Abstract
Synthetic analogs of the N7-methylated guanosine triphosphate cap at the 5′ end of eukaryotic mRNAs and snRNAs have played an important role in understanding their splicing, intracellular transport, translation, and turnover. We report here a new series of N7-benzylated dinucleoside tetraphosphate analogs, b7Gp4G, b7m3′-OGp4G, and b7m2Gp4G, that extend our knowledge of the role of the cap in translation. We used these novel analogs, along with 10 previously synthesized analogs, to explore five parameters: binding affinity to eIF4E, inhibition of cap-dependent translation in a rabbit reticulocyte lysate system, efficiency of incorporation into RNAs during in vitro transcription (% capping), orientation of the analog in the synthetic mRNA (% correct orientation), and in vitro translational efficiency of mRNAs capped with the analog. The 13 cap analogs differed in modifications of the first (distal) and second (proximal) guanine moieties, the first and second ribose moieties, and the number of phosphate residues. Among these were analogs of the naturally occurring cap m32,2,7Gp3G. These compounds varied by 61-fold in affinity for eIF4E, 146-fold in inhibition of cap-dependent translation, 1.4-fold in % capping, and 5.6-fold in % correct orientation. The most stimulatory analog enhanced translation 44-fold compared with uncapped RNA. mRNAs capped with b7m2Gp4G, m7Gp3m7G, b7m3′-OGp4G, and m7Gp4m7G were translated 2.5-, 2.6-, 2.8-, and 3.1-fold more efficiently than mRNAs capped with m7Gp3G, respectively. Relative translational efficiencies could generally be explained in terms of cap affinity for eIF4E, % capping, and % correct orientation. The measurement of all five parameters provides insight into factors that contribute to translational efficiency.
Keywords: cap-dependent translation, capping efficiency, eIF4E, in vitro transcription, T7 RNA polymerase, translational efficiency
INTRODUCTION
Post-transcriptional regulation of gene expression involves the recognition by protein factors of specific signals located throughout the mRNA. These signals include the N7-methylated guanosine triphosphate cap at the 5′ end and the poly(A) tract at the 3′ end (Flaherty et al. 1997). The cap structure, m7G(5′)p3(5′)N, is present in RNA polymerase II transcripts, including primary transcripts of all eukaryotic mRNAs (Muthukrishnan et al. 1975) and most U-type snRNAs (uridine-rich small nuclear RNAs; Mattaj 1986). The cap is added enzymatically to the RNA precursor in the nucleus during the initial phases of transcription and acts as one of the signals for nuclear export (Izaurralde et al. 1992). Whereas mRNAs remain in the cytosol, snRNAs bind Sm proteins, their m7Gp3N is further methylated to m32,2,7Gp3N, and they are then imported back into the nucleus, where they play a catalytic role in splicing of pre-mRNAs (Mattaj 1986; Moore et al. 1993). The cap of mRNAs is specifically recognized by the translational initiation factor eIF4E (Gingras et al. 1999; Rhoads 1999). Binding of eIF4E to the cap occurs during formation of the 48S initiation complex, which is thought to be rate-limiting for translational initiation under normal conditions. The cap structure both stimulates translation and protects mRNA against nucleolytic degradation.
A great variety of cap analogs have been synthesized and tested in vitro for their capacity to substitute for the native form of the cap. These have played important roles in elucidating splicing, intracellular transport, translation, and turnover, both as competitive inhibitors and as alternative structures at the 5′ ends of RNAs. Some analogs are more inhibitory for translation than the corresponding natural compounds (Darzynkiewicz et al. 1985, 1987, 1989). For example, addition of a methyl group at the N2 position of m7G produces a more inhibitory compound for analogs of the form m7GMP, m7GTP, m7Gp3G, and m7Gp4G (Cai et al. 1999). The greatest inhibition is observed with cap analogs in the dinucleoside tetra- and pentaphosphate series (Cai et al. 1999; Jemielity et al. 2003). The inhibitory properties of cap analogs generally correlate with their equilibrium binding affinity for eIF4E, although there are exceptions (Carberry et al. 1990; Niedzwiecka et al. 2002; Jemielity et al. 2003).
One motivation for discovering cap analogs with higher affinity for eIF4E is the potential of developing novel anti-cancer therapies. Overexpression of eIF4E has a profound effect on cell growth and phenotype, causing accelerated cell division and malignant transformation (for review, see De Benedetti and Graff 2004). eIF4E overexpression from transfected vectors also prevents apoptosis in growth-factor-restricted fibroblasts. Aggressive growth phenotypes can be reversed by lowering eIF4E levels via expression of antisense RNA and by lowering eIF4E availability via overexpression of the eIF4E-binding proteins 4E-BP1 and 4E-BP2. Also, introduction of peptides containing the eIF4E-binding motif of 4E-BP1 linked to the penetratin peptide-carrier sequence results in dose-dependent cell death with characteristics of apoptosis (Herbert et al. 2000). These studies suggest that novel, highly specific cap analog inhibitors of eIF4E might counteract elevated eIF4E levels in tumor cells.
Cap analogs are also used to synthesize capped RNA transcripts in vitro. In the presence of m7Gp3G, transcription is initiated with nucleophilic attack by the 3′-OH of Guo on the α-phosphate of the next nucleoside triphosphate specified by the DNA template, yielding the initial product m7G(5′)p3(5′)GpN. However, it was found that one-third to one-half of the caps are incorporated in the reverse orientation because the polymerase can also utilize the 3′-OH of m7Guo, yielding G(5′)p3(5′)m7GpN (Pasquinelli et al. 1995). A different type of cap analog was synthesized to prevent incorrect incorporation (Stepinski et al. 2001; Peng et al. 2002; Jemielity et al. 2003). These ‘anti-reverse’ cap analogs (ARCAs) have modifications in either the C2′ or C3′ positions of m7 Guo (e.g., m27,3′-O Gp3G), which allow incorporation in only the correct orientation during the transcription reaction. mRNAs capped with ARCAs are translated more efficiently than those capped with the natural cap. ARCAs in the tetraphosphate series (e.g., m2 7,3′-O Gp4G) provide the strongest translational enhancement of any cap analogs tested to date.
In the present work, we synthesized a new series of N7-benzylated dinucleoside tetraphosphate cap analogs, P1-7-benzylguanosine-5′ P4-guanosine-5′ tetraphosphate (b7Gp4G), P1-7-benzyl-3′-O-methylguanosine-5′ P4-guanosine-5′ tetraphosphate (b7m3′-OGp4G), and P1-7-benzyl-2-methylguanosine-5′ P4-guanosine-5′ tetraphosphate (b7m2Gp4G). Using these, as well as 10 previously synthesized cap analogs, we attempted to understand the dependence of translational efficiency on four parameters: affinity of the cap analog for eIF4E, ability of the cap analog to inhibit cell free translation, the degree to which RNAs synthesized in vitro are capped in the presence of various cap analogs, and the degree to which cap analogs are incorporated into RNA in the correct rather than reverse orientation. These studies revealed differences in the manner by which the various cap analogs are recognized by T7 RNA polymerase during in vitro transcription reactions. They also led to the identification of several cap analogs that, when incorporated into mRNA, produce high translational efficiencies.
RESULTS AND DISCUSSION
Synthesis of new cap analogs
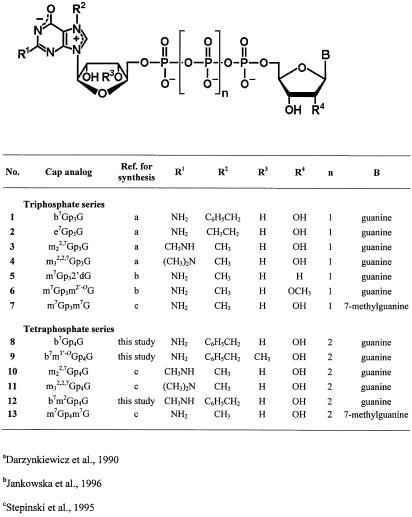
Three new cap analogs were synthesized for this study: b7Gp4G (8), b7m3′-OGp4G (9), and b7m2Gp4G (12; Fig. 1). These represented the combination of four modifications that were previously shown to produce cap analogs with superior translational properties: benzyl-for-methyl substitution at N7 (Darzynkiewicz et al. 1989), methoxy-for-hydroxyl substitution at the 3′-O position of the first Guo moiety (R3; Stepinski et al. 2001), addition of one methyl group at N2 of the same Guo moiety (R1; Cai et al. 1999), and addition of a fourth phosphate moiety (n = 2; Jemielity et al. 2003). Chemical synthesis of the new cap analogs was performed by a strategy similar to that developed previously (Stepinski et al. 2001; Jemielity et al. 2003). The solution conformations of the new cap analogs, as determined by NMR spectroscopy, were similar to one another and to those of previously investigated analogs (Stepinski et al. 2001; Jemielity et al. 2003).
FIGURE 1.
Structures of cap analogs used.
Binding affinity of new cap analogs for eIF4E
At least four factors determine the degree to which the cap increases translational efficiency of an mRNA: the binding affinity of the cap to eIF4E, the inhibition of cap-dependent translation, the percentage of RNAs containing a cap, and the orientation of the cap in the RNA (Stepinski et al. 2001; Jemielity et al. 2003). We tested the first of these parameters, binding to recombinant mouse eIF4E(28–217), for both new and previously synthesized cap analogs. This was determined by quenching of intrinsic Trp fluorescence (Niedzwiecka et al. 2002). The equilibrium association constants (KAS) for new and related cap analogs are shown in Table 1 (KAS).
TABLE 1.
Assay of five parameters for three novel cap analogs and 10 previously synthesized cap analogs
| No. | Cap analog | KAS × 10−6 (M-1)a | KI (μM)b | % Cappingc | % Correct orientationd | Relative translational efficiencye |
| Triphosphate series | ||||||
| m7Gp3G | 12.6 ± 0.3 | 17.1 ± 1.0i | 69 ± 6 | 58 ± 4 | 1.00 | |
| m27,3′-OGp3Gf | 10.2 ± 0.3 | 14.3 ± 1.3 | 72 | 100 | 1.88 ± 0.40 | |
| 1 | b7Gp3G | 14.6 ± 0.3 | 14.1 ± 1.3 | 79 ± 5 | 76 ± 2 | 1.87 ± 0.02 |
| 2 | e7Gp3G | 3.1 ± 0.0 | 32.5 ± 7.9 | 81 ± 5 | 79 ± 0 | 0.68 ± 0.03 |
| 3 | m22,7Gp3G | 18.4 ± 0.3 | 10.3 ± 0.9 | 91 ± 2 | 83 ± 1 | 1.23 ± 0.18 |
| 4 | m32,2,7Gp3G | NDg | >1000 | 66 ± 4 | 76 ± 3 | 0.37 ± 0.01 |
| 5 | m7Gp32′dG | NDg | 13.5 ± 2.9 | 81 ± 4 | 18 ± 2 | 0.46 ± 0.21 |
| 6 | m7Gp3m2′-OG | 8.0 ± 0.5h | 12.6 ± 0.2 | 86 ± 0 | 24 ± 1 | 0.08 ± 0.21 |
| 7 | m7Gp3m7G | 3.7 ± 0.6h | 12.5 ± 0.2 | 73 ± 2 | 100k | 2.66 ± 0.64 |
| Tetraphosphate series | ||||||
| m7Gp4G | 129.5 ± 2.3 | 10.8 ± 0.8j | NDg | NDg | 1.32 ± 0.08 | |
| 8 | b7Gp4G | 128.2 ± 3.4 | 16.0 ± 1.2 | 76 ± 0 | 84 ± 0 | 1.98 ± 0.40 |
| 9 | b7m3′-OGp4G | 70.7 ± 1.2 | 8.0 ± 0.1 | 64 ± 4 | 100l | 2.87 ± 0.38 |
| 10 | m22,7Gp4G | 107.0 ± 2.5 | 8.8 ± 1.2 | 92 ± 0 | 78 ± 5 | 1.29 ± 0.50 |
| 11 | m32,2,7Gp4G | NDg | 209 ± 23 | 70 ± 6 | 83 ± 2 | 0.52 ± 0.16 |
| 12 | b7m2Gp4G | 188.9 ± 7.7 | 7.5 ± 0.5 | 81 ± 4 | 86 ± 4 | 2.55 ± 0.86 |
| 13 | m7Gp4m7G | 47.0 ± 2.7h | 6.8 ± 0.9 | 74 ± 3 | 100k | 3.14 ± 0.24 |
aEquilibrium association constants for interaction of mouse eIF4E (28-217) with various cap analogs at 20°C. The final KAS value for each cap analog was calculated as a weighted average of independent titrations.
bInhibitory constants for inhibition of natural globin mRNA translation in a rabbit reticulocyte lysate system. For the triphosphate series, each value for KI was normalized by dividing with the value for KI for the cap analog standard m7Gp3G. For the tetraphosphate series, the KI for m7Gp4G was used for normalization.
cThe % of RNAs containing a cap, regardless of whether in the correct or reversed orientation, was calculated by comparing labeled products derived from capped mRNA (in the case of m7Gp3G-capped mRNA, these are m7Gp3Gp* + Gp3m7Gp*) to the total 5′-terminal products (p3Gp* + m7Gp3Gp* + Gp3m7Gp*).
dOrientation of the analog in synthetic luciferase mRNA. The % of RNAs for which the cap is oriented normally was calculated from labeled products derived from correct orientation (in the case of m7Gp3G-capped mRNA, this is pGp*) to total 5′-terminal labeled products (pGp* + pm7Gp*). Each value for pGp* was first corrected for the contribution from uncapped mRNA from the % Capping data.
eTranslational efficiency of luciferase mRNAs containing various cap analogs in rabbit reticulocyte lysate system. Between two and four syntheses of capped RNA and between four and eight translation reactions were performed for each cap analog. The standard cap (m7Gp3G) was used to normalize variation between experiments. Each value for overall translational efficiency was corrected for cap-independent translation by subtracting the value for overall translational efficiency of mRNA capped with Gp3G (Jemielity et al. 2003). Each value for cap-dependent translational efficiency was normalized by dividing by the value for cap-dependent translational efficiency of mRNA capped with m7Gp3G.
fThis compound is commercially available. All data for this compound come from Jemielity et al. (2003).
gNot determined.
hData from Niedzwiecka et al. (2002).
iData from Cai et al. (1999).
jData from Jemielity et al. (2003).
kThese are symmetrical molecules for which normal and reverse orientations are identical.
lThe b7m3′-O Guo moiety of this compound cannot form a 3′-5′ phosphodiester, whereas the Guo moiety can. This compound can therefore be incorporated only in the correct orientation. This was experimentally confirmed for other 3′-O-methyl dinucleoside tri- and tetraphosphates (Stepinski et al. 2001; Jemielity et al. 2003).
Substitution of benzyl for methyl at N7 of the first Guo moiety in the triphosphate series and adding a second methyl group at N2 slightly increases binding, whereas substitution of ethyl decreases it. A much greater increase in affinity is caused by adding a fourth phosphate, whether measured with 7-methyl compounds (10.3-fold; m7Gp4G versus m7Gp3G), 7-benzyl compounds [8.8-fold; b7Gp4G (8) versus b7Gp3G (1)], or 2,7-dimethyl compounds [5.8-fold; m22,7Gp4G (10) versus m22,7Gp3G (3)]. Within the tetraphosphate series, the 7-benzyl substituent binds no better than the 7-methyl substituent [b7Gp4G (8) versus m7Gp4G]. Addition of a methyl group at the 3′-O position of the first nucleoside residue causes a 1.8-fold reduction in affinity [b7m3′-OGp4G (12) versus b7Gp4G (8)]. When two favorable modifications to the guanine base are made (methyl at N2 and benzyl at N7), an increase in affinity can be detected even in the tetraphosphate series [b7m2Gp4G (12) versus m7Gp4G], suggesting that the guanine base interactions are significant even in the presence of strong phosphate interactions.
The influence of structural modifications in synthetic caps on binding to eIF4E can be rationalized on the basis of the structures of various eIF4E-cap complexes and interactions inside the eIF4E cap-binding slot (Marcotrigiano et al. 1997; Matsuo et al. 1997; Niedzwiecka et al. 2002; Tomoo et al. 2002) as well as dynamics of conformational rearrangements of the complexes (Blachut-Okrasinska et al. 2000; Miyoshi et al. 2002; Tomoo et al. 2003). Electrostatic interaction between the phosphate chain of the cap and the positively charged amino acid side chains at the entrance to the eIF4E cap binding slot determines a “molecular anchor” for the binding (Niedzwiecka et al. 2002), emphasizing the significance of the number of phosphate groups at early stages of the complex formation. This in turn enables sandwich stacking of 7-substituted guanine between two Trp rings (Trp-102 and Trp-56) and formation of hydrogen bonds as well as the rest of the stabilizing van der Waals contacts. Two counteracting effects probably result from replacement of the N7 methyl group with a larger substituent: steric hindrance and stronger hydrophobic interaction with Trp-166. The former prevails in the case of 7-ethyl, leading to a decrease of KAS, whereas the latter prevails in the case of 7-benzyl, due to efficient stacking with the indole ring of Trp-166. Methylation at N2 may enhance the sandwich stacking.
Inhibition of cap-dependent translation
Inhibition of cap-dependent translation was the second parameter that we explored. The cap analogs were assayed over a series of concentrations for inhibition of cap-dependent translation using the microccocal nuclease-treated rabbit reticulocyte lysate system (Pelham and Jackson 1976). Exogenous native β-globin mRNA was added back at a concentration known to be highly responsive to added cap analogs (Chu and Rhoads 1980). This provides a separate measure of binding affinity for eIF4E. Though less direct than binding in a purified system, it has the advantages of measuring competition between cap analog and mRNA in a system containing all the proteins and RNAs involved in protein synthesis as well as providing kinetic rather than static data. We calculated KI, the concentration at which inhibition is 50%, from a kinetic equation for translation as described (Cai et al. 1999). Inhibition of translation by all new cap analogs is shown in Table 1 (KI).
Substitution of benzyl for methyl at N7 of the first guanosine moiety in the dinucleoside triphosphate series slightly increases the effectiveness of the cap analog as a translational inhibitor but slightly decreases it in the dinucleoside tetraphosphate series. Alkyl substitution decreases the effectiveness of the cap analog as a translational inhibitor. N2 methylation in the first Guo slightly improves inhibitory activity in both the dinucleoside triphosphate and tetraphosphate series. Compounds with both N7 and N2 modifications are better inhibitors than those with either modification separately in both the 7-methyl and 7-benzyl series. Table 1 also presents translational inhibition results for analogs that possess two methyl groups at the N2 position of the first Guo. This modification is of particular interest because the mRNAs of some lower metazoans contain mRNAs of two types, those containing m7GTP and those containing m32,2,7GTP. mRNAs from ~70% of the genes in Caenhorabditis elegans contain the m32,2,7Gp3G cap due to trans-splicing (Liou and Blumenthal 1990; van Doren and Hirsh 1990; Zorio et al. 1994). In the present study, we found that m32,2,7Gp3G (4) in the nucleoside triphosphate series is such a poor inhibitor that we can determine only the lower limit for its KI value (>1000 μM). In the dinucleoside tetraphosphate series, addition of two N2 methyl groups decreases inhibitory activity by 19.4-fold [KI = 209 versus 10.8 μM for m32,2,7Gp4G (11) versus m7Gp4G]. Two symmetric cap analogs, m7Gp3m7G (7) and m7Gp4m7G (13), are more potent inhibitors than the respective parent compounds. The explanation may be that, because of the presence of two m7G moieties, their effective concentration in solution is twice that of the corresponding analogs with only one m7G moiety. Finally, we tested a new “anti-reverse” cap analog, b7m3′-OGp4G (9). As noted above, a benzyl substitution at N7 is favorable for some but not all cap analogs. Making both substitutions, 3′-O methyl and 7-benzyl, slightly decreased KI. Thus, the advantage conferred by 3′-O methyl substitution is partially cancelled out by the disadvantage of 7-benzyl substitution in the dinucleoside tetraphosphate series.
There is a qualitative but not quantitative correlation between KAS and 1/KI. For instance, despite the more than 10-fold increases in the KAS values for b7Gp4G (8), b7m2Gp4G (12), and m7Gp4G over the normal cap analog m7Gp3G, there are only small differences in KI values. One explanation may be that KAS determinations are made with eIF4E alone, but KI determinations are made in a complete translation system. Specifically, eIF4E acts in the presence of eIF4G and poly(A)-binding protein, both of which increase the affinity of eIF4E for caps (Haghighat and Sonenberg 1997; Wei et al. 1998). Another explanation of these qualitative and quantitative differences may be that the highly charged tetraphosphate cap analogs bind to additional proteins in the reticulocyte lysate, lowering their effective concentrations and decreasing their binding to eIF4E. Finally, KAS values represent an equilibrium measurement, whereas KI values represent a kinetic measurement. Thus, as eIF4E becomes more inhibited, some other step in the initiation of protein synthesis may become rate-limiting, making the addition of more cap analog less and less effective.
Efficiency of cap incorporation during in vitro transcription
The third parameter that we investigated was the degree to which various cap analogs are incorporated into RNAs during in vitro transcription (% capping). This was determined by incubating T7 RNA polymerase with a cap dinucleotide, all four ribonucleotide triphosphates, [α-32P]GTP, and a short DNA template in which G is the first ribonucleotide specified after the promoter (see Materials and Methods). Any nucleotide on the 5′ side of a G residue acquires a 32P-labeled 3′-phosphate group after RNase T2 digestion by nearest-neighbor transfer. Anion exchange chromatography is then used to resolve labeled nucleoside 3′-monophos-phates, resulting from internal positions in the RNA, from 5′-terminal products. The latter are of two types. Uncapped RNAs yield labeled guanosine 5′-triphosphate 3′-monophosphate (p3Gp*; in which * indicates the labeled phosphate group). Capped RNAs yield various 5′-terminal structures, depending on the nature of the cap analog used (m7Gp3Gp* and Gp3m7Gp* when the cap analog is m7Gp3G). The results for each type of analog are shown in Table 1 (% capping).
In the dinucleoside triphosphate series, the % capping varies widely, from 66% to 91%. This presumably reflects differences in the ability of T7 polymerase to misincorpo rate different dinucleoside triphosphates of the form Np3G in place of GTP. Most of the analogs tested are incorporated into RNA more efficiently than the parent m7Gp3G. The most efficiently incorporated dinucleoside triphosphate analog is m22,7Gp3G (3; 91%). In the tetraphosphate series, the m22,7G-containing compound is also the most efficiently incorporated [m22,7Gp4G (10); 92%]. However, adding a second methyl group at N2 abolishes this advantage: m32,2,7Gp3G (4) is incorporated no more efficiently than m7Gp3G. Such constructs may be of interest for those studying U-type snRNAs as well as trans-spliced mRNAs in nematodes. One must take into account, however, that 17%–24% of these analogs are incorporated in the reverse orientation (see below). A trimethylated ARCA cap dinucleotide would therefore be more useful.
Analysis of cap orientation
The fourth factor determining the contribution of the cap to overall translational efficiency is the orientation. The strategy for determining % correct orientation was similar, but not identical, to that described previously (Stepinski et al. 2001). Labeled RNAs were synthesized and digested with RNase T2. This digestion yields labeled internal nucleoside monophosphates and 5′-terminal products. To determine cap orientation, the RNAs were further digested with tobacco acid pyrophosphatase (TAP), which hydrolyzes pyrophosphate bonds between the first and second nucleoside moieties. Using the standard cap analog as an example, for those RNAs in which m7Gp3G is incorporated in the normal orientation, the labeled product of RNase T2 and TAP digestion is pGp*. For RNAs in which m7Gp3G is incorporated in the reverse orientation, the labeled product is pm7Gp*. These nucleoside diphosphates differ in both charge and mass, as the m7 group confers a positive charge on G (Hendler et al. 1970), and are separable by ion-exchange HPLC.
In an earlier study, we calculated % correct orientation from the ratio of pGp* to pm7Gp* (Stepinski et al. 2001). Unfortunately, we were unaware that % capping was incomplete because the highly charged RNase T2 digestion product p3Gp* did not elute from our anion exchange column under the gradient conditions used. We have since modified the gradient conditions to allow elution of p3Gp*. Accordingly, we now recognize that digestion with RNAse T2 and TAP produces pGp* from two sources: capped RNAs, terminated with m7Gp3Gp*, and uncapped RNAs, terminated with p3Gp*. Only the former of these should enter into the calculation of % correct orientation. The amount of pGp* derived only from m7Gp3Gp*-capped RNAs can be calculated from % capping (see footnote ‘d’ in Table 1). In the earlier study (Stepinski et al. 2001), % correct orientation was erroneously calculated, from the ratio pGp*/(pGp* + pm7Gp*), to be 67%. But because the % capping with m7Gp3G is only 69% (Table 1), only 69% of the pGp* is derived from the 5′ ends of capped RNAs, the remainder being from uncapped RNAs. Recalculating % correct orientation from the data of Stepinski et al. gives a result (54%) that is closer to what was observed in the present study (58%).
The results for all cap analogs are shown in the Table 1 (% correct orientation). This parameter varies from 18% to 86% for the various cap analogs. b7m3′-OGp4G (9) is presumed to be incorporated 100% in the correct orientation because the 3′-O of the first nucleoside moiety is unable to form a phosphodiester bond. This was previously verified experimentally with related compounds in the dinucleoside triphosphate (m27,3′-OGp3G; Stepinski et al. 2001), tetraphosphate (m2 7,3′-O Gp4G; Jemielity et al. 2003), and pentaphosphate (m2 7,3′-O Gp5G; Jemielity et al. 2003) series. Surprisingly, four compounds in the dinucleoside triphosphate series that are lacking the 3′-O modification, b7Gp3G ( 1), e7Gp3G (2), m22,7Gp3G (3), and m32,2,7Gp3G (4), are incorporated substantially more in the correct orientation than the parent compound, m7Gp3G.
In contrast to compounds modified in the first nucleoside moiety, those modified in the second nucleoside are incorporated considerably more in the reverse orientation than the parent compound m7Gp3G. Previously we showed that cap dinucleoside triphosphates modified at the 2′-O position of the first nucleoside moiety, m27,2′-O Gp3G and m72′dGp3G, were incorporated exclusively in the normal orientation (Jemielity et al. 2003). This presumably reflects the ability of T7 RNA polymerase to distinguish between NTPs and dNTPs in vivo. This result predicts that dinucleoside triphosphates modified at the 2′-O position of the second nucleoside moiety would be incorporated exclusively in the reverse orientation. We tested this with the dinucleoside triphosphates m7Gp32′dG (5) and m7Gp3m2′-OG (6). When confronted with m7Gp32′dG, T7 RNA polymerase could bind with either the m7GTP or dGTP moiety in the GTP binding site. One might expect the dGTP orientation to be strongly discriminated against. The data in Table 1 indicate that binding of the m7GTP moiety occurs ~fivefold more often than binding of the dGTP moiety (18% correct orientation). Similarly, m7Gp3m2′-OG (6) is incorporated primarily in the reverse orientation (24% correct orientation).
Translational activity of RNAs capped with novel cap analogs
Finally, we measured the efficiency with which transcripts capped with the various cap analogs are translated in vitro. Translation reactions were conducted under conditions in which luciferase production was linear with both time and mRNA concentration. Translational efficiency relative to m7Gp3G-capped mRNA was calculated as described (Jemielity et al. 2003). The results for all analogs are shown in Table 1 (relative translational efficiency).
The 7-benzyl analogs produce mRNAs that are more efficiently translated than their 7-methyl counterparts. For b7Gp3G-capped mRNA, the relative translational efficiency of 1.87 is higher than would be predicted from the 1.2-fold lower KI of b7Gp3G (1) compared to m7Gp3G. This is likely due to a higher % capping (79% versus 69%) and higher % correct orientation (76% versus 58%) of b7Gp3G (1) compared to m7Gp3G. Similarly, the relative translational efficiency of b7Gp4G-capped mRNA, 1.98, is higher than would be expected from the slightly lower KI of b7Gp4G (8) (16 μM) compared to m7Gp3G (17.1 μM), but the two other parameters are higher for b7Gp4G (8) than for m7Gp3G (76% vs. 69% for % capping, 84% vs. 58% for % correct orientation). Adding a favorable N2 methyl group to this cap analog to produce b7m2Gp4G (12) boosts relative translational efficiency even more, to 2.55. This presumably results from a combination of lower KI, higher % capping, and higher % correct orientation for b7m2Gp4G (12) compared to m7Gp3G, none of which alone could account for the magnitude of improvement in efficiency. Even more favorable is the addition of a 3′-O methyl group to the first nucleoside residue, forming a true ARCA, b7m3′-OGp4G (9), because incorporation of the cap analog in the reverse orientation is blocked. This improves relative translational efficiency to 2.87. In contrast, the relative translational efficiency of e7Gp3G-capped mRNA is only 0.68.
Cap analogs with N2 methyl substitutions do not improve translational efficiency as much as 7-benzyl substitutions. Adding a fourth phosphate to this analog to make m22,7Gp4G (10) improves relative translational efficiency only slightly. When a second methyl group is added at N2, forming m32,2,7Gp3G (4), the relative translational efficiency drops to dramatically, reflecting the poor binding to eIF4E (KAS and KI). Adding a fourth phosphate, to make m32,2,7Gp4G (11), improves the relative translational efficiency. Cap dinucleoside triphosphates modified in the second nucleoside residue diminish translational efficiency. The translational efficiencies of mRNAs capped with “two-headed” analogs are the highest in both the dinucleoside triphosphate and dinucleoside tetraphosphate series. A major contributor to this efficiency is the fact that, because they are symmetrical, they are incorporated 100% in the correct orientation. The efficiencies predicted by the product of 1/KI, % capping, and % correct orientation for m7Gp3m7G- and m7Gp4m7G-capped mRNAs are 2.49 and 4.59, respectively, which is in reasonable agreement with the observed 2.66 and 3.14. Interestingly the % capping for these compounds (73% and 74%, respectively) is lower than of several other of the cap analogs. If conditions could be devised to increase the % capping, mRNAs of even higher translational efficiency might result. Together with mRNAs capped with b7m2Gp4G (2.55) and b7m3′-OGp4G (2.87) discussed above, these translational efficiencies are higher than any previously reported cap analog.
The most straightforward use of these new analogs is to produce mRNAs of higher translational efficiency, in turn yielding more protein product. However, although we have tested these in an in vitro translation system, their behavior in cultured cells, for example, by RNA transfections, has not yet been explored. Conceivably, they could improve the stability of mRNAs in cultured cells (which is not a concern in the rabbit reticulocyte system), providing a higher protein yield than that predicted from the increased translational efficiency. One should be aware that rabbit reticulocytes are unusual in having very high levels of initiation factors compared with other cells (Rau et al. 1996), and thus extrapolation of the results reported here to transfected cells or other cell free translation systems such as wheat germ may not be warranted. One also must take into account that mRNAs with more 5′-UTR secondary structure are more dependent on the eIF4-factor-containing unwinding machinery (for review, see De Benedetti and Graff 2004), and therefore the novel cap analogs may have greater or lesser effects on translational efficiency depending on the mRNA used. Another factor is that caps act synergistically with the 3′-terminal poly(A) (Gallie 1991). Thus, the degree to which novel cap analogs stimulate translation could also depend on the state of mRNA polyadenylation.
These analogs may also be useful for the study of biochemical processes in addition to translation. For instance, synthetic capped pre-mRNAs are widely used in splicing experiments, and it would be interesting to test whether pre-mRNAs capped with novel analogs are spliced more efficiently than conventional pre-mRNAs or bind differently to the nuclear cap-binding complex (CBC 20/80). To date we have not determined KAS values for these cap analogs with CBC 20/80, but studies are in progress. Similarly, novel cap analogs may improve nucleocytoplasmic transport, as mRNAs with ‘reverse’ caps are poorly transported (Pasquinelli et al. 1995).
MATERIALS AND METHODS
Synthesis of cap analogs
b7Gp4G (8). Gp4G (TEA salt; 30 mg; 0.028 mmole; Stepinski et al. 1995) was dissolved in 0.5 mL of DMSO, and benzyl bromide (0.05 mL) was added. The mixture was stirred at room temperature for 1.5 h. Then 30 mL of cold water was added and the solution was extracted with diethyl ether (3 × 10 mL). The aqueous phase was concentrated to a small volume on a rotary evaporator. The product was isolated from the resulting mixture using semipreparative reverse phase HPLC (Supelcosil LC-18-T column with a linear 0%–50% gradient of methanol in 0.05 M ammonium acetate). Yield: 6 mg (ammonium salt, 21.1%).
b7m3′-OGp4G (9). 3′-O-methylguanosine-5′ diphosphate (TEA salt; 66 mg; 0.1 mmol; Stepinski et al. 2001), DMSO (3 mL) and benzyl bromide (0.1 mL) were magnetically stirred for 5 h. Then 60 mL of cold water was added and the solution was extracted with diethyl ether (3 × 20 mL). The product, 7-benzyl-3′-O-methylguanosine 5′-diphosphate, was isolated by column chromatography on DEAE-Sephadex (A-25, HCO3- form) using a linear gradient of triethylammonium bicarbonate (TEAB, water solution, pH 7.5, 0–1 M) at 4°C. Appropriate fractions were pooled and evaporated to dryness (bath temperature not exceeding 30°C, ethanol repeatedly added to remove the TEAB buffer). Next, the resulting 7-benzyl-3′-O-methylguanosine 5′-diphosphate (TEA salt) was stirred overnight with DMF (1 mL), guanosine 5′-diphosphate P2-imidazolide (50 mg; 0.09 mmol; Jemielity et al. 2003), and ZnCl2 (73 mg; 0.54 mmol). The reaction was quenched by addition of EDTA (170 mg) in 20 mL of water and neutralized with 1 M NaHCO3. Chromatographic isolation on DEAE Sephadex using a linear gradient of TEAB (0–1.1 M) gave the product as the TEA salt. The product was converted to its Na+ salt by ion-exchange chromatography (Dowex 50WX8, Na+ form). Finally compound 9 was obtained as an amorphous white powder. Yield: 9 mg (8.9% based on the amount of starting material 3′-O-methylguanosine 5′-diphosphate used).
b7m2Gp4G (12) was synthesized by coupling 7-benzyl-2-methylguanosine 5′-monophosphate (TEA salt; 22 mg; 0.04 mmol; Jankowska et al. 1993) and guanosine 5′-triphosphate P3-imidazolide (39 mg; 0.06 mmole; Jemielity et al. 2003) in DMF (1 mL) in the presence of ZnCl2 (69 mg; 0.5 mmole). The compound 12 was isolated and converted into its Na+ salt in the same manner as 9. Yield: 8 mg (19.8%).
NMR spectroscopy
The structures of newly synthesized cap analogs 8, 9, and 12 were confirmed by 1H and 31P spectra. Spectra were recorded on a Varian UNITYplus 400 MHz instrument in 2H2O at 20°C and at concentrations ranging from 1 to 2 mg/mL. Conformations of the sugar moieties were derived from the vicinal 1H-1H coupling constants (Haasnoot et al. 1980), and conformations of the α-phosphate groups about the C5′-O5′ bond from the vicinal 1H-31P coupling constants (Lankhorst et al. 1984).
b7Gp4G (8). 1H NMR (s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet). b7G: H8 deuterated, benzyl ring 7.33–7.43 ppm (m), benzyl CH2 5.63 (d) and 5.68 (d), H1′ 5.96 ppm (d), H2′ 4.62 ppm (t), H3′ 4.51 ppm (t), H4′,H5′, H5″ 4.20–4.38 ppm (m); G: H8 8.07 ppm (s), H1′ 5.81 ppm (d), H2′ 4.74 ppm (t), H3′ 4.53 ppm (q), H4′, H5′, H5″ 4.20–4.38 ppm (m); 31P NMR: α and δ phosphates -12.10 ppm (m), β and γ phosphates -23.70 ppm (m).
b7m3′-OGp4G (9). 1H NMR. b7m3′-OG: H8 deuterated, benzyl ring 7.33–7.43 ppm (m), benzyl CH2 5.65 (s), H1′ 5.93 ppm (d), H2′ 4.77 ppm (t), H3′ 4.17 ppm (t), H4′ 4.46 ppm (m), H5′, H5″ 4.20–4.36 ppm (m); G: H8 8.07 ppm (s), H1′ 5.79 ppm (d), H2′ 4.74 ppm (t), H3′ 4.52 ppm (q), H4′, H5′, H5″ 4.20–4.38 ppm (m); 31P NMR: α and δ phosphates -12.05 ppm (m), β and γ phosphates -23.65 ppm (m).
b7m2Gp4G (12). 1H NMR. m2b7G: H8 deuterated, benzyl ring 7.33–7.42 ppm (m), benzyl CH2 5.62 (d) and 5.67 (d), H1′ 6.00 ppm (d), H2′ 4.67 ppm (t), H3′ 4.49 ppm (m), H4′, H5′, H5″ 4.20–4.40 ppm (m); G: H8 8.09 ppm (s), H1′ 5.82 ppm (d), H2′ 4.75 ppm (t), H3′ 4.49 ppm (m), H4′,H5′, H5″ 4.20–4.40 ppm (m); N2CH3 1.92 ppm (s). 31P NMR: α and δ phosphates -12.10 ppm (m), β and γ phosphates -23.70 ppm (m).
Analysis of percent capping
The short RNAs (Strepinski et al. 2001) were digested with RNase T2 (Invitrogen) and analyzed by anion exchange HPLC on a 4.5 × 250-mm Partisil 10SAX/25 column (Whatman). The program of elution of nucleotides consisted of water for the first 5 min, a linear gradient of 0–1.5 M KH2PO4 at either pH 3.5 (for mRNAs containing triphosphate caps) or pH 4.5 (for mRNAs containing tetraphosphate caps) for 40 min, and isocratic elution at 1.5 M KH2PO4 for 2 min, all at the flow rate 1 mL/min. Fractions of 1 mL were collected, and the Cherenkov radiation was determined.
Analysis of cap orientation
The short RNAs were subjected to a two-step digestion with TAP (Epicentre Technologies) and RNase T2 (Invitrogen) followed by anion exchange HPLC on a 4.6 × 250-mm Partisil 10SAX/25 column (Whatman). The gradient consisted of water for the first 5 min, a linear gradient of 0–87.5 mM KH2PO4, pH 3.5, for 35 min, a linear gradient of 87.5–500 mM KH2PO4 for 35 min, and isocratic elution at 500 mM of KH2PO4 for 21 min, all at the flow rate 1 mL/min. Fractions of 1 mL were collected, and the Cherenkov radiation was determined.
Other methods
Mouse eIF4E (residues 28–217) was expressed in E. coli (Zuberek et al. 2003) and purified by ion-exchange chromatography on a Mono SP column (Edery et al. 1988). Fluorescence time-synchronized titrations were performed as described (Jemielity et al. 2003). RNAs either uncapped or capped with various analogs were synthesized by in vitro transcription in two lengths as described (Jemielity et al. 2003). The translation inhibition experiments were performed as described by Cai et al. (1999). Translational efficiency was measured using luciferase mRNAs capped with various analogs (Jemielity et al. 2003).
Acknowledgments
This project was supported by Grant No. 1 R03 TW006446 from the Fogarty International Center (NIH), grant #3 P04A 021 25 and #059/T09/10 from the Polish Committee for Scientific Research (KBN), and grant #GM20818 from the National Institute of General Medical Sciences (NIH). We thank Nahum Sonenberg for the mouse eIF4E(28–217) plasmid, Dorota Haber for excellent chemical assistance, and Joanna Zuberek for assistance with KAS determinations.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7380904.
REFERENCES
- Blachut-Okrasinska, E., Bojarska, E., Niedzwiecka, A., Chlebicka, L., Darzynkiewicz, E., Stolarski, R., Stepinski, J., and Antosiewicz, J.M. 2000. Stopped-flow and brownian dynamics studies of electrostatic effects in the kinetics of binding of 7-methyl-GpppG to the protein eIF4E. Eur. Biophys. J. 29: 487–498. [DOI] [PubMed] [Google Scholar]
- Cai, A., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 1999. Quantitative assessment of mRNA cap analogs as inhibitors of in vitro translation. Biochemistry 38: 8538–8547. [DOI] [PubMed] [Google Scholar]
- Carberry, S.E., Darzynkiewicz, E., Stepinski, J., Tahara, S.M., Rhoads, R.E., and Goss, D.J. 1990. A spectroscopic study of the binding of N-7-substituted cap analogs to human protein synthesis initiation factor 4E. Biochemistry 29: 3337–3341. [DOI] [PubMed] [Google Scholar]
- Chu, L.-Y. and Rhoads, R.E. 1980. Inhibition of cell-free messenger ribonucleic acid translation by 7-methylguanosine 5′-triphosphate: Effect of messenger ribonucleic acid concentration. Biochemistry 19: 184–191. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Tahara, S.M., Seliger, L.S., and Shatkin, A. 1985. Chemical synthesis and characterization of 7-methylguanosine cap analogues. Biochemistry 24: 1701–1707. [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Lassota, P., and Tahara, S.M. 1987. Inhibition of eukaryotic translation by analogues of messenger RNA 5′-cap: Chemical and biological consequences of 5′- phosphate modifications of 7-methylguanosine 5′-monophosphate. Biochemistry 26: 4372–4380. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Ekiel, I., Goyer, C., Sonenberg, N., Temeriusz, A., Jin, Y., Sijuwade, T., Haber, D., and Tahara, S.M. 1989. Inhibition of eukaryotic translation by nucleoside 5′-monophosphate analogues of mRNA 5′-cap: Changes in N7 substituent affect analogue activity. Biochemistry 28: 4771–4778. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Tahara, S.M., Stolarski, R., Ekiel, I., Haber, D., Neuvonen, K., Lehikoinen, P., Labadi, I., and Lönnberg, H. 1990. Synthesis, conformation and hydrolytic stability of 1,P3-dinucleoside triphosphates related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleoside monophosphate analogs. Nucleosides Nucleotides 9: 599–618. [Google Scholar]
- De Benedetti, A. and Graff, J.R. 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 23: 3189–3199. [DOI] [PubMed] [Google Scholar]
- Edery, I., Altmann, M., and Sonenberg, N. 1988. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene 74: 517–525. [DOI] [PubMed] [Google Scholar]
- Flaherty, S.M., Fortes, P., Izaurralde, E., Mattaj, I.W., and Gilmartin, G.M. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. 94: 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D.R. 1991. The cap and poly(A) function synergistically to regulate mRNA translation efficiency. Genes & Dev. 5: 2108–2116. [DOI] [PubMed] [Google Scholar]
- Gingras, A.-C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Haasnoot, CAG, deLeeuw, F.A.A.M., and Altona, C. 1980. The relationship between proton-proton nmr coupling constants and substituent electronegativities. Tetrahedron 36: 2783–2792. [Google Scholar]
- Haghighat, A. and Sonenberg, N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272: 21677–21680. [DOI] [PubMed] [Google Scholar]
- Hendler, S., Fürer, E., and Srinivasan, P.R. 1970. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial DNA or RNA polymerases. Biochemistry 9: 4141–4153. [DOI] [PubMed] [Google Scholar]
- Herbert, T.P., Fåhraeus, R., Prescott, A.R., Lane, D.P., and Proud, C.G. 2000. Rapid induction of apoptosis by peptides that bind initiation factor eIF4E. Curr. Biol. 10: 793–796. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Stepinski, J., Darzynkiewicz, E., and Mattaj, I.W. 1992. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 118: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska, M., Stepinski, J., Stolarski, R., Temeriusz, A., and Darzynkiewicz, E. 1993. Synthesis and properties of new NH2 and N7 substituted GMP and GTP 5′-mRNA cap analogues. Collect. Czech. Chem. Commun. 58: 138–141. [Google Scholar]
- Jankowska, M., Stepinski, J., Stolarski, R., Wieczorek, Z., Temeriusz, A., Haber, D., and Darzynkiewicz, E. 1996. 1H NMR and fluorescence studies of new mRNA 5′-cap analogues. Collect. Czech. Chem. Commun. 61: S197–S202. [Google Scholar]
- Jemielity, J., Fowler, T., Zuberek, J., Stepinski, J., Lewdorowicz, M., Niedzwiecka, A., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 2003. Novel “Anti-Reverse” cap analogues with superior translational properties. RNA 9: 1108–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankhorst, P.L., Haasnoot, C.A.G., Erkelens, C., and Altona, C. 1984. Carbon-13 NMR in conformational analysis of nucleic acid fragments. Reparametrization of the Karplus equation for vicinal NMR coupling constants in COOP and HCOP fragments. J. Biomolec. Struct. Dyn. 1: 1387–1405. [DOI] [PubMed] [Google Scholar]
- Liou, R.F. and Blumenthal, T. 1990. Trans-spliced Caenorhabditis elegans messenger RNAs retain trimethylguanosine caps. Mol. Cell Biol. 10: 1764–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.-C., Sonenberg, N., and Burley, S.K. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.-C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4: 717–724. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W. 1986. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46: 905–911. [DOI] [PubMed] [Google Scholar]
- Miyoshi, H., Dwyer, D.S., Keiper, B.D., Jankowska-Anyszka, M., Dar-zynkiewicz, E., and Rhoads, R.E. 2002. Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 21: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C., and Sharp, P.A. 1993. Splicing of precursors to mRNA by the spliceosome. In The RNA World (R.F. Gestland and J.F. Atkins), pp. 303–357. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Muthukrishnan, S., Both, G.W., Furuichi, Y., and Shatkin, A.J. 1975. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255: 33–37. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.-C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E., Dahlberg, J.E., and Lund, E. 1995. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA 1: 957–967. [PMC free article] [PubMed] [Google Scholar]
- Pelham, H.R.B. and Jackson, R.J. 1976. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 47: 247–256. [DOI] [PubMed] [Google Scholar]
- Peng, Z.-H., Sharma, V., Singleton, S.F., and Gershon, P.D. 2002. Synthesis and application of a chain-terminating dinucleotide mRNA cap analog. Organic Lett. 4: 161–164. [DOI] [PubMed] [Google Scholar]
- Rau, M., Ohlmann, T., Morley, S.J., and Pain, V.M. 1996. A re-evaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem. 271: 8983–8990. [DOI] [PubMed] [Google Scholar]
- Rhoads, R.E. 1999. Minireview: Signal transduction pathways that regulate eukaryotic protein synthesis. J. Biol. Chem. 274: 30337–30340. [DOI] [PubMed] [Google Scholar]
- Stepinski, J., Bretner, M., Jankowska, M., Felczak, K., Stolarski, R., Wieczorek, Z., Cai, A.-L., Rhoads, R.E., Temeriusz, A., Haber, D., et al. 1995. Synthesis and properties of P1, P2, P3- and P1,P4-dinucleoside di-, tri- and tetraphosphate mRNA 5′-cap analogues. Nucleosides Nucleotides 14: 717–721. [Google Scholar]
- Stepinski, J., Waddell, C., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 2001. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogues 7-methyl(3′-O-methyl)GpppG and 7-methyl(3′-deoxy)GpppG. RNA 7: 1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Ishida, T., Taniguchi, T., Hasegawa, H., Terashima, A., et al. 2002. Crystal structure of 7-methylguanosine 5′-triphosphate (m7GTP)- and P1–7-methylguanosine-P3-adenosine-5′,5′-tri-phosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem. J. 362: 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Sasaki, M., Taniguchi, T., Miyagawa, H., Kitamura, K., et al. 2003. Structural feature of human factor 4E, studied by X-ray crystal analysis and molecular dynamics simulations. J. Mol. Biol. 328: 365–383. [DOI] [PubMed] [Google Scholar]
- van Doren, K. and Hirsh, D. 1990. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5– termini. Mol. Cell Biol. 10: 1769–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.C., Balasta, M.L., Ren, J.H., and Goss, D.J. 1998. Wheat gern poly(A)-binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A.R., Cheng, N.S.N., Blumenthal, T., and Spieth, J. 1994. Operons as a common form of chromosomal organization in C. elegans. Nature 372: 270–272. [DOI] [PubMed] [Google Scholar]
- Zuberek, J., Wyslouch-Cieszynska, A., Niedzwiecka, A., Dadlez, M., Stepinski, J., Augustyniak, W., Gingras, A.-C., Zhang, Z., Burley, S.K., Sonenberg, N., et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]