Abstract
A simple procedure has been developed to quickly retrieve and validate the DNA sequence encoding the RNA subunit of ribonuclease P (RNase P RNA) from microbial genomes. RNase P RNA sequences were identified from 94% of bacterial and archaeal complete genomes where previously no RNase P RNA was annotated. A sequence was found in camelpox virus, highly conserved in all orthopoxviruses (including smallpox virus), which could fold into a putative RNase P RNA in terms of conserved primary features and secondary structure. New structure features of RNase P RNA that enable one to distinguish bacteria from archaea and eukarya were found. This RNA is yet another RNA that can be a molecular criterion to divide the living world into three domains (bacteria, archaea, and eukarya). The catalytic center of this RNA, and its detection from some environmental whole genome shotgun sequences, is also discussed.
Keywords: preprocessing programs, pattern recognition sequence, core structure, camelpox virus
INTRODUCTION
Transfer RNA (tRNA), the RNA subunit of ribonuclease P (RNase P RNA), and ribosomal RNA (rRNA) are three well-known, essential RNA species that exist in all domains of life. The tRNAs are simple and short, and they are conserved in both secondary structure and tertiary structure. Many elegant programs (tRNAscan, Fichant and Burks 1991; tRNAscan-SE, Lowe and Eddy 1997; RNAMotif, Macke et al. 2001; and ARAGORN, Laslett and Canback 2004) have been developed to find tRNA sequences in genomes with a high success rate. The rRNAs are usually encoded in an rrn operon (~5 kb), are much larger, and can be easily identified in the nonprotein coding areas of genomes. Small subunit (SSU) rRNAs have served as molecular criteria for systematic phylogeny. Thus, their sequences are usually determined before entire genomes get sequenced.
For RNase P RNA, ranging in size from 276 to ~500 nucleotides (nt), there is no gene identification tool to find its sequence absolutely reliably from a given genome. Consequently, RNase P RNA is not annotated in eight of 19 archaea and 52 of 146 bacteria for which complete genome sequences are available (as of June 23, 2004; http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html). Existing programs such as Blast could retrieve some RNase P RNA sequences from these complete genomes if one of their close species is available as a query sequence. However, within the archaeal genus Sulfolobus, as an example, the RNase P RNA sequences from S. tokodaii could not be found by Blastn, even with the available RNAs from S. acidocaldarius, S. shibatae, and S. solfataricus (Brown 1999) as query sequences (data not shown). The failure of Blast on searching for at least some RNase P RNAs from archaeal genomes may be attributed to the fact that RNA consists of a smaller and less informative alphabet (4 nt) than protein (20 amino acids): This program is based on similarity comparison of primary sequences.
Previous phylogenetic comparisons based on RNase P RNA has been useful (Pace et al. 1989; Frank et al. 2000; Harris et al. 2001). However, the difficulty in differentiating an archaeon from a bacterium from previous RNase P RNA analysis (Harris et al. 2001) and the lack of a general tool to annotate its gene from a microbial genome has significantly limited the scope of RNase P RNA-based phylogeny analysis.
Here we describe a simple procedure for finding and validating RNase P RNA in a matter of a few minutes from a microbial genomic sequence. This method includes two steps: first, retrieving a DNA fragment containing two conserved nucleotide regions from a genome; and second, the validation of sequence segment by a pattern-profiling program, RNAMotif (Macke et al. 2001). We also disclose the structure features of RNase P RNAs that allow us to distinguish bacteria from archaea in this study.
RESULTS
Structure inspection
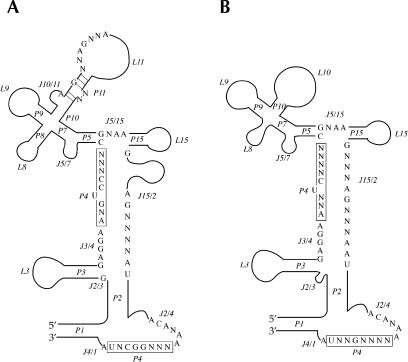
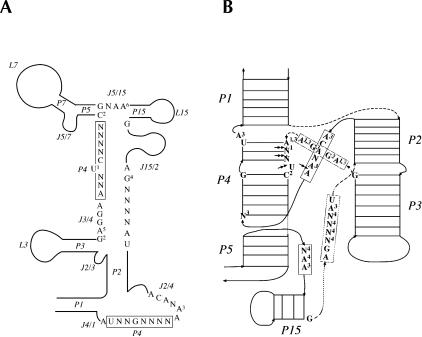
All of the RNase P RNA of bacteria and archaea from the RNase P database (http://www.mbio.ncsu.edu/RNaseP/, Brown 1999), and those annotated in GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) but not included in the RNase P database, were inspected and a new minimum consensus secondary structure was derived for either bacteria (Fig. 1A ▶) or archaea (Fig. 1B ▶). By extracting the information from these two consensus sequences, a new universal core structure of RNase P RNA for both bacteria and archaea (Fig. 2A ▶) was deduced. The core structure differs from a previously identified phylogenetic-minimum consensus structure (Chen and Pace 1997) in three aspects: First, the core structure includes both bacteria and archaea, and thus it has some advantages over previous attempts of this kind (see legend for Table 2 ▶, below); second, only the most conserved residues were presented (aside from seven single nucleotide exceptions in various organisms, 100% of the sequences were identical in all RNAs; Fig. 2A ▶); third, the loops and helices were thoroughly scrutinized and completely well defined in terms of their sequence size and mispairs allowed in helices (Table 1 ▶).
FIGURE 1.
The conserved structures of RNase P RNA from bacteria (A) and archaea (B). Helices are numbered from 5′ to 3′ according to the structure of the E. coli RNA and are designated with P (“pairing”, e.g., P1, P2; Chen and Pace 1997; Massire et al. 1998). The loop regions are designated with L/J (L3 linking the same P3 helix, J2/3 linking P2 and P3). For reasons of simplicity, some helices and loops may be simplified into one loop (i.e., L10 of archaea may contain P11, P12, L12, J11/12, and J12/11; Chen and Pace 1997; Massire et al. 1998; Harris et al. 2001).
FIGURE 2.
The core structure of RNase P RNA for both bacteria and archaea. (A) The conserved secondary structure of RNase P RNAs. There are some exceptions in the conserved regions in some microorganisms: (1) U→A in Sulfolobus tokadaii; (2) G→U, C→U in Phytoplasma asteris (onion yellows) and Phytoplasma sp. (periwinkle); (3) A→G in Shewanella putrefaciens and Shewanella oneidensis; (4) G→C in Mycoplasma pneumoniae; (5) A→G in Metallosphaera sedula; and (6) A→G in Herpetosiphon aurantiacus. (B) The schematic representation view of the catalytic domain of the core structure (based on previously established 3D modeling of bacterial RNase P RNA; Massire et al. 1998). This illustration attempts to render the respective spatial arrangement of helices and stacks in the 3D model. The arrows point to the 5′ to 3′ direction of the RNA. The structural elements have been implied to engage in catalysis: (1) polynuclear metal ion binding site in the catalytic domain by phosphorothioate modification and quantitative analysis of thiophilic metal ion rescue on catalysis (Christian et al. 2002); (2) metal ion specificity (C→U makes Ca2+ a better ion; Frank and Pace 1997); (3) nucleotides critical to catalysis identified by NAIM and site-specific modification (Kazantsev and Pace 1998; Kaye et al. 2002); the double arrows point to the locations of phosphate oxygen where sulfur substitution disrupts catalytic activity; and (4) the active site mapped by a photoaffinity agent coupled to the 5′-phosphate of a tRNA (Burgin and Pace 1990).
TABLE 2.
Microbial genomes with RNase P RNAs found in this study
| Microorganisms/sourcesa | Accessionb | B/Ac | Notes |
| Methanococcus maripaludis S2 | NC_005791 | A | |
| Methanopyrus kandleri AV19 | NC_003551 | A | |
| Methanosarcina acetivorans str. C2A | NC_003552 | A | |
| Methanosarcina mazei strain Goe1 | NC_003901 | A | |
| Picrophilus torridus DSM 9790 | NC_005877 | A | |
| Sulfolobus tokodaii | NC_003106 | A | A bulge in P4d |
| Bacillus anthracis A2012 | NC_003995 | B | |
| Bacillus anthracis str. Ames | NC_003997 | B | |
| Bacillus anthracis str. Ames 0581 | NC_007530 | B | |
| Bacillus cereus ATCC 10987 | NC_003909 | B | |
| Bacillus cereus ATCC 14579 | NC_004722 | B | |
| Bdellovibrio bacteriovorus | NC_005363 | B | |
| Bifidobacterium longum NCC2705 | NC_004307 | B | ACUUCCGGGe |
| Bordetella parapertussis | NC_002928 | B | |
| Brucella melitensis chromosome I | NC_003317 | B | |
| Brucella suis 1330 chromosome I | NC_004310 | B | |
| Chromobacterium violaceum ATCC 12472 | NC_005085 | B | |
| Clostridium perfringens | NC_003366 | B | |
| Corynebacterium efficiens YS-314 | NC_004369 | B | AAGUCUGAAe |
| Corynebacterium glutamicum ATCC 13032 | NC_003450 | B | |
| Enterococcus faecalis V583 | NC_004668 | B | |
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | NC_003454 | B | |
| Haemophilus ducreyi 35000HP | NC_002940 | B | |
| Helicobacter hepaticus ATCC 51449 | NC_004917 | B | |
| Lactobacillus johnsonii NCC 533 | NC_005362 | B | |
| Lactobacillus plantarum WCFS1 | NC_004567 | B | |
| Lactococcus lactis subsp. lactis | NC_002662 | B | |
| Leptospira interrogans serovar | NC_005823 | B | |
| Copenhageni str. Fiocruz L1–130 chromosome I | |||
| Leptospira interrogans serovar lai str. 56601 chromosome I | NC_004342 | B | |
| Listeria innocua Clip11262 | NC_003212 | B | |
| Listeria monocytogenes strain EGD | NC_003210 | B | |
| Mesorhizobium loti | NC_002678 | B | |
| Mycoplasma gallisepticum R | NC_004829 | B | |
| Mycoplasma mycoides subsp. mycoides SC | NC_005364 | B | AACUCCACGe |
| Mycoplasma penetrans | NC_004432 | B | |
| Mycoplasma pulmonis | NC_002771 | B | |
| Oceanobacillus iheyensis HTE831 | NC_004193 | B | |
| Parachlamydia sp. UWE25 | NC_005681 | B | |
| Pasteurella multocida | NC_002663 | B | |
| Photorhabdus luminescens subsp. laumondii TTO1 | NC_005126 | B | |
| Pirellula sp. | NC_005027 | B | |
| Pseudomonas syringae pv. tomato str. DC3000 | NC_004578 | B | |
| Ralstonia solanacearum | NC_003295 | B | |
| Shewanella oneidensis MR-1 | NC_004347 | B | ACAGGAd |
| Sinorhizobium meliloti 1021 | NC_003047 | B | |
| Streptococcus agalactiae 2603V/R | NC_004116 | B | |
| Streptococcus agalactiae NEM316 | NC_004368 | B | |
| Streptomyces avermitilis MA-4680 | NC_003155 | B | |
| Thermoanaerobacter tengcongensis strain MB4T | NC_004113 | B | |
| Treponema denticola ATCC 35405 | NC_002967 | B | |
| Tropheryma whipplei TW08/27 | NC_004551 | B | |
| Vibrio parahaemolyticus RIMD 2210633 chromosome I | NC_004603 | B | |
| Vibrio vulnificus CMCP6 chromosome I | NC_004459 | B | |
| Wigglesworthia glossinidia endosymbiont of Glossina brevipalpis | NC_004344 | B | |
| Wolinella succinogenes | NC_005090 | B | |
| Xanthomonas axonopodis pv. citri. str. 306 | NC_003919 | B | |
| Xanthomonas campestris pv. campestris str. ATCC 33913 | NC_003902 | B | |
| Environmental sequence AMC_Cont212 | AADL01000 212 | B | Leptospirillum group IIf |
| Environmental sequence AMC_Cont1374 | AADL01001 374 | A | Ferroplasma acidarmanusg |
| Environmental sequence AMC_Cont1488 | AADL01001 488 | A | Ferroplasmah |
| Environmental sequence AMC_Cont1899 | AADL01001 899 | A | Unidentified archaeoni |
| Environmental sequence IBEA_CTG_2115020 | AACY01000 258 | B | Unidentified bacteriumj |
| Environmental sequence IBEA_CTG_2145348 | AACY01000 579 | B | Unidentified bacteriumj |
aIf there are two chromosomes in an organism, both are searched and only the one with an RNase P RNA sequence is presented; the contig numbers are provided for the environmental whole genome shotgun sequencing.
bGenBank accession number.
cBacteria (B) or archaea (A).
dSome unconserved residues disclosed in this study (Fig. 2A ▶).
eThe RNase P RNAs from these bacteria represent the advantage of the universality of our core structure for both bacteria and archaea. If the conserved region ANGUCCNNN (P4, as in Fig. 1A ▶) instead of ANNUCNNNN (Fig. 2A ▶) was used, these sequences could not be found in the first place.
f–iSequences from the biofilm project.
f93% identical to L. ferrooxidans strain MK in the RNase P database (Brown 1999).
g100% identical to a sequence from F. acidarmanus (www.jgi.doe.gov).
h90% identical to a sequence from F. acidarmanus (www.jgi.doe.gov).
iNo significant hits against all RNase P RNAs; may represent a novel archaeon.
jNo significant hits against all RNase P RNAs (including Prochlorococcus and Synechococcus, the two dominating bacterial species in sea water); may represent a novel bacterium. The sequences are from the Sargasso project.
TABLE 1.
Core structure of RNase PRNA
| P/La | Length (bp or nt) | Mispairs allowed | Conserved sequenceb |
| P2 | 6–7 | 1 | — |
| J2/3 | 1 or 3–4 | — | “G” if length = 1 |
| P3 | 2–7 | 1 if length > 5 | — |
| L3 | 3–72 | — | — |
| End of P3 | G$ | ||
| J3/4 | 4 | — | AGGA |
| P4-A | 3 | 1 | ANN |
| P4_UBULGE | 1 | — | U |
| P4_B | 5 | 1 | CNNNN |
| P5 | 4–5 | 1 | ^C |
| J5/7 | 1–27 | — | — |
| P7 | 3–7 | 1 | — |
| L7 | 50–300 | — | — |
| End of P7 | |||
| End of P5 | |||
| J5/15 | 3 | — | NAA |
| P15 | 2–4 | — | — |
| L15 | 4–95 | — | — |
| End of P15 | |||
| J15/2 | 11–77 | — | ^G, AGNNNNAU$ |
| End of P2 | |||
| J2/4 | 8–66 | — | ACANAA$ |
| End of P4_B | |||
| End of P4_A | |||
| J4/1 | 1 | — | A |
aThe structure elements are traced from 5′ to 3′ (top to bottom) according to the consensus structure shown in Fig. 2A ▶. P1 is not included because it is difficult to define in bacteria such as Caulobacter crescentus (Brown 1999); L7 is simplified here to include the whole cruciform because there are few constraints left for the cruciform (P7, P8, P9, and P10/P11) once P8 and P11 are compromised in some archaea (Brown 1999; Harris et al. 2001).
b^N means starting with this nucleotide; N$ means ending with N (one or more nucleotides).
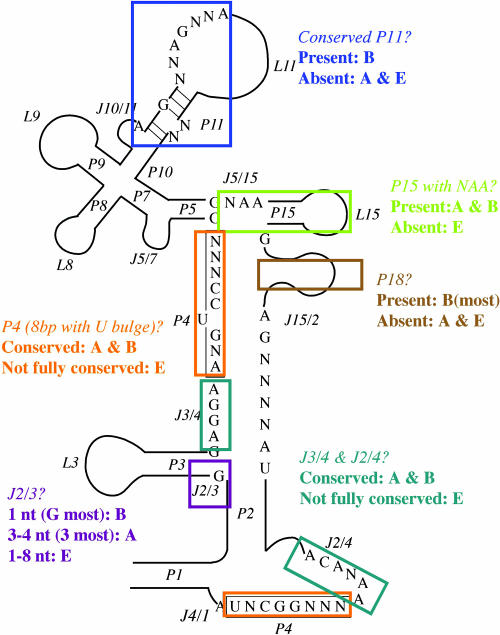
From the sequence comparison of RNase P RNAs from bacteria and archaea, we found that their major differences rely on J2/3 and P11 (Haas et al. 1996; Figs. 1 ▶, 3 ▶). Archaea may have P11 (Harris et al. 2001), but they do not possess the entire conserved feature as do bacteria (P11 is always 5 bp flanking with an A of J10/J11 while disrupted by 2 nt [mostly AA], and one mispair is allowed in its 3-bp half that begins with a G and is followed by the conserved NNAGNNA; N means any nucleotide). The distinctive structures of eukaryotic RNase P RNA were also analyzed and depicted in Figure 3 ▶.
FIGURE 3.
Comparison of the secondary structures of RNase P RNA from archaea (A), bacteria (B), and eukarya (E). The conserved structure of a bacterial RNA is depicted as a backbone. Different features are labeled by rectangles and descriptions with different colors.
Search procedure
There are two conserved sequence segments in the core structure: GAGGAANNUCNNNNC (previous designated as conserved region I, CR I; Chen and Pace 1997) and AGNNNNAU…{10–60 nt}…ACANAANNNNGNNUA (CR IV and CR V; Chen and Pace 1997). A new short sequence pattern recognition program was written in PERL as a preprocessor to retrieve ~600-nt DNA sequence(s) containing these two segments in one region from various genomes (Supplementary Fig. 1). This new preprocessing program reduces the amount of “junk” DNA containing no conserved RNase P RNA sequences and limits the amount of time to a few minutes for the entire search program.
The core structure (Fig. 2B ▶) is then exposed to the rules of a descriptor of RNAMotif (Macke et al. 2001), thereby leading to a general descriptor. This descriptor allows a search of significant features defined in RNase P RNA. Stems and loops are precisely defined by their lengths; mispairs are allowed in helices and sequence conservations (Table 1 ▶). The new descriptor is provided in Supplementary Figure 2 ▶.
Finally, the output sequence(s) from the PERL script are validated by RNAMotif with the new general descriptor for both bacteria and archaea.
Application to microbial genomes
All of the complete microbial genomes with no RNase P RNA annotated (eight archaea and 52 bacteria) are applied to the search procedure. Most genomes yield one to three sequences from the preprocessor (>90% of them yield just one sequence), but only one of these sequences can be validated by RNAMotif with the novel general descriptor. The validated sequence could form a reliable structure of RNase P RNA, as those previously identified sequences in the RNase P database (Brown 1999).
For some genomes (Sulfolobus tokodaii and Shewanella oneidensis) that did not yield any output by the PERL script, a strategy was attempted in which one of the conserved residues (in the conserved sequences) is mutated to N in the preprocessing step program: RNase P RNAs were immediately found after validating all of the resultant output sequences. It should be noted that an atypical A, instead of U, is bulged in the P4 helix of Sulfolobus tokodaii (Fig. 2A ▶).
Overall, we were able to identify RNase P RNA by this preprocessing-validating procedure in six of eight archaea, and 51 of 52 bacteria in their complete genome sequences where previously no RNase P RNA is annotated (Table 2 ▶; see Supplementary Figs. 3, 4).
There are three genomes (Aquifex aeolicus, Nanoarchaeum equitans, and Pyrobaculum aerophilum) for which no reliable RNase P RNA sequence could be found even by mutating three or four of the conserved residues. RNase P activities could not be demonstrated even in cell extracts of A. aeolicus (Willkomm et al. 2002). N. equitans is the only known archaeal parasite: It lives only when cocultured with another archaeon, Ignicoccus (Waters et al. 2003). Consequently, it might share the RNase P enzyme from its host. No success was obtained with P. aerophilium. Schemes in which two separate pieces of a genomic sequence resulted in a single, complex RNase P RNA were not analyzed.
Application to environmental genomes
Two recent sequence projects were performed with environmentally pooled DNA samples from an acid mine drainage microbial biofilm (Tyson et al. 2004) or the Sargasso Sea near Bermuda (Venter et al. 2004). The DNAs of a population of bacteria and/or archaea instead of individual colonies were sequenced. Some RNase P RNAs (four from the biofilm project and two from the Sargasso project) were derived using our new procedure from the deposited contig sequences. (Table 2 ▶; Supplementary Figs. 5, 6). Some novel sequence with no significant hits against all other RNase P RNA may represent a new probable form of a bacterium or archaeon (Table 2 ▶). One bacterial RNA from the Sargasso project is of special interest: Its J2/3 is an A instead of a G as in all known other bacteria (Supplementary Fig. 6).
Application to viral genomes
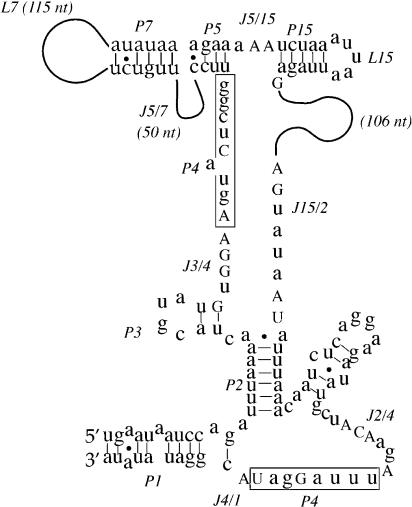
It is noteworthy that some viruses (mimivirus; La Scola et al. 2003) have genomes (734 kb) larger than the smallest bacterium (Mycoplasma genitalium, 580 kb; Fraser et al. 1995), and a mycobacteriophage (Bxz1 phage) possesses as many as 26 tRNA genes (Pedulla et al. 2003). To our knowledge, there is no rRNA found in any virus. We used our procedure to search viral genomes larger than 100 kb and failed to retrieve any sequence similar to RNase P RNA. However, when all of the sequence variations of bacteria and archaea were included in the preprocessor (Fig. 2A ▶; a few conserved nucleotides were allowed as N), one sequence was found in camelpox virus (Gubser and Smith 2002) that could be folded into a putative RNase P RNA (Fig. 4 ▶). This sequence segment has 98% homology with all other orthopoxviruses (smallpox, vaccinia, rabbitpox, cowpox, monkeypox, and mousepox virus; see Supplementary Fig. 7).
FIGURE 4.
A sequence from camelpox virus that forms a structure similar to the core structure of RNase P RNA from archaea and bacteria. The conserved residues, like those in Figure 2 ▶, are in uppercase letters. The L7 region could fold into a cruciform without the conserved P11 as in bacteria. The sequence (413 bp from 155,800 to 156,212 in camelpox genome) covers the intergenic region between a hypothetical ORF (155,444–155,689) and a gene coding for a DNA ligase (155,961–157,619), as well as part of the coding sequence for the ligase (Gubser and Smith 2002).
DISCUSSION
Identification of RNase P RNA
An efficient method to identify RNase P RNA from microbial genomes described in this study is comparable to tRNA-finding methods in terms of speed and accuracy. This search approach is so sensitive that it can pick up every true positive identified in reference sequences of bacteria and archaea from the RNase P database (Brown 1999) because the core structure engaged is deduced directly from those sequences. The core structure feature used for searching RNase P RNA contains the true catalysis domain of RNase P RNA (see below). Therefore, our method is also extremely selective and it never misidentifies any unrelated sequence as true RNase P RNA. In general, the RNase P RNA sequence has been detected from the vast majority (95%, 57 of 60) of microbial genomes where it had not been annotated previously. Only in three microorganisms (A. aeolicus, N. equitans, and P. aerophilum) is there no RNase P RNA sequence detected. Certain unique RNase P enzymes might exist in these microbes that are beyond the scope of our theoretical approach or even conventional experiments (as in the case of A. aeolicus; Willkomm et al. 2002).
The procedure identifying RNase P RNA can also be applied to other genomes containing putative prokaryotic RNase P RNA. Some RNase P RNAs were found in the contig sequences of environmental genomes (total 76 Mb and 1045 Mb nonredundant base pairs from biofilm and Sargasso project, respectively). In the biofilm project, four RNase P RNAs were found. This is acceptable considering that only two near-complete genomes and three partial genomes were constructed from the sequencing data, and rRNA analysis from the environmental sample indicated the presence of three bacterial and three archaeal lineages (Tyson et al. 2004). However, in the Sargasso project, only two RNase P RNAs were found in their DNA samples where over 1000 distinct SSU rRNA genes or fragments were identified (Venter et al. 2004). The outcome could partially be due to the lower quality of assembling from the much larger DNA pool of the Sargasso project compared with the bio-film project. We also found one sequence from camelpox virus (Gubser and Smith 2002) that could be folded into a putative RNase P RNA (Fig. 4 ▶). This sequence segment has 98% homology with all other orthopoxviruses. Further efforts need to be undertaken to find more RNase P RNA genes in the environmental genomes and to investigate the function of this putative RNA in orthopoxviruses that have no tRNA gene in their genomes.
It should be noted that most but not all of the RNase P RNAs found by this study could be retrieved by traditional sequence homology search tools such as Blast if the right query sequence is available. However, no other method could match our search procedure in terms of selectivity and sensitivity (Blast will generate some false positives/outputs). Moreover, our method is intended to solve the problem of microbes first, so it is not suitable to find RNase P RNA from eukaryotes. Because eukaryal RNase P RNAs have lower sequence conservation and eukaryal genomes are much larger, further efforts are needed to annotate RNase P RNAs from their complete genomes.
Structure of RNase P RNA
An archaeon can be easily differentiated from a bacterium according to their RNase P RNA sequences (Fig. 3 ▶). The highly conserved P18 is absent from all archaea but present in most bacteria (Harris et al. 2001). There are two previously known bacteria (Haas et al. 1994), and one more disclosed in this study (Chlorobium perfrigens; see Supplementary Fig. 4) that have no P18. The relevant feature identified previously (Haas et al. 1996) and confirmed in this study is much simpler and straightforward (Fig. 1A, B ▶): The joint region J2/3 linking P2 and P3 is always a single “G” in all bacteria, whereas in archaea, it is 3–4 nt long (J2/3 of 4 nt only appears in Halobacterium cutirubrum (Brown 1999) and Methanopyrus kandleri (this study; Fig. 3 ▶; see Supplementary Fig. 3). A new feature we found is that the conserved P11 helix region of 5 bp, disrupted by 2 nt and flanked by an A and the conserved NNAGNNA, exists in all bacteria but not in any archaeon.
The catalytic core of RNase P RNA is conserved in both bacteria and archaea. At least some of the archaeal RNase P RNA from methanobacteria, thermococci, and halobacteria are catalytically active in the absence of protein (Pannucci et al. 1999). A universal core structure was drawn according to the bacterial three-dimensional (3D) model (Fig. 2B ▶). All of the elements of bacterial RNase P RNA involved in catalysis (Burgin and Pace 1990; Frank and Pace 1997; Kazantsev and Pace 1998; Christian et al. 2002; Kaye et al. 2002) with supporting concrete data are present in this core structure (Fig. 2B ▶), indicating the catalytic center of archaea is similar to that of bacteria. The novel core structure does indeed contain the actual catalysis domain of the RNase P for both bacteria and archaea. The weaker catalytic activity of archaeal RNase P RNAs compared with that of bacterial ones may be due to the difference of the P11 region around the cruciform, which is believed to be involved in the binding of the TψC loop of ptRNAs in bacteria (Loria and Pan 1997; Fig. 1 ▶)
Information regarding why eukarya nuclear RNase P RNA is not catalytically active on its own may be derived from the results shown here (Fig. 3 ▶). First, the 8-bp P4 helix with a U bulge is missing in all cases (either <8 bp or without U bulge); second, the AGGA (J3/4) and ACANAA (J2/4) before P4 are no longer fully conserved; third, P15 is absent and the flanking NAA is missing in nearly every case in eukaryotes. The alteration in the core structure may significantly alter the microenvironment around the catalysis center and results in various protein subunits that are indispensable for enzyme activity. This speculation can be reflected in some properties of the bacterial enzyme: for example, an A65 deletion of the sequence AGGA65 (J3/4) in Escherichia coli RNase P RNA leads to an inactive RNA molecule, but catalysis could generally be rescued by the protein cofactor, C5 protein (C. Guerrier-Takada and S. Altman, unpubl.).
Phylogeny by RNase P RNA
Because RNase P RNA from archaea, bacteria, and eukarya are easily distinguishable from each other (Fig. 3 ▶), they could serve as a perfect molecular criterion for systematic phylogeny. Recent studies have indicated that RNase P RNA is suitable for phylogenetic analysis of closely related bacterial taxa and has potential as a tool for species discrimination. In the genus Streptococcus, RNase P RNA is a better criterion than SSU rRNAs (Tapp et al. 2003). The 16S rRNAs of Haemobartonella canis and Mycoplasma haemofelis were nearly identical (homology of 99.3%–99.7%); in contrast, RNase P RNAs have a lower degree (94.3%–95.5%) of sequence homology (Birkenheuer et al. 2002). One distinctive feature of RNase P RNA is that there is only one copy of its gene per genome instead of the multiple copies of rrn operons that are in most bacteria. This singleton feature of RNase P RNA may make its gene less likely to be compromised by interspecies lateral gene transfer and enable it to serve as a more reliable molecular chronometer. In the case of environmental genomes in the Sargasso project, large amounts of variation in copy number of rRNA genes between species has led to unreliable and doubtful determinations of species diversity and abundance based on rRNA (Venter et al. 2004).
Some RNase P RNAs from mitochondria and chloroplasts possess all of the conserved features of bacterial ones, though no activities could be demonstrated (E. Seif, pers. comm.; B.F. Lang, pers. comm.; S. Altman, pers. comm.), whereas others, such as those from ascomycete fungi, have no P2, P3, or P15 helices (Seif et al. 2003). The lack of specific RNase P protein subunits in individual tests in vitro may explain the absence of RNase P activity among these latter results.
The simple method to identify RNase P RNA from microbial genomes and the uncovered structure features to distinguish archaea, bacteria, and eukarya in this study will undoubtedly turn systematic phylogeny by RNase P RNA into a more extensive and compelling approach to analyze species diversity and evolution.
[Note: Readers interested in browsing through the Supplementary Material may contact the authors at sidney.altman@yale.edu.]
Acknowledgments
We are grateful to Dr. Tom Macke for the RNAMotif program and his valuable suggestions on descriptor composing. This research was supported by grant GM-19422 of the USPHS and a grant from the Provost of Yale University to S.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7970404.
REFERENCES
- Birkenheuer, A.J., Breitschwerdt, E.B., Alleman, A.R., and Pitulle, C. 2002. Differentiation of Haemobartonella canis and Mycoplasma haemofelis on the basis of comparative analysis of gene sequences. Am. J. Vet. Res. 63: 1385–1388. [DOI] [PubMed] [Google Scholar]
- Brown, J.W. 1999. The Ribonuclease P Database. Nucleic Acids Res. 27: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin, A.B. and Pace, N.R. 1990. Mapping the active site of ribo-nuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 9: 4111–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.L. and Pace, N.R. 1997. Identification of the universally conserved core of ribonuclease P RNA. RNA 3: 557–560. [PMC free article] [PubMed] [Google Scholar]
- Christian, E.L., Kaye, N.M., and Harris, M.E. 2002. Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA. EMBO J: 21: 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichant, G.A. and Burks, C. 1991. Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol. 220: 659–671. [DOI] [PubMed] [Google Scholar]
- Frank, D.N. and Pace, N.R. 1997. In vitro selection for altered divalent metal specificity in the RNase P RNA. Proc. Natl. Acad. Sci. 94: 14355–14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N., Adamidi, C., Ehringer, M.A., Pitulle, C., and Pace, N.R. 2000. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA 6: 1895–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, C.M., Gocayne, J.D., White, O., Adams, M.D., Clayton, R.A., Fleischmann, R.D., Bult, C.J., Kerlavage, A.R., Sutton, G.G., Kelley, J.M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270: 397–403. [DOI] [PubMed] [Google Scholar]
- Gubser, C. and Smith, G.L. 2002. The sequence of camelpox virus shows it is most closely related to variola virus, the cause of smallpox. J. Gen. Virol. 83: 855–872. [DOI] [PubMed] [Google Scholar]
- Haas, E.S., Brown, J.W., Pitulle, C., and Pace, N.R. 1994. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl. Acad. Sci. 91: 2527–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, E.S., Armbruster, D.W., Vucson, B.M., Daniels, C.J., and Brown, J.W. 1996. Comparative analysis of ribonuclease P RNA structure in Archaea. Nucleic Acids Res. 24: 1252–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J.K., Haas, E.S., Williams, D., Frank, D.N., and Brown, J.W. 2001. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA 7: 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye, N.M., Christian, E.L., and Harris, M.E. 2002. NAIM and site-specific functional group modification analysis of RNase P RNA: Magnesium dependent structure within the conserved P1-P4 multihelix junction contributes to catalysis. Biochemistry 41: 4533–4545. [DOI] [PubMed] [Google Scholar]
- Kazantsev, A.V. and Pace, N.R. 1998. Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA 4: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola, B., Audic, S., Robert, C., Jungang, L., de Lamballerie, X., Drancourt, M., Birtles, R., Claverie, J.M., and Raoult, D. 2003. A giant virus in amoebae. Science 299: 2033. [DOI] [PubMed] [Google Scholar]
- Laslett, D. and Canback, B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria, A. and Pan, T. 1997. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 36: 6317–6325. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke, T.J., Ecker, D.J., Gutell, R.R., Gautheret, D., Case, D.A., and Sampath, R. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 29: 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massire, C., Jaeger, L., and Westhof, E. 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279: 773–793. [DOI] [PubMed] [Google Scholar]
- Pace, N.R., Smith, D.K., Olsen, G.J., and James, B.D. 1989. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA—A review. Gene 82: 65–75. [DOI] [PubMed] [Google Scholar]
- Pannucci, J.A., Haas, E.S., Hall, T.A., Harris, J.K., and Brown, J.W. 1999. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. 96: 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla, M.L., Ford, M.E., Houtz, J.M., Karthikeyan, T., Wadsworth, C., Lewis, J.A., Jacobs-Sera, D., Falbo, J., Gross, J., Pannunzio, N.R., et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113: 171–182. [DOI] [PubMed] [Google Scholar]
- Seif, E.R., Forget, L., Martin, N.C., and Lang, B.F. 2003. Mitochondrial RNase P RNAs in ascomycete fungi: Lineage-specific variations in RNA secondary structure. RNA 9: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapp, J., Thollesson, M., and Herrmann, B. 2003. Phylogenetic relationships and genotyping of the genus Streptococcus by sequence determination of the RNase P RNA gene, rnpB. Int. J. Syst. Evol. Microbiol. 53: 1861–1871. [DOI] [PubMed] [Google Scholar]
- Tyson, G.W., Chapman, J., Hugenholtz, P., Allen, E.E., Ram, R.J., Richardson, P.M., Solovyev, V.V., Rubin, E.M., Rokhsar, D.S., and Banfield, J.F. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Remington, K., Heidelberg, J.F., Halpern, A.L., Rusch, D., Eisen, J.A., Wu, D., Paulsen, I., Nelson, K.E., Nelson, W.,et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74. [DOI] [PubMed] [Google Scholar]
- Waters, E., Hohn, M.J., Ahel, I., Graham, D.E., Adams, M.D., Barn-stead, M., Beeson, K.Y., Bibbs, L., Bolanos, R., Keller, M.,et al. 2003. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. 100: 12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willkomm, D.K., Feltens, R., and Hartmann, R.K. 2002. tRNA maturation in Aquifex aeolicus. Biochimie 84: 713–722. [DOI] [PubMed] [Google Scholar]