide 10791) from genomic DNA of two gld-2 mutants, gld-2(q497) and gld-2(h292). These two mutants have nucleotide-substitution mutations elsewhere in the gld-2 gene, and were confirmed in the sequence analysis (see Wang et al. 2002). However, both DNAs possessed a C at position 10791, corroborating the C at position 10791 in the cosmid sequence. mRNA: Sequences of germline and somatic mRNAs were derived in three ways. The first was derived from the sequence of a gld-2 cDNA clone isolated from a phage cDNA library. The library was prepared using total mRNA extracted from mixed-stage worms (including both germline and somatic tissues). A T was detected at position 10791, indicating editing from C to U in the mRNA. The second analysis (“germline-specific mRNAs [PCR1]”) detected germline-specific transcripts. It was performed using the PCR1 reaction (B), using primers in exons 1 and 11. Two germline-specific transcripts exist, designated 4.7L and 4.7S, differing in the use of an exon 3 splice site and inclusion or exclusion of exon 4. The sequence of these PCR products includes a T at position 10791. The third analysis (“somatic mRNAs [PCR2]”) specifically identifies the 4.6-kb somatic mRNA, which carries a C at position 10791, matching genomic DNA. PCR was performed using a primer in the first exon of the somatic mRNA (exon 1 [4.6]), which is missing in germline-specific mRNAs. Underline, AlwNI site. AlwNI cleaves the CAGNNNCTG sequence, which is found in the region of cDNAs but not genomic DNA. Arrowhead, position of cleavage by AlwNI. (B) gld-2 exon/intron structure: two germline-specific transcripts and one soma-enriched transcript. The two germline-specific transcripts, 4.7L and 4.7S, are detected in the PCR1 reaction. This PCR reaction used primers in exons 1 and 11 (open arrows). Exon 1 is present only in germline-specific mRNAs. Exon 1′ (4.6) is present only in the 4.6-kb RNA that is highly enriched in somatic cells. For PCR1 and PCR2 reactions, the primer indicated by a black arrow (in exon 19) was used to prime the reverse transcriptase on total mRNA from mixed-stage worms and q224 worms (germline-less worms), respectively (see Materials and Methods for details). Open boxes, exons; thin lines, introns; arrows, PCR primers; black arrowhead, position of nucleotide 10791 in exon 10. All the gld-2 mRNAs are trans-spliced to SL1. The lengths of the predicted PCR products derived from mRNAs are indicated below the lines. For additional information on mRNA analysis, see Wang et al. (2002). (C) Schematic diagram of the GLD-2 protein sequence. GL-specific, germline-specific region, encoded by exons specific to the germline transcripts corresponding to the 4.7S and 4.7L RNAs (Wang et al. 2002). Black and dark gray, central and catalytic domains of the GLD-2 nucleotidyltransferase (Aravind and Koonin 1999; Wang et al. 2002); light gray, region of GLD-2 protein present only in the germline.
In contrast, a cDNA insert isolated from phage cDNA library λRB1 (gift from Dr. Robert Barstead, Oklahoma Medical Research Foundation, Oklahoma City) carried a T instead of a C at position 10791 (Fig. 1A). This library was constructed using mRNA isolated from worms at mixed stages, and includes mRNAs from both somatic and germ-line tissues. The discrepancy between genomic and cDNA sequences suggested that a C-to-U RNA-editing event modified gld-2 mRNA.
To further test this idea, we determined the sequence of germline-specific and soma-enriched mRNAs in the vicinity of position 10791 by RT-PCR. Primers were designed to amplify the “edited” region of the two germline-specific 4.7-kb transcripts (4.7L and 4.7S), and the soma-enriched 4.6-kb transcript (Fig. 1B; see Materials and Methods). The two germline-specific transcripts, 4.7S and 4.7L, differ in the choice of alternative 3′ splice sites in exon 3, and in the exclusion/inclusion of exon 4. The soma-enriched transcript possesses a different 5′ terminal exon [exon 1′ (4.6)] located within an intron of the germline transcripts (Fig. 1B). Primers were designed that are specific for either the germline-specific mRNAs (PCR1) or the soma-enriched mRNAs (PCR2), exploiting the difference in their first exons (Fig. 1B). The sequence in the relevant region of the PCR products is shown in Figure 1A. Germline-specific mRNAs yielded a T at position 10791, whereas soma-enriched mRNAs yielded a C. These data strongly support the hypothesis that C-to-U editing occurs at position 10791, and that it is germline-specific.
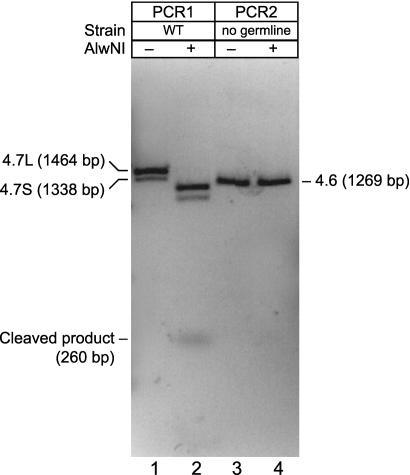
To quantify the fraction of mRNAs that undergo editing, we took advantage of the fact that the C-to-U change in the gld-2 RNA would introduce an AlwNI restriction endonuclease site in the RT-PCR product amplified from the edited RNA (Fig. 1A). If the RNA were not edited, the PCR product would have the same sequence as genomic DNA, without an AlwNI site, and so would not be cleaved by AlwNI. The predicted length of PCR products of germline-specific transcripts are 1464 bp and 1338 bp (PCR1 reaction, Fig. 1B), whereas the predicted length of PCR products of soma-enriched mRNAs is 1269 bp (PCR2 reaction, Fig. 1B). We analyzed poly(A+) RNA from wild-type C. elegans strain and from the mutant strain q224, which does not have a germline, and so contains only soma-enriched transcripts.
The PCR products amplified from germline-specific transcripts were cleaved by AlwNI (Fig. 2, lanes 1,2). Cleavage of the 1464- and 1338-bp bands was predicted to generate bands of 1204, 1078, and 260 bp; each was detected after AlwNI treatment, but not before. Because virtually all of the DNA was cleaved, editing of germline transcripts appears to be very efficient. In contrast, the PCR products from the soma-enriched transcript were not cleaved, demonstrating that somatic mRNAs are not edited detectably (Fig. 2, lanes 3,4). If they were, bands of 1009 bp and 260 bp would have been observed. Therefore, this C-to-U editing is confined to the germline, where it is very efficient.
FIGURE 2.
Editing of gld-2 mRNA is efficient and germline-specific. Reverse transcriptase reactions were performed with the same primer (indicated as a black arrow) on poly(A+) RNA from wild-type mixed-stage and q224 worms. q224 mutants possess essentially no germline (Kodoyianni et al. 1992). PCR products of 1464 and 1338 bp were obtained from the germline-specific mRNAs, corresponding to the mRNAs designated 4.7L and 4.7S in Figure 1B. A PCR product of 1269 bp was obtained from the soma-enriched transcripts, corresponding to the mRNA designated 4.6 in Figure 1B. The PCR products corresponding to the germline mRNAs are digested completely by AlwNI, demonstrating that they have been edited. In contrast, most (if not all) of the 1269-bp product is not cleaved, and so is not edited.
DISCUSSION
The apparent C-to-U editing of apoB mRNA was inferred from a discrepancy between cDNA and genomic DNA sequences (Powell et al. 1987). Other modifications that would change the base-pairing capacity of position 10791 from that of C to that of U could have the same effect. For example, the C of the CUA anticodon of tRNAIle in bacteria is modified to a 2-lysyl cytidine (lysidine), which enables that residue to base pair as would a U (Grosjean and Bjork 2004). However, lysidine modification has not been found in eukaryotic nuclear tRNAs, nor in the mRNAs of any species (see Grosjean and Bjork 2004). Direct RNA analysis, such as mass spectrometry or fingerprinting of authentic gld-2 mRNA, will be required to identify the modified base in gld-2 mRNA definitively. For simplicity, we will refer here to the event we have detected as C-to-U editing.
Editing of apoB and NF1 mRNAs introduces a stop codon. In contrast, the C-to-U nucleotide change in gld-2 mRNA is predicted to cause a proline-to-leucine change in GLD-2 protein. This proline, encoded by the nonedited mRNA, is located outside the conserved nucleotidyltransferase domain of GLD-2, at amino acid position 400 (P400L, Fig. 1C; see also Wang et al. 2002, Fig. 2). We have not detected a difference in the poly(A) polymerase activity of the P400 and L400 proteins translated in vitro (data not shown). However, it is possible that folding of the GLD-2 protein encoded by the edited mRNA may differ kinetically, or result in a locally different conformation, because proline cis-trans isomerization is considered a rate-limiting step during protein folding (Gothel and Marahiel 1999). It also may affect interaction of the protein partners, such as GLD-3 (Wang et al. 2002).
apoB and NF1 editing require an 11-nucleotide sequence 4–6 nucleotides downstream of the edited site, termed the “mooring sequence” (Backus and Smith 1991; Shah et al. 1991). Additional sequences and secondary structure also affect editing efficiency (Hersberger and Innerarity 1998; Hersberger et al. 1999). We compared the 30-nucleotide sequences containing the editing site of gld-2 mRNA to sequences near the editing site of apoB mRNA. No significant homology was identified. In particular, sequences near the editing site in gld-2 mRNA are not significantly related to the mooring sequence. The dissimilarity is not completely unexpected, given the apparent absence of an APOBEC-1 ortholog in C. elegans. Thus the unidentified cytidine deaminase, together with its auxiliary factors, may recognize sequence elements different from those of apoB mRNA.
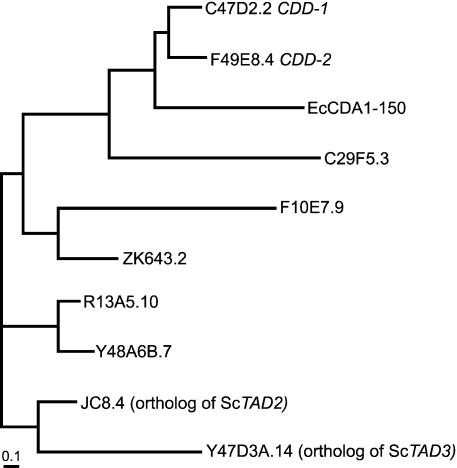
Nine putative cytidine deaminases in C. elegans have been identified by sequence analysis (Harris et al. 2003). A phylogenetic tree was built upon the alignment of these nine proteins with the E. coli cytidine deaminase (Fig. 3), using the Superfamily program (Gough et al. 2001). Two of the putative deaminases, CDD-1 and CDD-2, possess RNA-binding activity, and deaminate deoxycytidine in vitro (Thompson et al. 2002). Another two candidate proteins, JC8.4 and Y47D3A.14, appear to be orthologs of budding yeast Tad2p and Tad3p, respectively (Gerber and Keller 2001; Harris et al. 2003). These proteins appear to be positioned evolutionarily between the cytidine and adenosine deaminase families, and deaminate an adenosine in the anticodon of certain yeast tRNAs (Gerber and Keller 1999). RNAi against each of the nine genes depicted in Figure 3 yielded no mutant phenotypes (Gönczy et al. 2000; Kamath et al. 2003), suggesting functional redundancy; embryonic defects may be associated with cdd-2(RNAi), however (Thompson et al. 2002). Identification of the cytidine deaminase that edits gld-2 mRNA is now essential, but may be complicated by redundancy.
FIGURE 3.
Putative cytidine deaminases acting on RNA in C. elegans. An unrooted tree is presented consisting of nine putative cytidine deaminases of C. elegans and the E. coli cytidine deaminase. Transcript numbers of all nine C. elegans genes are given. EcCDA1–150, amino acid 1–150 of E. coli cytidine deaminase. First a multiple sequence alignment of the cytidine deaminase domains was generated by the Superfamily alignment program (http://supfam.org/SUPERFAMILY/). The tree was then built with a phylogenetic tree program (http://www.ebi.ac.uk/clustalw/) using the neighbor joining method, setting on Kimura correction of distances. 0.1, 0.1 substitutions per 100 residues.
In principle, the tissue-specific C-to-U editing event in gld-2 could occur at the DNA rather than RNA level. However, because somatic cells arise from the germ line, a DNA modification would have to occur quantitatively in germ-line DNA, and then be quantitatively reversed in somatic cells. Furthermore, we directly sequenced the high-fidelity PCR product from genomic DNA of whole adult worms (which have comparable numbers of somatic and germline nuclei), and no trace of T was detected at position 10791 in the sequencing chromatogram (data not shown). We conclude that the modification occurs at the RNA level.
Our finding that gld-2 mRNA is a target of tissue-specific C-to-U editing in C. elegans presents an unusual opportunity to examine C-to-U editing using molecular genetics. With its completely sequenced, annotated genome and effective molecular tools, C. elegans now may enable rapid progress in the analysis of C-to-U editing and CDARs, much as it has facilitated the analysis of A-to-I editing and ADARs (Tonkin et al. 2002).
MATERIALS AND METHODS
RNA analysis, PCR, and sequence determination
Poly(A+) RNAs were purified from worms using a Poly(A)Pure Kit (Ambion). Reverse transcriptase reactions were performed with SuperScript II reverse transcriptase (Invitrogen) using poly(A+) RNAs from N2 mixed-stage worms and q224 germline-less worms. The primer used for reverse transcriptase, LWp7, is complementary to sequences in exon 19, present in both germline and somatic gld-2 mRNAs (Fig. 1B, black arrow). PCR reactions were then prepared in parallel with LWp11 and LWp13 (PCR1) and LWp54 and LWp13 (PCR2; see Fig. 1B). The predicted sizes of PCR products are 1464 bp and 1338 bp for PCR1, and 1269 bp for PCR2. Sequences were determined by standard methods.
PCR products were phenol/chloroform extracted, ethanol precipitated, resuspended in water, and split into two halves. One was digested by AlwNI (NEB) overnight at 37°C, and the other was incubated without enzyme. After digestion, reactions were run on a 2% agarose gel and stained with ethidium bromide. Images were taken and analyzed by NucleoVision (NucleoTech).
Oligonucleotide primers
The sequences of PCR primers are given below.
LWp7, 5′-TCATTGAGATACATTTGATGA -3′
LWp11, 5′-ACATGCCATGGTTATGGCTCAACAGC -3′
LWp13, 5′-CATGACCATGGTTAGTTCAAAGCCTTCTCCTCC -3′
LWp54, 5′-CGGGATCCAACCTAAATTCGATCAATTAGC -3′
Acknowledgments
We thank Andrei Petcherski for helpful discussions, and Laura vanderploeg for assistance with figures. J.K. is an investigator with the Howard Hughes Medical Institute, and M.W. is supported by the National Institutes of Health (NIH).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7570804.
REFERENCES
- Aravind, L. and Koonin, E.V. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: Justification of three new families, classification and evolutionary history. Nucleic Acids Res. 27: 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus, J.W. and Smith, H.C. 1991. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 19: 6781–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. 1993. RNA editing: New uses for old players in the RNA world. In The RNA world (eds. R. Gesteland and J. Atkins), pp. 383–418. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Benne, R., Van den Burg, J., Brakenhoff, J.P., Sloof, P., Van Boom, J.H., and Tromp, M.C. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826. [DOI] [PubMed] [Google Scholar]
- Blanc, V. and Davidson, N.O. 2003. C-to-U RNA editing: Mechanisms leading to genetic diversity. J. Biol. Chem. 278: 1395–1398. [DOI] [PubMed] [Google Scholar]
- Dance, G.S., Beemiller, P., Yang, Y., Mater, D.V., Mian, I.S., and Smith, H.C. 2001. Identification of the yeast cytidine deaminase CDD1 as an orphan C→U RNA editase. Nucleic Acids Res. 29: 1772–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandy, A. 2003. Activation-induced cytidine deaminase: A dual role in class-switch recombination and somatic hypermutation. Eur. J. Immunol. 33: 2069–2073. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P. and Keller, W. 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286: 1146–1149. [DOI] [PubMed] [Google Scholar]
- ———. 2001. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 26: 376–384. [DOI] [PubMed] [Google Scholar]
- Giege, P. and Brennicke A. 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. 96: 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336. [DOI] [PubMed] [Google Scholar]
- Gothel, S.F. and Marahiel, M.A. 1999. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol. Life Sci. 55: 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, J., Karplus, K., Hughey, R., and Chothia, C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313: 903–919. [DOI] [PubMed] [Google Scholar]
- Grosjean, H. and Bjork, G.R. 2004. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial tRNAII3—An alternative way of RNA editing. Trends Biochem. Sci. 29: 165–168. [DOI] [PubMed] [Google Scholar]
- Harris, T.W., Lee, R., Schwarz, E., Bradnam, K., Lawson, D., Chen, W., Blasier, D., Kenny, E., Cunningham, F., Kishore, R., et al. 2003. WormBase: A cross-species database for comparative genomics. Nucleic Acids Res. 31: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersberger, M. and Innerarity, T.L. 1998. Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J. Biol. Chem. 273: 9435–9442. [DOI] [PubMed] [Google Scholar]
- Hersberger, M., Patarroyo-White, S., Arnold, K.S., and Innerarity, T.L. 1999. Phylogenetic analysis of the apolipoprotein B mRNA-editing region. Evidence for a secondary structure between the mooring sequence and the 3′ efficiency element. J. Biol. Chem. 274: 34590–34597. [DOI] [PubMed] [Google Scholar]
- Kadyk, L.C. and Kimble, J. 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125: 1803–1813. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kodoyianni, V., Maine, E.M., and Kimble, J. 1992. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3: 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, D., Anant, S., Lee, R.M., Kennedy, S., Viskochil, D., and Davidson, N.O. 2002. C–U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am. J. Hum. Genet. 70: 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, I., Yoshikawa, K., Kinoshita, K., Muramatsu, M., Nagaoka, H., and Honjo, T. 2003. Activation-induced cytidine deaminase links class switch recombination and somatic hypermutation. Ann. N.Y. Acad. Sci. 987: 1–8. [DOI] [PubMed] [Google Scholar]
- Powell, L.M., Wallis, S.C., Pease, R.J., Edwards, Y.H., Knott, T.J., and Scott, J. 1987. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell 50: 831–840. [DOI] [PubMed] [Google Scholar]
- Shah, R.R., Knott, T.J., Legros, J.E., Navaratnam, N., Greeve, J.C., and Scott, J. 1991. Sequence requirements for the editing of apolipoprotein B mRNA. J. Biol. Chem. 19: 16301–16304. [PubMed] [Google Scholar]
- Simpson, L. 1999. RNA Editing—An evolutionary perspective, In The RNA world, 2nd ed., pp. 585–608. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Skuse, G.R., Cappione, A.J., Sowden, M., Metheny, L.J., and Smith, H.C. 1996. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 24: 478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, B., Burant, C.F., and Davidson, N.O. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260: 1816–1819. [DOI] [PubMed] [Google Scholar]
- Thompson, F.J., Britton, C., Wheatley, I., Maitland, K., Walker, G., Anant, S., Davidson, N.O., and Devaney, E. 2002. Biochemical and molecular characterization of two cytidine deaminases in the nematode Caenorhabditis elegans. Biochem. J. 365: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A., Saccomanno, L., Morse, D.P., Brodigan, T., Krause, M., and Bass, B.L. 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21: 6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Eckmann, C.R., Kadyk, L.C., Wickens, M., and Kimble, J. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Yamanaka, S., Poksay, K.S., Arnold, K.S., and Innerarity, T.L. 1997. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes & Dev. 11: 321–333. [DOI] [PubMed] [Google Scholar]